This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 110 genes and 10 molecular subtypes across 284 patients, 14 significant findings detected with P value < 0.05 and Q value < 0.25.
-
EGFR mutation correlated to 'CN_CNMF', 'RPPA_CNMF', and 'RPPA_CHIERARCHICAL'.
-
IDH1 mutation correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'MIR_CNMF', 'MIR_CHIERARCHICAL', and 'CN_CNMF'.
-
TP53 mutation correlated to 'MRNA_CNMF' and 'CN_CNMF'.
-
ATRX mutation correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'MIR_CHIERARCHICAL', and 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between mutation status of 110 genes and 10 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 14 significant findings detected.
|
Clinical Features |
MRNA CNMF |
MRNA CHIERARCHICAL |
MIR CNMF |
MIR CHIERARCHICAL |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| IDH1 | 14 (5%) | 270 |
1.13e-06 (0.000969) |
4.41e-07 (0.00038) |
0.000227 (0.193) |
1.24e-05 (0.0106) |
1.68e-05 (0.0143) |
0.00121 (1.00) |
0.00352 (1.00) |
0.0203 (1.00) |
0.000712 (0.601) |
0.00263 (1.00) |
| ATRX | 16 (6%) | 268 |
0.000227 (0.192) |
0.000201 (0.171) |
0.00455 (1.00) |
0.000137 (0.117) |
6.23e-05 (0.0532) |
0.155 (1.00) |
0.00605 (1.00) |
0.0121 (1.00) |
0.00195 (1.00) |
0.0421 (1.00) |
| EGFR | 74 (26%) | 210 |
0.000485 (0.41) |
0.0656 (1.00) |
0.242 (1.00) |
0.217 (1.00) |
7.49e-07 (0.000643) |
0.0108 (1.00) |
6.37e-05 (0.0543) |
1.46e-06 (0.00125) |
0.0413 (1.00) |
0.357 (1.00) |
| TP53 | 79 (28%) | 205 |
4.16e-05 (0.0356) |
0.000502 (0.424) |
0.000313 (0.265) |
0.0272 (1.00) |
0.000166 (0.141) |
0.0083 (1.00) |
0.0384 (1.00) |
0.00968 (1.00) |
0.00184 (1.00) |
0.0375 (1.00) |
| PIK3R1 | 32 (11%) | 252 |
0.96 (1.00) |
0.779 (1.00) |
0.467 (1.00) |
0.205 (1.00) |
0.115 (1.00) |
0.84 (1.00) |
0.988 (1.00) |
0.273 (1.00) |
0.406 (1.00) |
0.649 (1.00) |
| BRAF | 6 (2%) | 278 |
0.395 (1.00) |
0.567 (1.00) |
0.616 (1.00) |
0.175 (1.00) |
0.586 (1.00) |
1 (1.00) |
0.0781 (1.00) |
0.791 (1.00) |
1 (1.00) |
|
| PTEN | 87 (31%) | 197 |
0.458 (1.00) |
0.366 (1.00) |
0.0452 (1.00) |
0.594 (1.00) |
0.584 (1.00) |
0.95 (1.00) |
0.378 (1.00) |
0.389 (1.00) |
0.27 (1.00) |
0.196 (1.00) |
| PIK3CA | 30 (11%) | 254 |
0.689 (1.00) |
0.195 (1.00) |
0.571 (1.00) |
0.785 (1.00) |
0.501 (1.00) |
0.627 (1.00) |
0.754 (1.00) |
0.281 (1.00) |
0.262 (1.00) |
0.838 (1.00) |
| RB1 | 24 (8%) | 260 |
0.00652 (1.00) |
0.0265 (1.00) |
0.0653 (1.00) |
0.357 (1.00) |
0.0873 (1.00) |
0.027 (1.00) |
0.493 (1.00) |
0.691 (1.00) |
0.0818 (1.00) |
0.0754 (1.00) |
| NF1 | 29 (10%) | 255 |
0.0435 (1.00) |
0.071 (1.00) |
0.0897 (1.00) |
0.501 (1.00) |
0.00154 (1.00) |
0.00396 (1.00) |
0.23 (1.00) |
0.12 (1.00) |
0.566 (1.00) |
0.915 (1.00) |
| SPTA1 | 26 (9%) | 258 |
