This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 37 genes and 8 molecular subtypes across 217 patients, 37 significant findings detected with P value < 0.05 and Q value < 0.25.
-
IDH1 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
TP53 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
PIK3CA mutation correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
ATRX mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
FUBP1 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
CIC mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
NOTCH1 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
PTEN mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MIRSEQ_CNMF'.
-
NF1 mutation correlated to 'METHLYATION_CNMF' and 'MRNASEQ_CNMF'.
-
EGFR mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
Table 1. Get Full Table Overview of the association between mutation status of 37 genes and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 37 significant findings detected.
|
Clinical Features |
MRNA CNMF |
MRNA CHIERARCHICAL |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| TP53 | 113 (52%) | 104 |
0.113 (1.00) |
0.157 (1.00) |
2.78e-23 (6.47e-21) |
6.42e-42 (1.52e-39) |
9.59e-16 (2.19e-13) |
5.01e-12 (1.12e-09) |
0.000891 (0.18) |
0.0181 (1.00) |
| CIC | 41 (19%) | 176 |
0.0502 (1.00) |
0.0625 (1.00) |
6.22e-19 (1.43e-16) |
1.53e-22 (3.55e-20) |
4.98e-13 (1.12e-10) |
5.58e-15 (1.26e-12) |
1.43e-06 (0.000304) |
0.00468 (0.913) |
| NOTCH1 | 19 (9%) | 198 |
0.615 (1.00) |
0.458 (1.00) |
1.01e-05 (0.00212) |
6.03e-06 (0.00127) |
0.000143 (0.0297) |
2.65e-05 (0.00552) |
0.00323 (0.632) |
0.000941 (0.189) |
| ATRX | 93 (43%) | 124 |
0.0256 (1.00) |
0.022 (1.00) |
3.04e-15 (6.9e-13) |
3.48e-29 (8.14e-27) |
1.17e-16 (2.67e-14) |
1.15e-08 (2.49e-06) |
0.00191 (0.378) |
0.0485 (1.00) |
| FUBP1 | 22 (10%) | 195 |
0.0231 (1.00) |
0.0459 (1.00) |
1.06e-09 (2.31e-07) |
8.13e-10 (1.79e-07) |
1.98e-08 (4.27e-06) |
2e-07 (4.31e-05) |
0.0194 (1.00) |
0.222 (1.00) |
| PTEN | 12 (6%) | 205 |
6.19e-09 (1.35e-06) |
4.63e-10 (1.02e-07) |
2.43e-11 (5.39e-09) |
0.00194 (0.382) |
0.000294 (0.0602) |
0.0174 (1.00) |
||