0.705 (1.00) |
0.0354 (1.00) |
0.543 (1.00) |
0.913 (1.00) |
0.335 (1.00) |
0.223 (1.00) |
0.508 (1.00) |
1 (1.00) |
0.018 (1.00) |
0.573 (1.00) |
| KRTAP4-11 | 9 (3%) | 275 |
0.113 (1.00) |
0.0873 (1.00) |
1 (1.00) |
0.658 (1.00) |
0.693 (1.00) |
0.343 (1.00) |
0.116 (1.00) |
0.474 (1.00) |
0.209 (1.00) |
0.0823 (1.00) |
| GABRA6 | 11 (4%) | 273 |
0.126 (1.00) |
0.426 (1.00) |
0.506 (1.00) |
0.486 (1.00) |
0.365 (1.00) |
0.943 (1.00) |
0.318 (1.00) |
0.474 (1.00) |
0.124 (1.00) |
1 (1.00) |
| KEL | 15 (5%) | 269 |
0.162 (1.00) |
0.49 (1.00) |
1 (1.00) |
1 (1.00) |
0.394 (1.00) |
0.179 (1.00) |
0.376 (1.00) |
0.911 (1.00) |
0.408 (1.00) |
0.908 (1.00) |
| CDH18 | 11 (4%) | 273 |
0.0384 (1.00) |
0.0166 (1.00) |
0.104 (1.00) |
0.253 (1.00) |
0.853 (1.00) |
0.258 (1.00) |
0.791 (1.00) |
0.776 (1.00) |
0.318 (1.00) |
0.049 (1.00) |
| PRB2 | 6 (2%) | 278 |
0.284 (1.00) |
1 (1.00) |
0.441 (1.00) |
0.634 (1.00) |
0.772 (1.00) |
|||||
| RPL5 | 7 (2%) | 277 |
0.528 (1.00) |
0.174 (1.00) |
0.388 (1.00) |
0.679 (1.00) |
1 (1.00) |
0.882 (1.00) |
0.843 (1.00) |
0.532 (1.00) |
0.465 (1.00) |
1 (1.00) |
| SEMA3C | 11 (4%) | 273 |
0.0448 (1.00) |
0.296 (1.00) |
0.147 (1.00) |
0.383 (1.00) |
0.293 (1.00) |
0.339 (1.00) |
0.532 (1.00) |
0.103 (1.00) |
0.649 (1.00) |
|
| TPTE2 | 8 (3%) | 276 |
0.908 (1.00) |
1 (1.00) |
0.318 (1.00) |
0.289 (1.00) |
0.271 (1.00) |
0.506 (1.00) |
0.624 (1.00) |
0.409 (1.00) |
0.376 (1.00) |
0.741 (1.00) |
| ZNF844 | 6 (2%) | 278 |
0.693 (1.00) |
0.874 (1.00) |
0.143 (1.00) |
0.767 (1.00) |
0.192 (1.00) |
|||||
| OR8K3 | 7 (2%) | 277 |
0.625 (1.00) |
0.376 (1.00) |
0.762 (1.00) |
0.462 (1.00) |
0.0783 (1.00) |
0.765 (1.00) |
0.905 (1.00) |
0.821 (1.00) |
1 (1.00) |
0.764 (1.00) |
| OR5AR1 | 7 (2%) | 277 |
0.241 (1.00) |
0.312 (1.00) |
0.388 (1.00) |
0.679 (1.00) |
0.433 (1.00) |
0.882 (1.00) |
0.514 (1.00) |
0.858 (1.00) |
||
| STAG2 | 12 (4%) | 272 |
0.85 (1.00) |
0.204 (1.00) |
0.21 (1.00) |
0.132 (1.00) |
0.756 (1.00) |
0.676 (1.00) |
0.00694 (1.00) |
0.184 (1.00) |
0.479 (1.00) |
0.771 (1.00) |
| SEMG1 | 8 (3%) | 276 |
0.493 (1.00) |
0.117 (1.00) |
0.762 (1.00) |
1 (1.00) |
0.146 (1.00) |
1 (1.00) |
0.764 (1.00) |
|||
| CDC27 | 6 (2%) | 278 |
0.241 (1.00) |
0.376 (1.00) |
0.261 (1.00) |
0.29 (1.00) |
0.449 (1.00) |
0.138 (1.00) |
0.278 (1.00) |
0.325 (1.00) |
||
| PDGFRA | 11 (4%) | 273 |
0.333 (1.00) |
0.223 (1.00) |
0.729 (1.00) |
0.846 (1.00) |
0.548 (1.00) |
1 (1.00) |
0.851 (1.00) |
0.0265 (1.00) |
0.129 (1.00) |
|
| ADAM29 | 9 (3%) | 275 |
0.82 (1.00) |
0.73 (1.00) |
0.701 (1.00) |
0.794 (1.00) |
0.835 (1.00) |
0.489 (1.00) |
0.673 (1.00) |
0.0559 (1.00) |
0.384 (1.00) |
|
| SULT1B1 | 6 (2%) | 278 |
0.0442 (1.00) |
0.617 (1.00) |
0.27 (1.00) |
0.37 (1.00) |
0.289 (1.00) |