| IDH1 | 165 (76%) | 52 |
0.225 (1.00) |
0.329 (1.00) |
1.02e-12 (2.28e-10) |
1.4e-32 (3.3e-30) |
1.8e-19 (4.15e-17) |
0.0595 (1.00) |
0.0684 (1.00) |
0.106 (1.00) |
| EGFR | 10 (5%) | 207 |
7.43e-06 (0.00156) |
3.01e-07 (6.45e-05) |
0.000501 (0.102) |
0.45 (1.00) |
0.162 (1.00) |
0.835 (1.00) |
||
| PIK3CA | 19 (9%) | 198 |
0.000509 (0.103) |
0.000193 (0.0398) |
0.0642 (1.00) |
0.345 (1.00) |
0.4 (1.00) |
0.246 (1.00) |
||
| NF1 | 16 (7%) | 201 |
0.0451 (1.00) |
1.94e-06 (0.000412) |
0.00101 (0.202) |
0.774 (1.00) |
0.426 (1.00) |
0.551 (1.00) |
||
| IDH2 | 9 (4%) | 208 |
0.103 (1.00) |
0.075 (1.00) |
0.0501 (1.00) |
0.00158 (0.314) |
0.0207 (1.00) |
0.509 (1.00) |
||
| IL32 | 6 (3%) | 211 |
0.0609 (1.00) |
0.0214 (1.00) |
0.33 (1.00) |
0.304 (1.00) |
0.357 (1.00) |
0.28 (1.00) |
||
| PIK3R1 | 14 (6%) | 203 |
0.615 (1.00) |
0.594 (1.00) |
0.00526 (1.00) |
0.0921 (1.00) |
0.5 (1.00) |
0.451 (1.00) |
0.875 (1.00) |
1 (1.00) |
| ZNF844 | 6 (3%) | 211 |
1 (1.00) |
0.534 (1.00) |
0.442 (1.00) |
0.628 (1.00) |
0.185 (1.00) |
1 (1.00) |
||
| TCF12 | 9 (4%) | 208 |
0.907 (1.00) |
0.703 (1.00) |
0.571 (1.00) |
0.193 (1.00) |
0.39 (1.00) |
0.812 (1.00) |
||
| ZBTB20 | 10 (5%) | 207 |
0.415 (1.00) |
0.339 (1.00) |
0.562 (1.00) |
0.908 (1.00) |
0.911 (1.00) |
0.45 (1.00) |
||
| TIMD4 | 6 (3%) | 211 |
0.577 (1.00) |
0.871 (1.00) |
0.711 (1.00) |
0.858 (1.00) |
0.554 (1.00) |
0.754 (1.00) |
||
| CREBZF | 4 (2%) | 213 |
0.161 (1.00) |
0.248 (1.00) |
0.356 (1.00) |
0.0581 (1.00) |
0.343 (1.00) |
0.0769 (1.00) |
||
| ZNF57 | 6 (3%) | 211 |
0.131 (1.00) |
0.218 (1.00) |
0.0976 (1.00) |
0.085 (1.00) |
0.0274 (1.00) |
0.382 (1.00) |
||
| ARID1A | 13 (6%) | 204 |
0.866 (1.00) |
0.651 (1.00) |
0.381 (1.00) |
0.277 (1.00) |
0.339 (1.00) |
0.745 (1.00) |
||
| EMG1 | 4 (2%) | 213 |
0.683 (1.00) |
0.681 (1.00) |
0.97 (1.00) |
0.642 (1.00) |
0.226 (1.00) |
0.68 (1.00) |
||
| PRAMEF11 | 6 (3%) | 211 |
1 (1.00) |
0.328 (1.00) |
0.213 (1.00) |
0.242 (1.00) |
0.222 (1.00) |
0.0269 (1.00) |
||
| PRDM9 | 6 (3%) | 211 |
0.659 (1.00) |
0.218 (1.00) |
0.305 (1.00) |
0.858 (1.00) |
0.437 (1.00) |
1 (1.00) |
||
| NOX4 | 5 (2%) | 212 |
0.102 (1.00) |