0.496 (1.00) |
0.588 (1.00) |
0.466 (1.00) |
0.802 (1.00) |
|
| ABCC9 | 11 (4%) | 273 |
0.12 (1.00) |
0.316 (1.00) |
0.246 (1.00) |
0.735 (1.00) |
0.503 (1.00) |
0.0162 (1.00) |
0.453 (1.00) |
0.661 (1.00) |
0.336 (1.00) |
0.37 (1.00) |
| NLRP5 | 12 (4%) | 272 |
0.861 (1.00) |
1 (1.00) |
0.929 (1.00) |
0.377 (1.00) |
0.548 (1.00) |
0.477 (1.00) |
0.913 (1.00) |
0.221 (1.00) |
0.72 (1.00) |
0.568 (1.00) |
| LZTR1 | 10 (4%) | 274 |
0.706 (1.00) |
0.795 (1.00) |
0.754 (1.00) |
1 (1.00) |
0.917 (1.00) |
0.138 (1.00) |
0.839 (1.00) |
1 (1.00) |
0.173 (1.00) |
0.536 (1.00) |
| CALCR | 8 (3%) | 276 |
0.253 (1.00) |
0.874 (1.00) |
0.93 (1.00) |
0.736 (1.00) |
0.208 (1.00) |
1 (1.00) |
0.262 (1.00) |
0.506 (1.00) |
||
| QKI | 5 (2%) | 279 |
0.0442 (1.00) |
0.312 (1.00) |
1 (1.00) |
0.454 (1.00) |
0.738 (1.00) |
0.173 (1.00) |
0.496 (1.00) |
0.776 (1.00) |
||
| ZPBP | 5 (2%) | 279 |
0.166 (1.00) |
0.287 (1.00) |
0.0929 (1.00) |
0.454 (1.00) |
0.23 (1.00) |
0.609 (1.00) |
1 (1.00) |
|||
| PSPH | 5 (2%) | 279 |
0.329 (1.00) |
0.533 (1.00) |
0.869 (1.00) |
0.462 (1.00) |
0.0138 (1.00) |
0.106 (1.00) |
0.093 (1.00) |
0.588 (1.00) |
1 (1.00) |
0.536 (1.00) |
| UGT2A3 | 6 (2%) | 278 |
1 (1.00) |
0.533 (1.00) |
0.762 (1.00) |
0.462 (1.00) |
0.673 (1.00) |
0.788 (1.00) |
0.575 (1.00) |
0.532 (1.00) |
||
| WNT2 | 5 (2%) | 279 |
0.0209 (1.00) |
0.00332 (1.00) |
1 (1.00) |
0.789 (1.00) |
1 (1.00) |
0.0416 (1.00) |
0.423 (1.00) |
|||
| OR5D18 | 6 (2%) | 278 |
0.217 (1.00) |
1 (1.00) |
0.475 (1.00) |
0.555 (1.00) |
0.515 (1.00) |
0.531 (1.00) |
0.728 (1.00) |
0.532 (1.00) |
1 (1.00) |
0.764 (1.00) |
| OR5W2 | 5 (2%) | 279 |
0.312 (1.00) |
0.184 (1.00) |
0.328 (1.00) |
0.617 (1.00) |
1 (1.00) |
0.439 (1.00) |
0.195 (1.00) |
|||
| ABCB1 | 10 (4%) | 274 |
0.64 (1.00) |
0.0873 (1.00) |
0.309 (1.00) |
0.23 (1.00) |
0.435 (1.00) |
0.613 (1.00) |
0.931 (1.00) |
0.851 (1.00) |
1 (1.00) |
0.0828 (1.00) |
| COL1A2 | 11 (4%) | 273 |
0.666 (1.00) |
0.333 (1.00) |
0.447 (1.00) |
0.817 (1.00) |
0.859 (1.00) |
0.829 (1.00) |
0.489 (1.00) |
1 (1.00) |
0.855 (1.00) |
0.0126 (1.00) |
| C1ORF150 | 3 (1%) | 281 |
0.379 (1.00) |
0.613 (1.00) |
0.892 (1.00) |
0.776 (1.00) |
||||||
| LRRC55 | 6 (2%) | 278 |
0.329 (1.00) |
0.449 (1.00) |
0.539 (1.00) |
0.143 (1.00) |
0.328 (1.00) |
|||||
| DCAF12L2 | 8 (3%) | 276 |
0.868 (1.00) |
1 (1.00) |
0.631 (1.00) |
0.679 (1.00) |
0.311 (1.00) |
0.697 (1.00) |
0.583 (1.00) |
1 (1.00) |
0.466 (1.00) |
1 (1.00) |
| LRFN5 | 7 (2%) | 277 |
0.896 (1.00) |
0.619 (1.00) |
0.426 (1.00) |
1 (1.00) |
1 (1.00) |
0.817 (1.00) |
0.292 (1.00) |
0.434 (1.00) |
1 (1.00) |
0.848 (1.00) |
| LUM | 5 (2%) | 279 |
0.312 (1.00) |
0.474 (1.00) |
0.507 (1.00) |
1 (1.00) |
0.33 (1.00) |
|||||
| MMP13 | 6 (2%) | 278 |
0.743 (1.00) |
0.726 (1.00) |
0.504 (1.00) |
0.452 (1.00) |
0.88 (1.00) |
0.204 (1.00) |
0.195 (1.00) |
0.336 (1.00) |
0.37 (1.00) |
|
| HEATR7B2 | 11 (4%) | 273 |
0.135 (1.00) |
0.712 (1.00) |
0.344 (1.00) |
0.342 (1.00) |
0.594 (1.00) |
0.355 (1.00) |
0.346 (1.00) |
0.851 (1.00) |
0.72 (1.00) |
1 (1.00) |
| UGT2B28 | 6 (2%) | 278 |
1 (1.00) |
0.67 (1.00) |
1 (1.00) |
0.252 (1.00) |
0.515 (1.00) |
|||||
| OR5D13 | 5 (2%) | 279 |
0.0767 (1.00) |
1 (1.00) |
0.291 (1.00) |
0.831 (1.00) |
0.33 (1.00) |
0.439 (1.00) |
0.0781 (1.00) |
|||
| ZNF99 | 6 (2%) | 278 |
1 (1.00) |
0.821 (1.00) |
0.382 (1.00) |
0.617 (1.00) |
0.385 (1.00) |
0.402 (1.00) |
0.246 (1.00) |
0.3 (1.00) |
||
| SLC5A7 | 7 (2%) | 277 |
0.102 (1.00) |
0.204 (1.00) |
0.352 (1.00) |
0.831 (1.00) |
0.385 (1.00) |
0.765 (1.00) |
0.682 (1.00) |
0.277 (1.00) |
||
| SEMA3E | 8 (3%) | 276 |
0.166 (1.00) |
0.171 (1.00) |
0.504 (1.00) |
0.37 (1.00) |
0.819 (1.00) |
0.0529 (1.00) |
0.00942 (1.00) |
0.0408 (1.00) |
0.336 (1.00) |
0.058 (1.00) |
| TCHH | 16 (6%) | 268 |
0.274 (1.00) |
0.131 (1.00) |
0.43 (1.00) |
0.137 (1.00) |
0.446 (1.00) |
0.354 (1.00) |
0.525 (1.00) |
0.593 (1.00) |
0.322 (1.00) |
0.135 (1.00) |
| SLC26A3 | 7 (2%) | 277 |
0.45 (1.00) |
0.874 (1.00) |
0.554 (1.00) |
0.878 (1.00) |
0.801 (1.00) |
0.167 (1.00) |
0.0104 (1.00) |
0.156 (1.00) |
0.163 (1.00) |
|
| GABRB2 | 6 (2%) | 278 |
0.693 (1.00) |
0.0944 (1.00) |
0.111 (1.00) |
1 (1.00) |
0.112 (1.00) |
0.153 (1.00) |
0.0209 (1.00) |
0.3 (1.00) |
0.572 (1.00) |
0.12 (1.00) |
| CFHR4 | 5 (2%) | 279 |
0.743 (1.00) |
0.853 (1.00) |
0.828 (1.00) |
0.679 (1.00) |
0.524 (1.00) |
0.411 (1.00) |
1 (1.00) |
0.609 (1.00) |
1 (1.00) |
|
| IL18RAP | 6 (2%) | 278 |
0.329 (1.00) |
0.0595 (1.00) |
0.227 (1.00) |
0.121 (1.00) |
0.88 (1.00) |
0.138 (1.00) |
0.532 (1.00) |
0.673 (1.00) |
0.465 (1.00) |
0.764 (1.00) |
| OGDH | 3 (1%) | 281 |
1 (1.00) |
0.792 (1.00) |
1 (1.00) |
0.791 (1.00) |
0.286 (1.00) |
|||||
| PCDH11X | 9 (3%) | 275 |
0.544 (1.00) |
0.533 (1.00) |
0.172 (1.00) |
0.0354 (1.00) |
0.116 (1.00) |
0.0251 (1.00) |
0.0203 (1.00) |
0.663 (1.00) |
0.312 (1.00) |
|
| SPRYD5 | 6 (2%) | 278 |
0.874 (1.00) |
1 (1.00) |
0.214 (1.00) |
0.511 (1.00) |
0.586 (1.00) |
1 (1.00) |
0.434 (1.00) |
0.523 (1.00) |
0.705 (1.00) |
|
| TGFA | 4 (1%) | 280 |
0.688 (1.00) |
1 (1.00) |
0.434 (1.00) |
|||||||
| OR5P2 | 4 (1%) | 280 |
0.652 (1.00) |
0.792 (1.00) |
0.382 (1.00) |
1 (1.00) |
0.193 (1.00) |
|||||
| UGT2B4 | 5 (2%) | 279 |
0.0305 (1.00) |
0.26 (1.00) |
0.328 (1.00) |
0.789 (1.00) |
0.524 (1.00) |
|||||
| FOXR2 | 5 (2%) | 279 |
1 (1.00) |
0.685 (1.00) |
0.754 (1.00) |
0.0385 (1.00) |
0.39 (1.00) |
|||||
| PSG8 | 6 (2%) | 278 |
1 (1.00) |
1 (1.00) |
0.828 (1.00) |
0.364 (1.00) |
0.673 (1.00) |
0.788 (1.00) |
0.703 (1.00) |