0.916 (1.00) |
0.594 (1.00) |
0.238 (1.00) |
0.865 (1.00) |
0.323 (1.00) |
||
| ANKRD30A | 8 (4%) | 209 |
0.529 (1.00) |
0.409 (1.00) |
0.841 (1.00) |
1 (1.00) |
0.503 (1.00) |
0.302 (1.00) |
||
| MUC7 | 5 (2%) | 212 |
0.24 (1.00) |
0.256 (1.00) |
0.401 (1.00) |
0.57 (1.00) |
0.805 (1.00) |
0.72 (1.00) |
||
| PAX4 | 5 (2%) | 212 |
0.453 (1.00) |
0.916 (1.00) |
0.757 (1.00) |
0.705 (1.00) |
0.473 (1.00) |
0.72 (1.00) |
||
| ZNF845 | 6 (3%) | 211 |
0.278 (1.00) |
0.929 (1.00) |
0.711 (1.00) |
0.628 (1.00) |
0.839 (1.00) |
0.535 (1.00) |
||
| C15ORF2 | 8 (4%) | 209 |
0.239 (1.00) |
0.339 (1.00) |
0.0132 (1.00) |
0.318 (1.00) |
0.69 (1.00) |
0.302 (1.00) |
||
| SPDYE5 | 4 (2%) | 213 |
0.0526 (1.00) |
0.0338 (1.00) |
0.0565 (1.00) |
0.286 (1.00) |
0.0712 (1.00) |
0.25 (1.00) |
||
| SCAF1 | 4 (2%) | 213 |
1 (1.00) |
1 (1.00) |
0.424 (1.00) |
0.373 (1.00) |
0.0853 (1.00) |
1 (1.00) |
||
| FSTL5 | 6 (3%) | 211 |
0.0609 (1.00) |
0.304 (1.00) |
0.118 (1.00) |
0.0256 (1.00) |
0.0875 (1.00) |
0.382 (1.00) |
||
| C9ORF79 | 8 (4%) | 209 |
1 (1.00) |
0.467 (1.00) |
0.657 (1.00) |
0.779 (1.00) |
0.792 (1.00) |
0.801 (1.00) |
||
| ZCCHC12 | 3 (1%) | 214 |
0.0543 (1.00) |
0.0816 (1.00) |
0.546 (1.00) |
0.144 (1.00) |
0.434 (1.00) |
0.191 (1.00) |
||
| SMARCA4 | 13 (6%) | 204 |
0.714 (1.00) |
0.612 (1.00) |
0.5 (1.00) |
0.676 (1.00) |
0.35 (1.00) |
0.412 (1.00) |
||
| MYH4 | 5 (2%) | 212 |
0.453 (1.00) |
0.916 (1.00) |
0.418 (1.00) |
0.107 (1.00) |
0.00846 (1.00) |
0.72 (1.00) |
||
| TRDN | 4 (2%) | 213 |
0.448 (1.00) |
1 (1.00) |
0.97 (1.00) |
0.642 (1.00) |
1 (1.00) |
0.427 (1.00) |
P value = 1.02e-12 (Fisher's exact test), Q value = 2.3e-10
Table S1. Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 90 | 49 | 76 |
| IDH1 MUTATED | 78 | 17 | 69 |
| IDH1 WILD-TYPE | 12 | 32 | 7 |
Figure S1. Get High-res Image Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
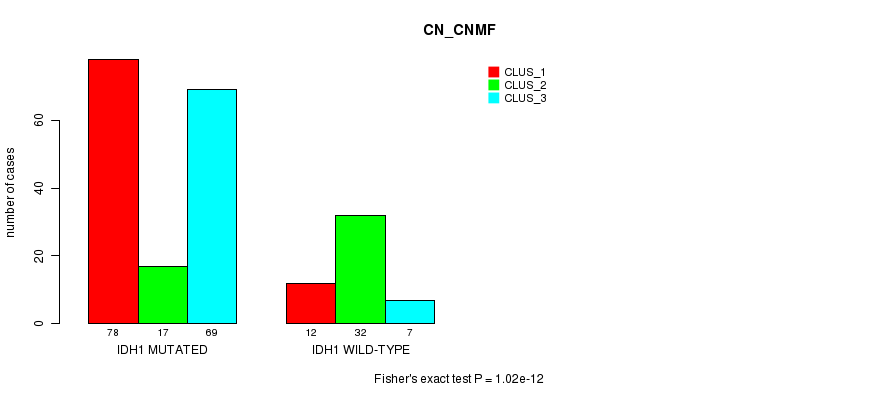
P value = 1.4e-32 (Fisher's exact test), Q value = 3.3e-30
Table S2. Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 98 | 32 | 59 | 23 |
| IDH1 MUTATED | 96 | 0 | 54 | 11 |
| IDH1 WILD-TYPE | 2 | 32 | 5 | 12 |