1 (1.00) |
0.72 (1.00) |
0.848 (1.00) |
| CNTNAP2 | 13 (5%) | 271 |
0.815 (1.00) |
0.553 (1.00) |
0.17 (1.00) |
0.514 (1.00) |
0.756 (1.00) |
0.676 (1.00) |
0.861 (1.00) |
0.713 (1.00) |
0.685 (1.00) |
1 (1.00) |
| PIK3C2G | 9 (3%) | 275 |
0.226 (1.00) |
0.0355 (1.00) |
0.891 (1.00) |
0.577 (1.00) |
0.266 (1.00) |
0.703 (1.00) |
0.761 (1.00) |
0.709 (1.00) |
||
| SCN9A | 11 (4%) | 273 |
0.686 (1.00) |
0.498 (1.00) |
0.963 (1.00) |
0.31 (1.00) |
0.687 (1.00) |
0.761 (1.00) |
0.851 (1.00) |
0.523 (1.00) |
0.705 (1.00) |
|
| GABRA1 | 6 (2%) | 278 |
0.202 (1.00) |
0.726 (1.00) |
0.703 (1.00) |
0.37 (1.00) |
0.88 (1.00) |
0.439 (1.00) |
0.195 (1.00) |
1 (1.00) |
0.802 (1.00) |
|
| DYNC1I1 | 7 (2%) | 277 |
0.377 (1.00) |
0.0441 (1.00) |
0.47 (1.00) |
0.212 (1.00) |
1 (1.00) |
0.147 (1.00) |
0.821 (1.00) |
|||
| PODXL | 3 (1%) | 281 |
1 (1.00) |
1 (1.00) |
0.507 (1.00) |
0.789 (1.00) |
||||||
| HCN1 | 11 (4%) | 273 |
0.922 (1.00) |
0.921 (1.00) |
0.134 (1.00) |
0.0435 (1.00) |
0.687 (1.00) |
0.953 (1.00) |
1 (1.00) |
0.536 (1.00) |
||
| POTEF | 5 (2%) | 279 |
0.0305 (1.00) |
0.792 (1.00) |
1 (1.00) |
0.789 (1.00) |
0.0709 (1.00) |
|||||
| CXORF22 | 8 (3%) | 276 |
0.307 (1.00) |
0.795 (1.00) |
0.111 (1.00) |
1 (1.00) |
0.385 (1.00) |
0.506 (1.00) |
0.581 (1.00) |
0.574 (1.00) |
0.572 (1.00) |
0.802 (1.00) |
| AFM | 6 (2%) | 278 |
1 (1.00) |
0.533 (1.00) |
0.809 (1.00) |
0.452 (1.00) |
0.515 (1.00) |
0.791 (1.00) |
0.588 (1.00) |
|||
| KRTAP20-2 | 3 (1%) | 281 |
0.312 (1.00) |
0.792 (1.00) |
1 (1.00) |
0.617 (1.00) |
0.281 (1.00) |
0.439 (1.00) |
0.0781 (1.00) |
|||
| OTC | 3 (1%) | 281 |
0.417 (1.00) |
0.792 (1.00) |
0.194 (1.00) |
|||||||
| TRAT1 | 4 (1%) | 280 |
0.469 (1.00) |
0.685 (1.00) |
0.762 (1.00) |
0.789 (1.00) |
0.827 (1.00) |
0.254 (1.00) |
0.764 (1.00) |
|||
| KLK6 | 4 (1%) | 280 |
0.00654 (1.00) |
1 (1.00) |
0.434 (1.00) |
|||||||
| OR4D5 | 4 (1%) | 280 |
0.272 (1.00) |
0.465 (1.00) |
0.352 (1.00) |
1 (1.00) |
0.313 (1.00) |
0.466 (1.00) |
0.163 (1.00) |
|||
| GPX5 | 3 (1%) | 281 |
0.776 (1.00) |
0.339 (1.00) |
0.498 (1.00) |
0.093 (1.00) |
0.588 (1.00) |
1 (1.00) |
0.536 (1.00) |
|||
| KRTAP4-7 | 3 (1%) | 281 |
0.208 (1.00) |
0.26 (1.00) |
0.379 (1.00) |
0.292 (1.00) |
0.434 (1.00) |
|||||
| CDH9 | 8 (3%) | 276 |
0.402 (1.00) |
0.129 (1.00) |
0.616 (1.00) |
0.577 (1.00) |
0.801 (1.00) |
0.335 (1.00) |
0.247 (1.00) |
0.261 (1.00) |
0.233 (1.00) |
|
| CDHR3 | 3 (1%) | 281 |
0.417 (1.00) |
1 (1.00) |
0.194 (1.00) |
|||||||
| OR4P4 | 4 (1%) | 280 |
0.193 (1.00) |
|||||||||
| OR52M1 | 4 (1%) | 280 |
0.469 (1.00) |
1 (1.00) |
0.183 (1.00) |
0.0587 (1.00) |
0.106 (1.00) |
1 (1.00) |
0.764 (1.00) |
|||
| KLF17 | 5 (2%) | 279 |
0.625 (1.00) |
0.449 (1.00) |
0.631 (1.00) |
0.831 (1.00) |
0.738 (1.00) |