Figure S2. Get High-res Image Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
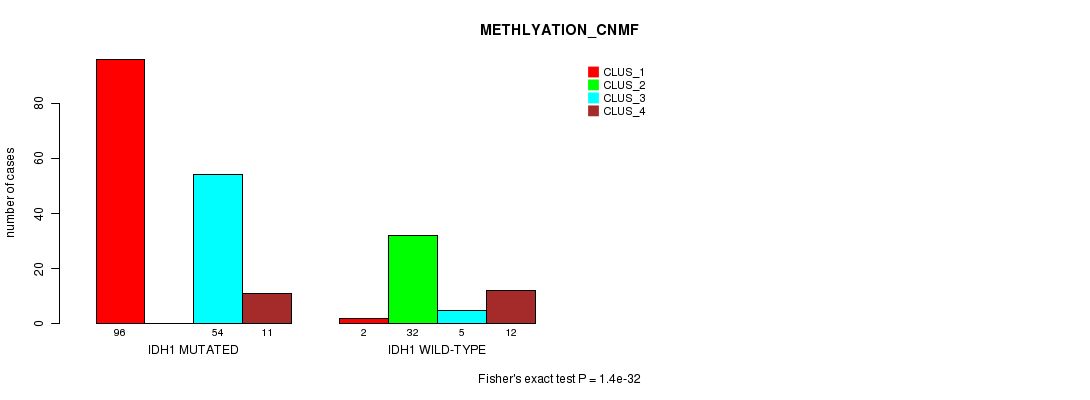
P value = 1.8e-19 (Chi-square test), Q value = 4.1e-17
Table S3. Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 58 | 40 | 36 | 71 | 10 |
| IDH1 MUTATED | 58 | 8 | 33 | 56 | 9 |
| IDH1 WILD-TYPE | 0 | 32 | 3 | 15 | 1 |
Figure S3. Get High-res Image Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
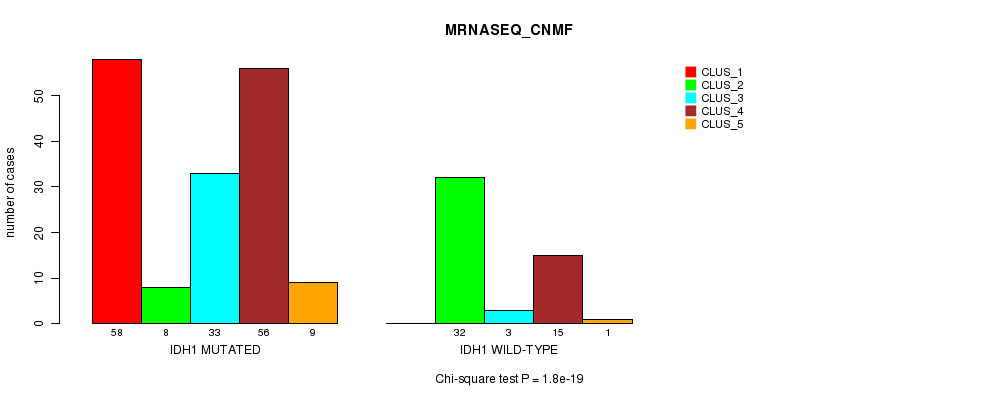
P value = 2.78e-23 (Fisher's exact test), Q value = 6.5e-21
Table S4. Gene #4: 'TP53 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 90 | 49 | 76 |
| TP53 MUTATED | 80 | 22 | 11 |
| TP53 WILD-TYPE | 10 | 27 | 65 |
Figure S4. Get High-res Image Gene #4: 'TP53 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
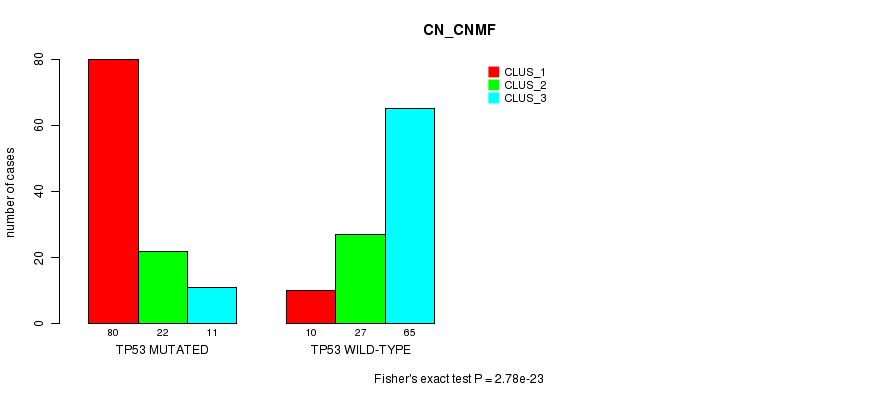
P value = 6.42e-42 (Fisher's exact test), Q value = 1.5e-39
Table S5. Gene #4: 'TP53 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 98 | 32 | 59 | 23 |
| TP53 MUTATED | 96 | 6 | 3 | 7 |
| TP53 WILD-TYPE | 2 | 26 | 56 | 16 |
Figure S5. Get High-res Image Gene #4: 'TP53 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
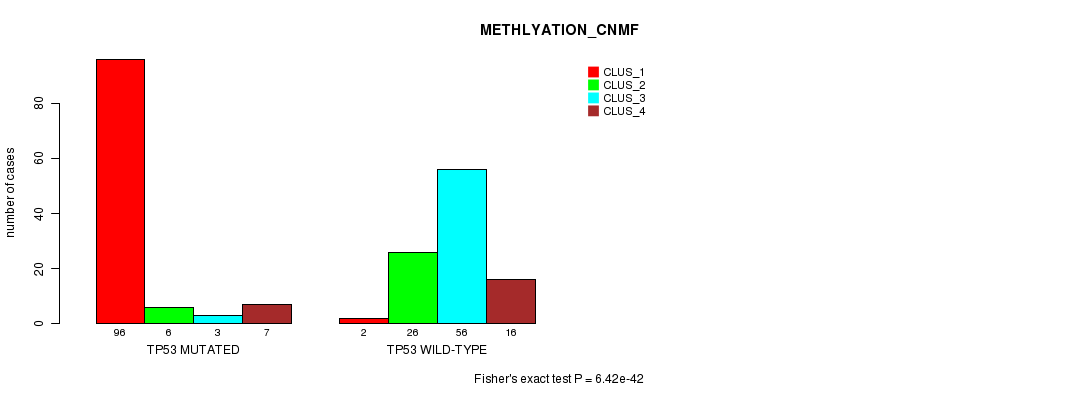
P value = 9.59e-16 (Chi-square test), Q value = 2.2e-13
Table S6. Gene #4: 'TP53 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 58 | 40 | 36 | 71 | 10 |
| TP53 MUTATED | 56 | 14 | 4 | 32 | 6 |
| TP53 WILD-TYPE | 2 | 26 | 32 | 39 | 4 |
Figure S6. Get High-res Image Gene #4: 'TP53 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
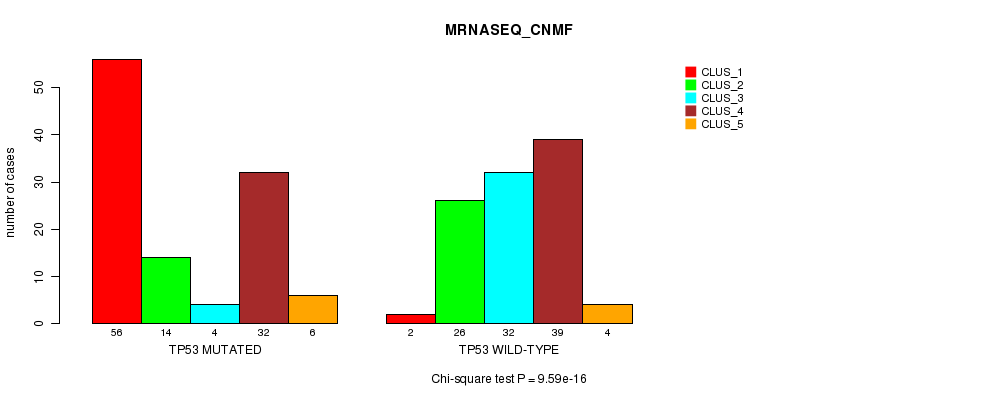
P value = 5.01e-12 (Fisher's exact test), Q value = 1.1e-09
Table S7. Gene #4: 'TP53 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 112 | 70 |
| TP53 MUTATED | 2 | 80 | 30 |
| TP53 WILD-TYPE | 31 | 32 | 40 |
Figure S7. Get High-res Image Gene #4: 'TP53 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
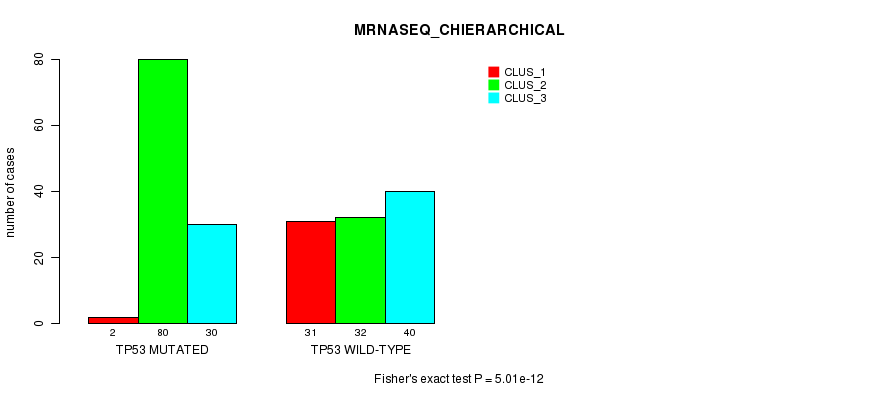
P value = 0.000891 (Fisher's exact test), Q value = 0.18
Table S8. Gene #4: 'TP53 MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 55 | 33 | 82 | 46 |
| TP53 MUTATED | 35 | 12 | 33 | 32 |
| TP53 WILD-TYPE | 20 | 21 | 49 | 14 |
Figure S8. Get High-res Image Gene #4: 'TP53 MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
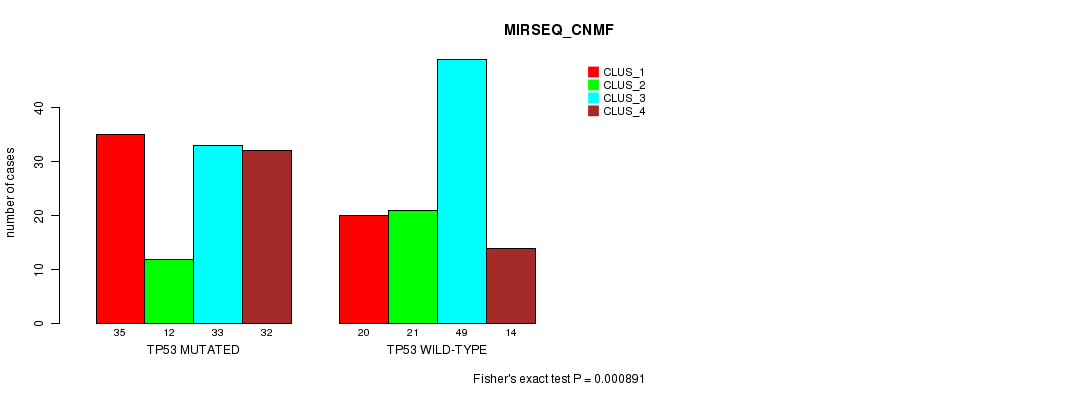
P value = 0.000509 (Fisher's exact test), Q value = 0.1
Table S9. Gene #6: 'PIK3CA MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 90 | 49 | 76 |
| PIK3CA MUTATED | 1 | 5 | 13 |
| PIK3CA WILD-TYPE | 89 | 44 | 63 |
Figure S9. Get High-res Image Gene #6: 'PIK3CA MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
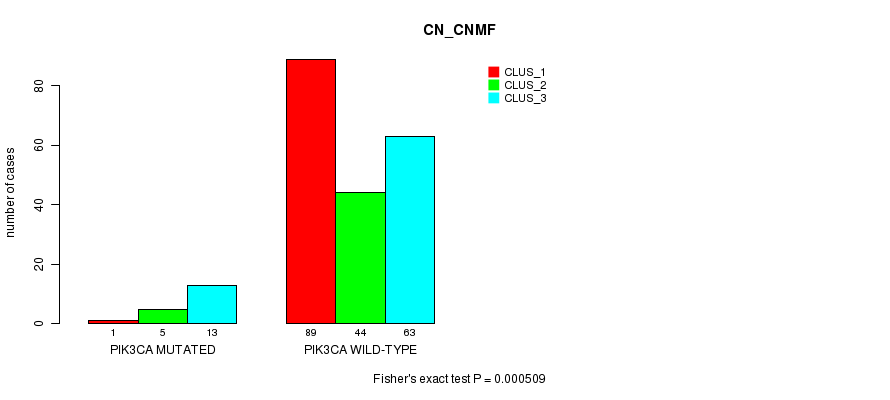
P value = 0.000193 (Fisher's exact test), Q value = 0.04
Table S10. Gene #6: 'PIK3CA MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 98 | 32 | 59 | 23 |
| PIK3CA MUTATED | 1 | 5 | 11 | 2 |
| PIK3CA WILD-TYPE | 97 | 27 | 48 | 21 |
Figure S10. Get High-res Image Gene #6: 'PIK3CA MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
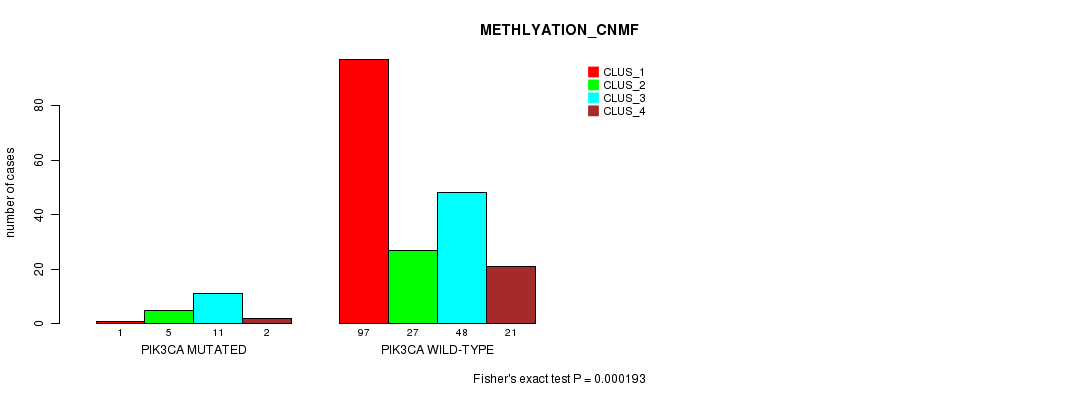
P value = 3.04e-15 (Fisher's exact test), Q value = 6.9e-13
Table S11. Gene #7: 'ATRX MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 90 | 49 | 76 |
| ATRX MUTATED | 66 | 17 | 10 |
| ATRX WILD-TYPE | 24 | 32 | 66 |
Figure S11. Get High-res Image Gene #7: 'ATRX MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
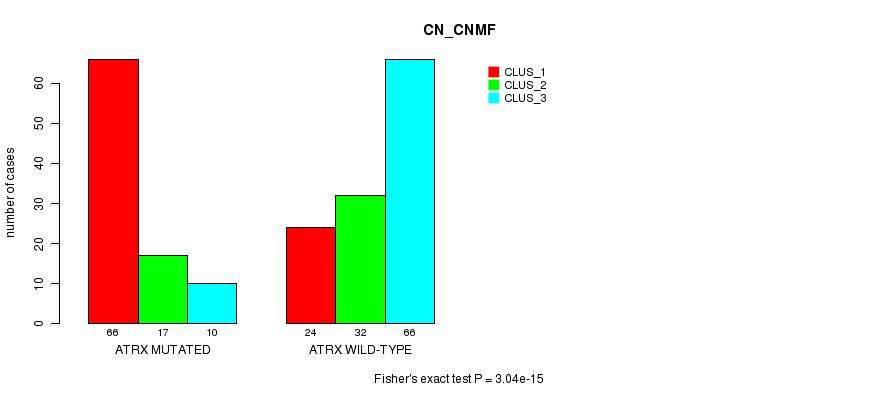
P value = 3.48e-29 (Fisher's exact test), Q value = 8.1e-27
Table S12. Gene #7: 'ATRX MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 98 | 32 | 59 | 23 |
| ATRX MUTATED | 81 | 4 | 2 | 5 |
| ATRX WILD-TYPE | 17 | 28 | 57 | 18 |
Figure S12. Get High-res Image Gene #7: 'ATRX MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
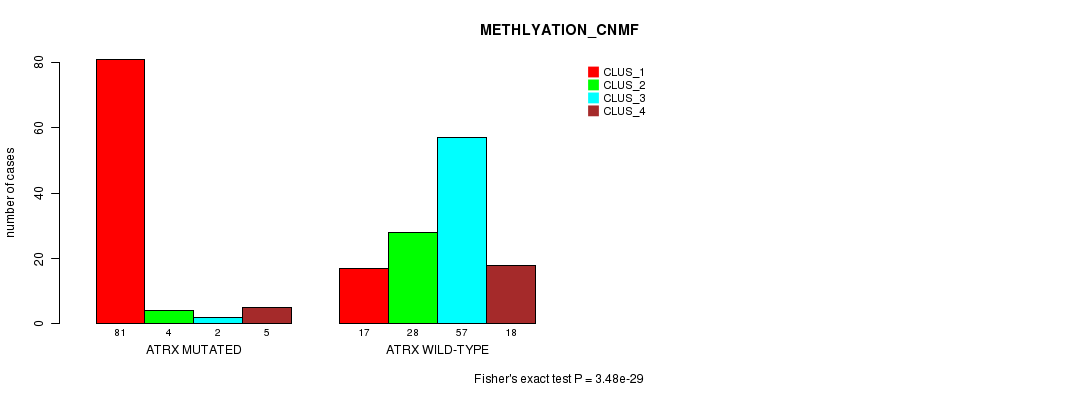
P value = 1.17e-16 (Chi-square test), Q value = 2.7e-14
Table S13. Gene #7: 'ATRX MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 58 | 40 | 36 | 71 | 10 |
| ATRX MUTATED | 52 | 10 | 2 | 26 | 2 |
| ATRX WILD-TYPE | 6 | 30 | 34 | 45 | 8 |
Figure S13. Get High-res Image Gene #7: 'ATRX MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
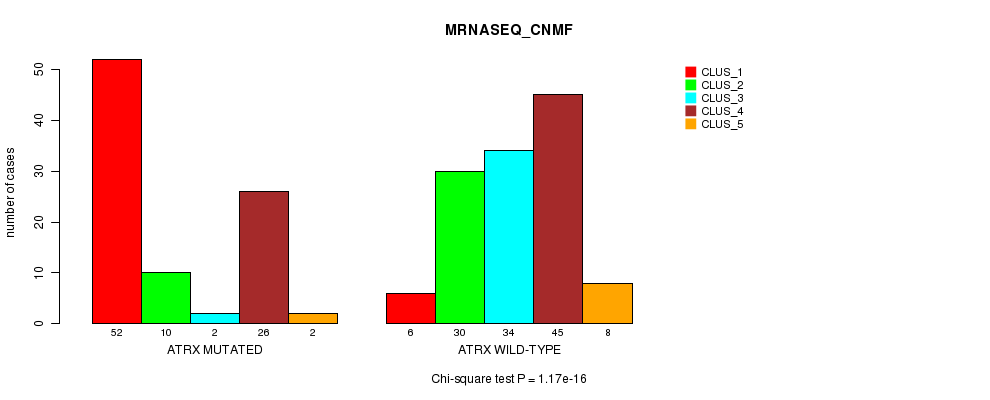
P value = 1.15e-08 (Fisher's exact test), Q value = 2.5e-06
Table S14. Gene #7: 'ATRX MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 112 | 70 |