0.785 (1.00) |
0.673 (1.00) |
|||
| OR8J3 | 4 (1%) | 280 |
0.693 (1.00) |
0.821 (1.00) |
0.291 (1.00) |
0.174 (1.00) |
0.194 (1.00) |
0.496 (1.00) |
0.434 (1.00) |
|||
| AP3S1 | 3 (1%) | 281 |
0.534 (1.00) |
1 (1.00) |
0.589 (1.00) |
1 (1.00) |
0.633 (1.00) |
|||||
| ST6GAL2 | 6 (2%) | 278 |
1 (1.00) |
0.874 (1.00) |
0.374 (1.00) |
0.878 (1.00) |
0.862 (1.00) |
0.208 (1.00) |
0.163 (1.00) |
|||
| CD3EAP | 3 (1%) | 281 |
1 (1.00) |
0.792 (1.00) |
1 (1.00) |
0.617 (1.00) |
1 (1.00) |
0.609 (1.00) |
1 (1.00) |
|||
| F9 | 4 (1%) | 280 |
0.776 (1.00) |
1 (1.00) |
0.144 (1.00) |
0.183 (1.00) |
1 (1.00) |
|||||
| DPP10 | 7 (2%) | 277 |
0.706 (1.00) |
0.129 (1.00) |
0.267 (1.00) |
0.658 (1.00) |
0.88 (1.00) |
0.647 (1.00) |
0.168 (1.00) |
0.609 (1.00) |
1 (1.00) |
|
| TCN1 | 4 (1%) | 280 |
0.693 (1.00) |
0.567 (1.00) |
0.673 (1.00) |
0.121 (1.00) |
0.391 (1.00) |
0.0529 (1.00) |
0.336 (1.00) |
0.37 (1.00) |
||
| TRPV6 | 7 (2%) | 277 |
0.874 (1.00) |
0.766 (1.00) |
0.238 (1.00) |
0.338 (1.00) |
0.673 (1.00) |
0.164 (1.00) |
0.588 (1.00) |
0.0941 (1.00) |
0.465 (1.00) |
|
| PAN3 | 6 (2%) | 278 |
0.868 (1.00) |
0.617 (1.00) |
0.539 (1.00) |
0.634 (1.00) |
0.772 (1.00) |
|||||
| CDKN2C | 3 (1%) | 281 |
0.652 (1.00) |
0.792 (1.00) |
0.673 (1.00) |
0.617 (1.00) |
0.781 (1.00) |
0.465 (1.00) |
0.286 (1.00) |
|||
| AZGP1 | 5 (2%) | 279 |
0.868 (1.00) |
0.617 (1.00) |
0.109 (1.00) |
0.174 (1.00) |
1 (1.00) |
|||||
| DRD5 | 7 (2%) | 277 |
0.896 (1.00) |
0.619 (1.00) |
0.595 (1.00) |
0.385 (1.00) |
0.433 (1.00) |
0.374 (1.00) |
0.532 (1.00) |
0.791 (1.00) |
1 (1.00) |
|
| FGA | 6 (2%) | 278 |
0.241 (1.00) |
0.617 (1.00) |
0.0809 (1.00) |
0.0647 (1.00) |
1 (1.00) |
0.465 (1.00) |
0.764 (1.00) |
|||
| FGG | 5 (2%) | 279 |
0.217 (1.00) |
0.287 (1.00) |
0.738 (1.00) |
1 (1.00) |
0.209 (1.00) |
0.325 (1.00) |
||||
| OR10G8 | 4 (1%) | 280 |
0.842 (1.00) |
1 (1.00) |
0.869 (1.00) |
1 (1.00) |
0.827 (1.00) |
0.703 (1.00) |
1 (1.00) |
|||
| PCMTD1 | 3 (1%) | 281 |
1 (1.00) |
0.792 (1.00) |
0.781 (1.00) |
0.791 (1.00) |
0.588 (1.00) |
|||||
| PROKR2 | 6 (2%) | 278 |
0.533 (1.00) |
0.211 (1.00) |
0.864 (1.00) |
0.847 (1.00) |
0.112 (1.00) |
0.209 (1.00) |
0.233 (1.00) |
|||
| SHB | 3 (1%) | 281 |
0.312 (1.00) |
0.474 (1.00) |
1 (1.00) |
|||||||
| CYP3A5 | 5 (2%) | 279 |
1 (1.00) |
0.474 (1.00) |
0.862 (1.00) |
0.765 (1.00) |
0.622 (1.00) |
0.132 (1.00) |
0.609 (1.00) |
1 (1.00) |
||
| FRMD7 | 6 (2%) | 278 |
0.693 (1.00) |
1 (1.00) |
0.673 (1.00) |
0.789 (1.00) |
0.449 (1.00) |
0.402 (1.00) |
0.411 (1.00) |
0.673 (1.00) |
0.791 (1.00) |
0.536 (1.00) |
| KCNB2 | 6 (2%) | 278 |
0.451 (1.00) |
0.312 (1.00) |
0.144 (1.00) |
0.679 (1.00) |
0.189 (1.00) |
0.788 (1.00) |
0.439 (1.00) |
0.0781 (1.00) |
0.609 (1.00) |
1 (1.00) |
| KDR | 8 (3%) | 276 |
0.256 (1.00) |
0.304 (1.00) |
0.134 (1.00) |
0.0017 (1.00) |
0.399 (1.00) |
1 (1.00) |
0.144 (1.00) |
0.851 (1.00) |
P value = 7.49e-07 (Fisher's exact test), Q value = 0.00064
Table S1. Gene #3: 'EGFR MUTATION STATUS' versus Clinical Feature #5: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 87 | 108 | 79 |
| EGFR MUTATED | 16 | 47 | 9 |
| EGFR WILD-TYPE | 71 | 61 | 70 |
Figure S1. Get High-res Image Gene #3: 'EGFR MUTATION STATUS' versus Clinical Feature #5: 'CN_CNMF'

P value = 6.37e-05 (Fisher's exact test), Q value = 0.054
Table S2. Gene #3: 'EGFR MUTATION STATUS' versus Clinical Feature #7: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 41 | 38 | 31 | 33 |
| EGFR MUTATED | 13 | 5 | 16 | 2 |
| EGFR WILD-TYPE | 28 | 33 | 15 | 31 |
Figure S2. Get High-res Image Gene #3: 'EGFR MUTATION STATUS' versus Clinical Feature #7: 'RPPA_CNMF'

P value = 1.46e-06 (Fisher's exact test), Q value = 0.0013
Table S3. Gene #3: 'EGFR MUTATION STATUS' versus Clinical Feature #8: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 47 | 28 | 68 |
| EGFR MUTATED | 24 | 6 | 6 |
| EGFR WILD-TYPE | 23 | 22 | 62 |
Figure S3. Get High-res Image Gene #3: 'EGFR MUTATION STATUS' versus Clinical Feature #8: 'RPPA_CHIERARCHICAL'

P value = 1.13e-06 (Fisher's exact test), Q value = 0.00097
Table S4. Gene #4: 'IDH1 MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 76 | 85 | 80 |
| IDH1 MUTATED | 0 | 13 | 0 |
| IDH1 WILD-TYPE | 76 | 72 | 80 |
Figure S4. Get High-res Image Gene #4: 'IDH1 MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'

P value = 4.41e-07 (Fisher's exact test), Q value = 0.00038
Table S5. Gene #4: 'IDH1 MUTATION STATUS' versus Clinical Feature #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 103 | 59 | 79 |
| IDH1 MUTATED | 0 | 0 | 13 |
| IDH1 WILD-TYPE | 103 | 59 | 66 |
Figure S5. Get High-res Image Gene #4: 'IDH1 MUTATION STATUS' versus Clinical Feature #2: 'MRNA_CHIERARCHICAL'

P value = 0.000227 (Fisher's exact test), Q value = 0.19
Table S6. Gene #4: 'IDH1 MUTATION STATUS' versus Clinical Feature #3: 'MIR_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 57 | 72 | 30 | 44 |
| IDH1 MUTATED | 0 | 10 | 0 | 0 |
| IDH1 WILD-TYPE | 57 | 62 | 30 | 44 |
Figure S6. Get High-res Image Gene #4: 'IDH1 MUTATION STATUS' versus Clinical Feature #3: 'MIR_CNMF'

P value = 1.24e-05 (Fisher's exact test), Q value = 0.011
Table S7. Gene #4: 'IDH1 MUTATION STATUS' versus Clinical Feature #4: 'MIR_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 73 | 81 | 49 |
| IDH1 MUTATED | 0 | 1 | 9 |
| IDH1 WILD-TYPE | 73 | 80 | 40 |
Figure S7. Get High-res Image Gene #4: 'IDH1 MUTATION STATUS' versus Clinical Feature #4: 'MIR_CHIERARCHICAL'

P value = 1.68e-05 (Fisher's exact test), Q value = 0.014