| ATRX MUTATED | 1 | 64 | 27 |
| ATRX WILD-TYPE | 32 | 48 | 43 |
Figure S14. Get High-res Image Gene #7: 'ATRX MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
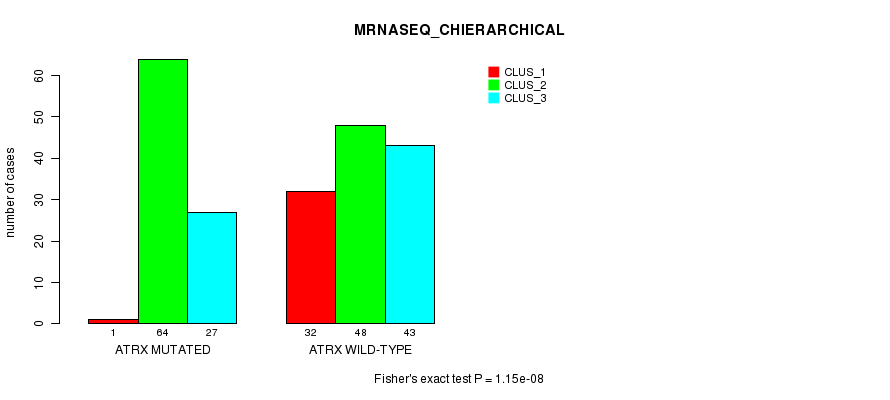
P value = 1.06e-09 (Fisher's exact test), Q value = 2.3e-07
Table S15. Gene #8: 'FUBP1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 90 | 49 | 76 |
| FUBP1 MUTATED | 0 | 1 | 21 |
| FUBP1 WILD-TYPE | 90 | 48 | 55 |
Figure S15. Get High-res Image Gene #8: 'FUBP1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
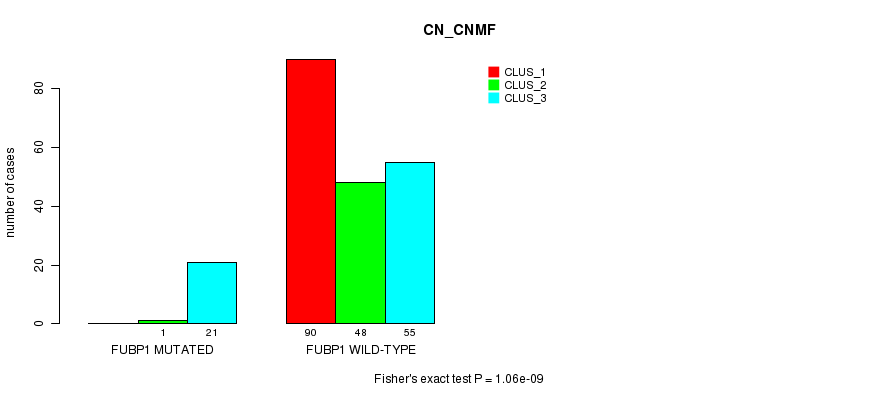
P value = 8.13e-10 (Fisher's exact test), Q value = 1.8e-07
Table S16. Gene #8: 'FUBP1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 98 | 32 | 59 | 23 |
| FUBP1 MUTATED | 0 | 1 | 18 | 0 |
| FUBP1 WILD-TYPE | 98 | 31 | 41 | 23 |
Figure S16. Get High-res Image Gene #8: 'FUBP1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
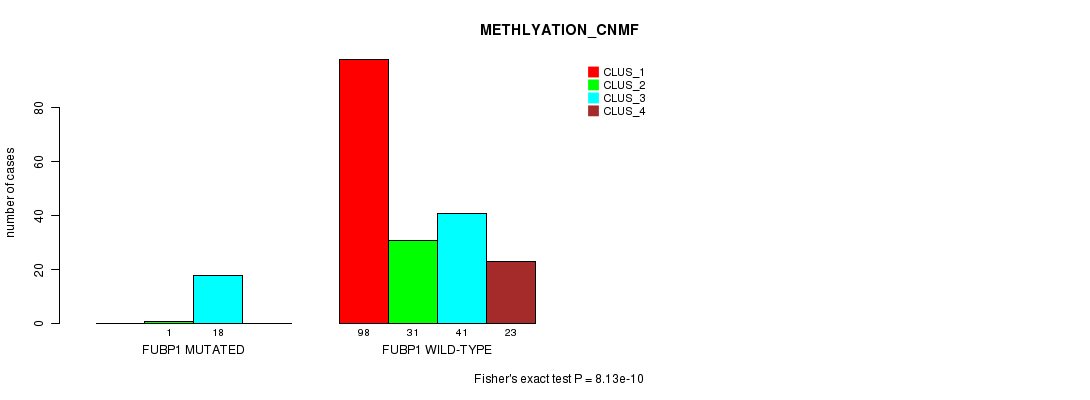
P value = 1.98e-08 (Chi-square test), Q value = 4.3e-06
Table S17. Gene #8: 'FUBP1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 58 | 40 | 36 | 71 | 10 |
| FUBP1 MUTATED | 0 | 1 | 14 | 6 | 1 |
| FUBP1 WILD-TYPE | 58 | 39 | 22 | 65 | 9 |
Figure S17. Get High-res Image Gene #8: 'FUBP1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
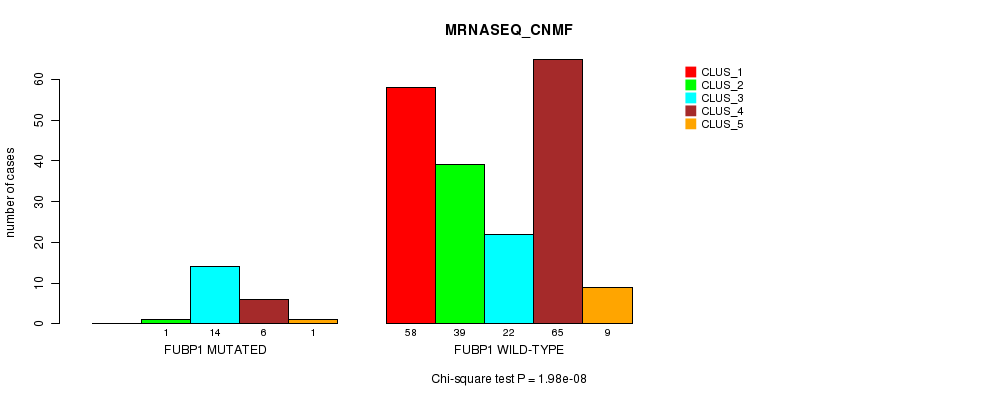
P value = 2e-07 (Fisher's exact test), Q value = 4.3e-05
Table S18. Gene #8: 'FUBP1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 112 | 70 |
| FUBP1 MUTATED | 13 | 3 | 6 |
| FUBP1 WILD-TYPE | 20 | 109 | 64 |
Figure S18. Get High-res Image Gene #8: 'FUBP1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
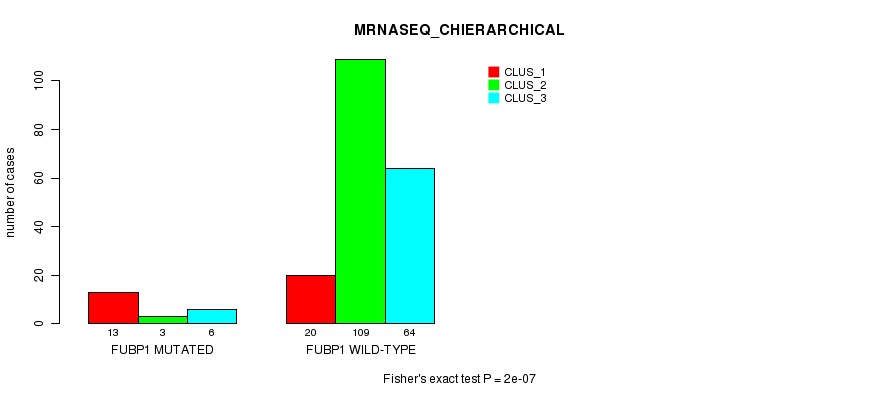
P value = 6.22e-19 (Fisher's exact test), Q value = 1.4e-16
Table S19. Gene #9: 'CIC MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 90 | 49 | 76 |
| CIC MUTATED | 1 | 1 | 39 |
| CIC WILD-TYPE | 89 | 48 | 37 |
Figure S19. Get High-res Image Gene #9: 'CIC MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
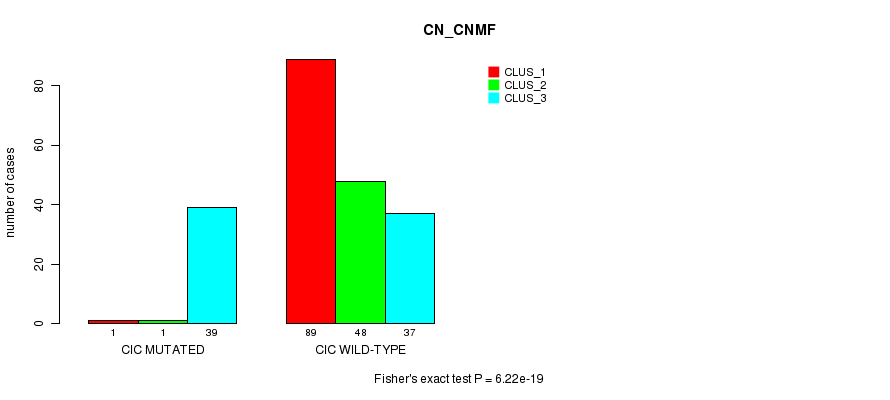
P value = 1.53e-22 (Fisher's exact test), Q value = 3.6e-20