Table S8. Gene #4: 'IDH1 MUTATION STATUS' versus Clinical Feature #5: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 87 | 108 | 79 |
| IDH1 MUTATED | 2 | 0 | 11 |
| IDH1 WILD-TYPE | 85 | 108 | 68 |
Figure S8. Get High-res Image Gene #4: 'IDH1 MUTATION STATUS' versus Clinical Feature #5: 'CN_CNMF'

P value = 4.16e-05 (Fisher's exact test), Q value = 0.036
Table S9. Gene #6: 'TP53 MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 76 | 85 | 80 |
| TP53 MUTATED | 19 | 38 | 11 |
| TP53 WILD-TYPE | 57 | 47 | 69 |
Figure S9. Get High-res Image Gene #6: 'TP53 MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'

P value = 0.000166 (Fisher's exact test), Q value = 0.14
Table S10. Gene #6: 'TP53 MUTATION STATUS' versus Clinical Feature #5: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 87 | 108 | 79 |
| TP53 MUTATED | 34 | 16 | 28 |
| TP53 WILD-TYPE | 53 | 92 | 51 |
Figure S10. Get High-res Image Gene #6: 'TP53 MUTATION STATUS' versus Clinical Feature #5: 'CN_CNMF'

P value = 0.000227 (Fisher's exact test), Q value = 0.19
Table S11. Gene #41: 'ATRX MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 76 | 85 | 80 |
| ATRX MUTATED | 2 | 11 | 0 |
| ATRX WILD-TYPE | 74 | 74 | 80 |
Figure S11. Get High-res Image Gene #41: 'ATRX MUTATION STATUS' versus Clinical Feature #1: 'MRNA_CNMF'

P value = 0.000201 (Fisher's exact test), Q value = 0.17
Table S12. Gene #41: 'ATRX MUTATION STATUS' versus Clinical Feature #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 103 | 59 | 79 |
| ATRX MUTATED | 2 | 0 | 11 |
| ATRX WILD-TYPE | 101 | 59 | 68 |
Figure S12. Get High-res Image Gene #41: 'ATRX MUTATION STATUS' versus Clinical Feature #2: 'MRNA_CHIERARCHICAL'
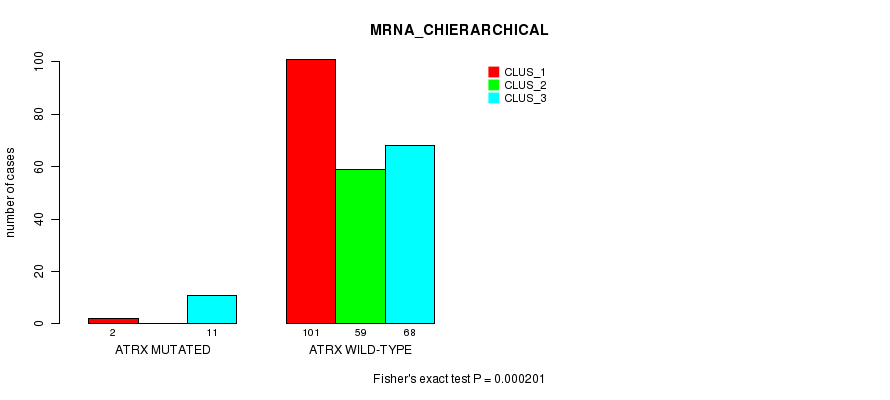
P value = 0.000137 (Fisher's exact test), Q value = 0.12
Table S13. Gene #41: 'ATRX MUTATION STATUS' versus Clinical Feature #4: 'MIR_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 73 | 81 | 49 |
| ATRX MUTATED | 0 | 2 | 8 |
| ATRX WILD-TYPE | 73 | 79 | 41 |
Figure S13. Get High-res Image Gene #41: 'ATRX MUTATION STATUS' versus Clinical Feature #4: 'MIR_CHIERARCHICAL'

P value = 6.23e-05 (Fisher's exact test), Q value = 0.053
Table S14. Gene #41: 'ATRX MUTATION STATUS' versus Clinical Feature #5: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 87 | 108 | 79 |
| ATRX MUTATED | 2 | 1 | 12 |
| ATRX WILD-TYPE | 85 | 107 | 67 |
Figure S14. Get High-res Image Gene #41: 'ATRX MUTATION STATUS' versus Clinical Feature #5: 'CN_CNMF'

-
Mutation data file = GBM-TP.mutsig.cluster.txt
-
Molecular subtypes file = GBM-TP.transferedmergedcluster.txt
-
Number of patients = 284
-
Number of significantly mutated genes = 110
-
Number of Molecular subtypes = 10
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.