Table S20. Gene #9: 'CIC MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 98 | 32 | 59 | 23 |
| CIC MUTATED | 0 | 1 | 37 | 2 |
| CIC WILD-TYPE | 98 | 31 | 22 | 21 |
Figure S20. Get High-res Image Gene #9: 'CIC MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
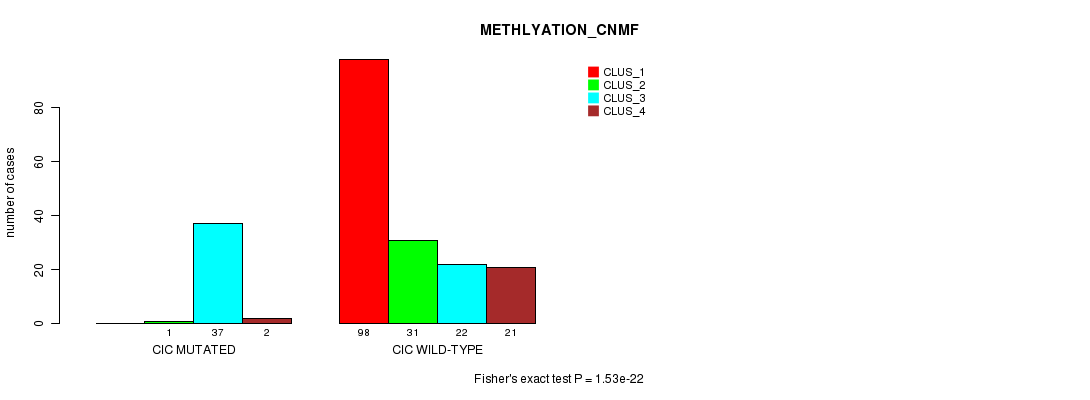
P value = 4.98e-13 (Chi-square test), Q value = 1.1e-10
Table S21. Gene #9: 'CIC MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 58 | 40 | 36 | 71 | 10 |
| CIC MUTATED | 0 | 1 | 22 | 17 | 1 |
| CIC WILD-TYPE | 58 | 39 | 14 | 54 | 9 |
Figure S21. Get High-res Image Gene #9: 'CIC MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
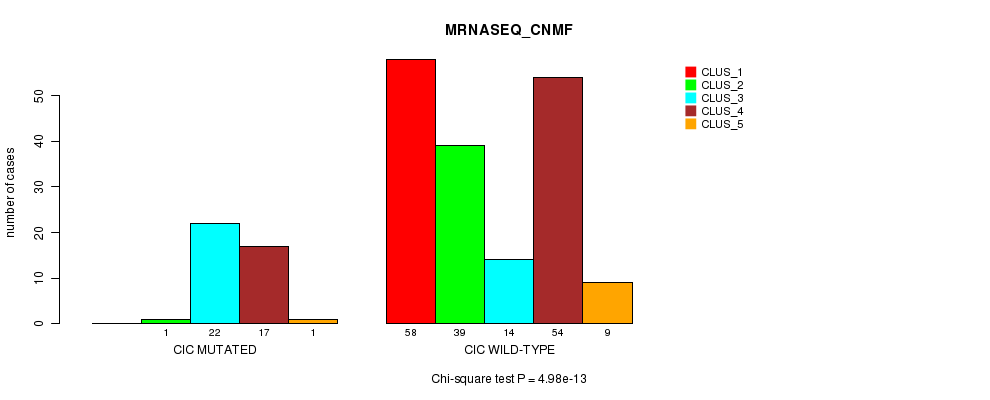
P value = 5.58e-15 (Fisher's exact test), Q value = 1.3e-12
Table S22. Gene #9: 'CIC MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 112 | 70 |
| CIC MUTATED | 22 | 3 | 16 |
| CIC WILD-TYPE | 11 | 109 | 54 |
Figure S22. Get High-res Image Gene #9: 'CIC MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
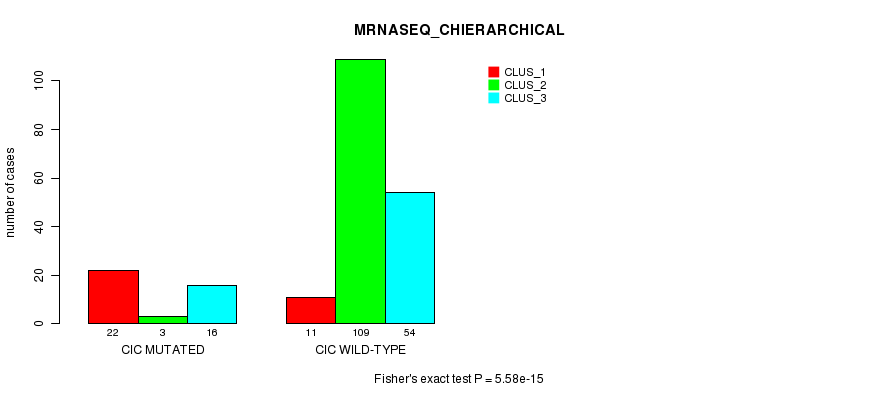
P value = 1.43e-06 (Fisher's exact test), Q value = 3e-04
Table S23. Gene #9: 'CIC MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 55 | 33 | 82 | 46 |
| CIC MUTATED | 6 | 16 | 18 | 1 |
| CIC WILD-TYPE | 49 | 17 | 64 | 45 |
Figure S23. Get High-res Image Gene #9: 'CIC MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
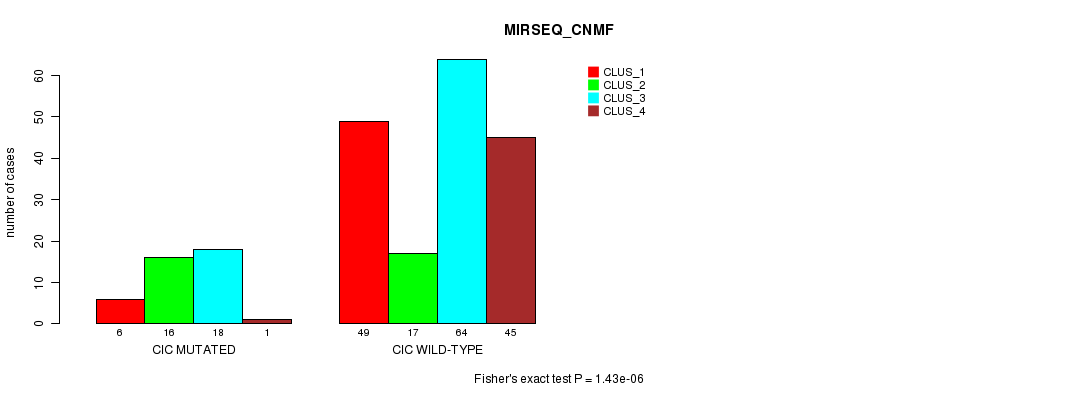
P value = 1.01e-05 (Fisher's exact test), Q value = 0.0021
Table S24. Gene #10: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 90 | 49 | 76 |
| NOTCH1 MUTATED | 1 | 2 | 16 |
| NOTCH1 WILD-TYPE | 89 | 47 | 60 |
Figure S24. Get High-res Image Gene #10: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
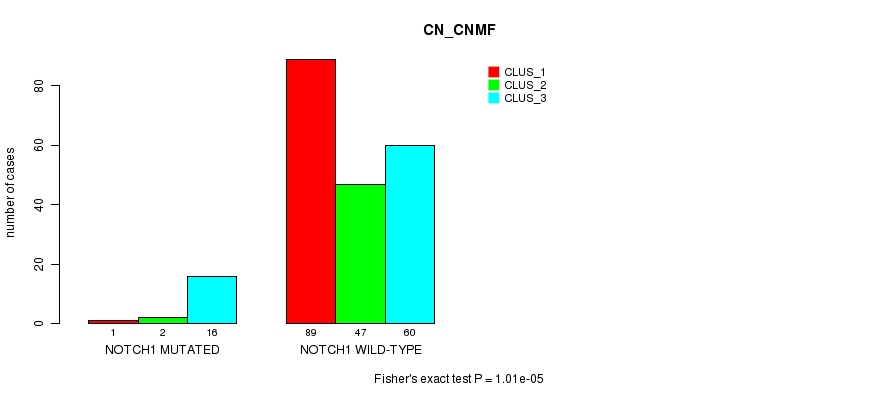
P value = 6.03e-06 (Fisher's exact test), Q value = 0.0013
Table S25. Gene #10: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 98 | 32 | 59 | 23 |
| NOTCH1 MUTATED | 2 | 1 | 15 | 0 |
| NOTCH1 WILD-TYPE | 96 | 31 | 44 | 23 |
Figure S25. Get High-res Image Gene #10: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
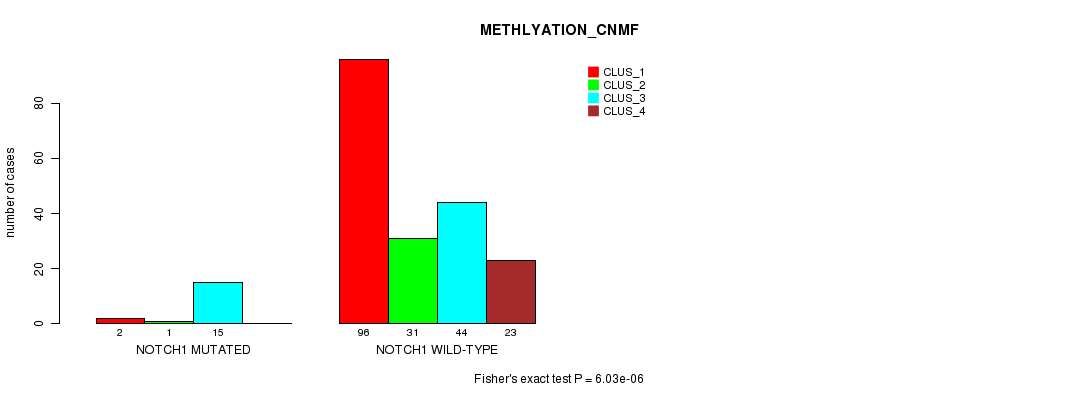
P value = 0.000143 (Chi-square test), Q value = 0.03
Table S26. Gene #10: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 58 | 40 | 36 | 71 | 10 |
| NOTCH1 MUTATED | 1 | 1 | 10 | 7 | 0 |
| NOTCH1 WILD-TYPE | 57 | 39 | 26 | 64 | 10 |
Figure S26. Get High-res Image Gene #10: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
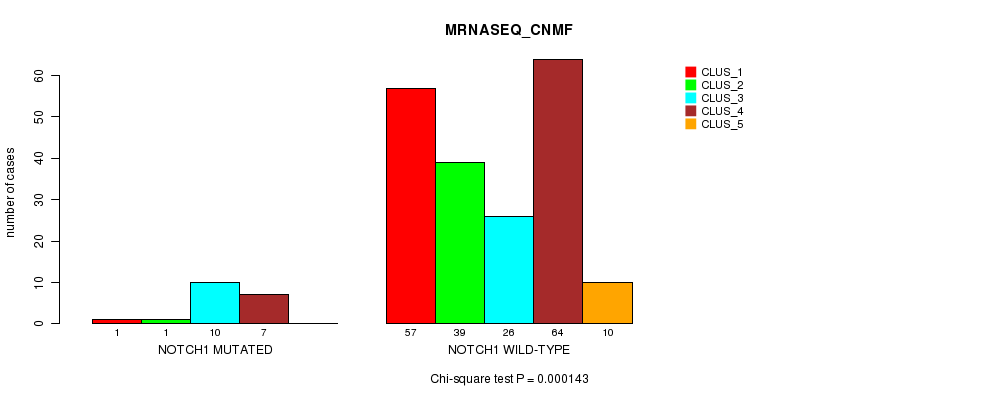
P value = 2.65e-05 (Fisher's exact test), Q value = 0.0055
Table S27. Gene #10: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 112 | 70 |
| NOTCH1 MUTATED | 9 | 2 | 8 |
| NOTCH1 WILD-TYPE | 24 | 110 | 62 |
Figure S27. Get High-res Image Gene #10: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #6: 'MRNASEQ_CHIERARCHICAL'
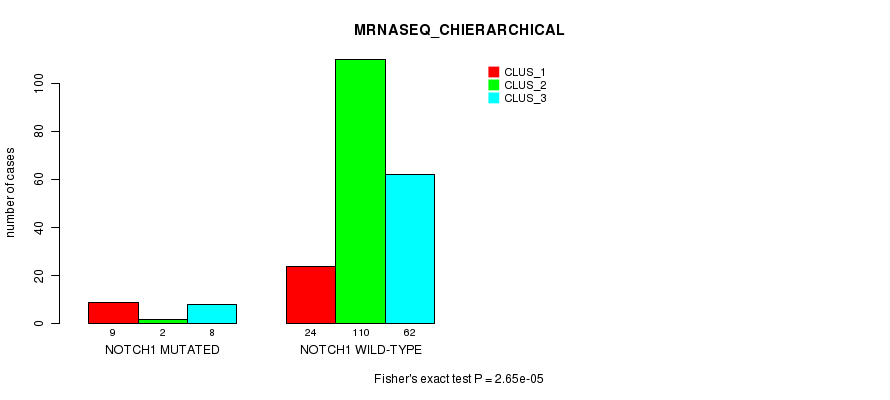
P value = 0.000941 (Fisher's exact test), Q value = 0.19
Table S28. Gene #10: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 8 | 95 | 113 |
| NOTCH1 MUTATED | 0 | 16 | 3 |
| NOTCH1 WILD-TYPE | 8 | 79 | 110 |
Figure S28. Get High-res Image Gene #10: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #8: 'MIRSEQ_CHIERARCHICAL'
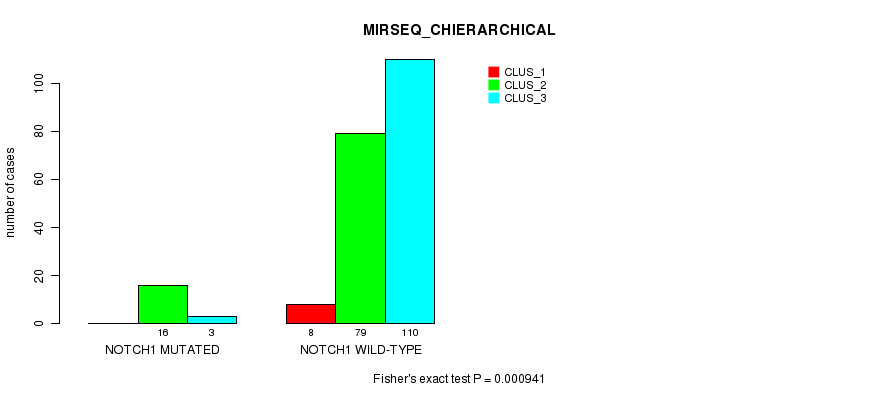
P value = 6.19e-09 (Fisher's exact test), Q value = 1.3e-06
Table S29. Gene #11: 'PTEN MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 90 | 49 | 76 |
| PTEN MUTATED | 0 | 12 | 0 |
| PTEN WILD-TYPE | 90 | 37 | 76 |
Figure S29. Get High-res Image Gene #11: 'PTEN MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
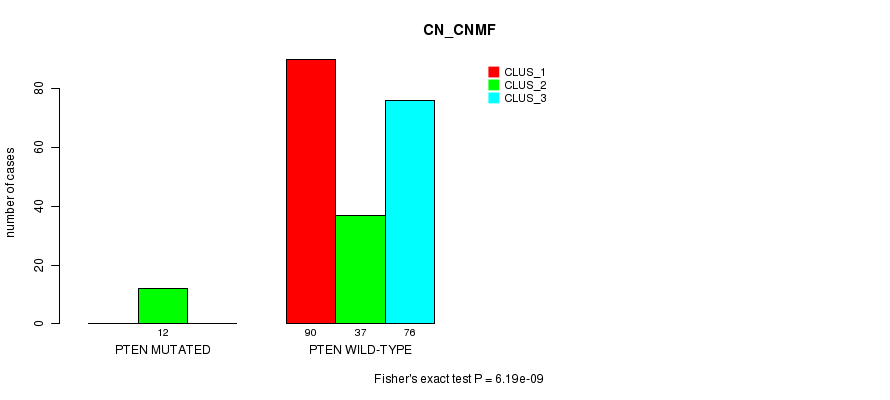
P value = 4.63e-10 (Fisher's exact test), Q value = 1e-07
Table S30. Gene #11: 'PTEN MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 98 | 32 | 59 | 23 |
| PTEN MUTATED | 0 | 11 | 0 | 0 |
| PTEN WILD-TYPE | 98 | 21 | 59 | 23 |
Figure S30. Get High-res Image Gene #11: 'PTEN MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
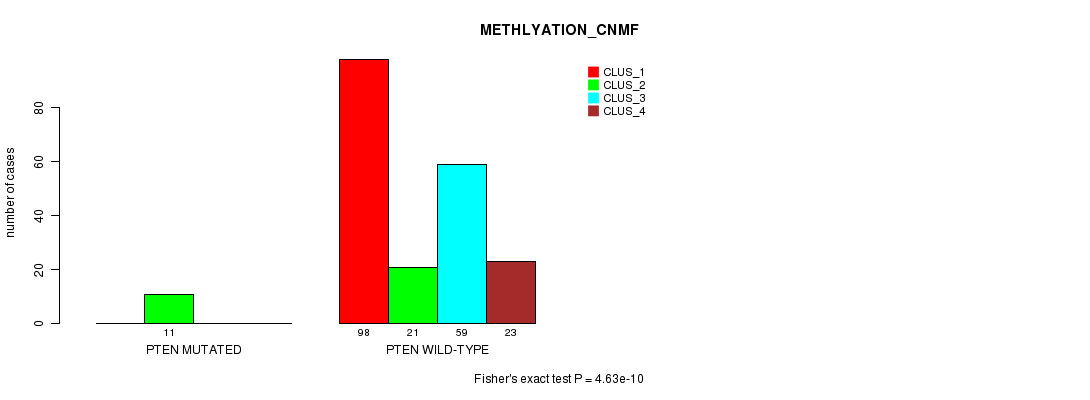
P value = 2.43e-11 (Chi-square test), Q value = 5.4e-09
Table S31. Gene #11: 'PTEN MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 58 | 40 | 36 | 71 | 10 |
| PTEN MUTATED | 0 | 12 | 0 | 0 | 0 |
| PTEN WILD-TYPE | 58 | 28 | 36 | 71 | 10 |
Figure S31. Get High-res Image Gene #11: 'PTEN MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
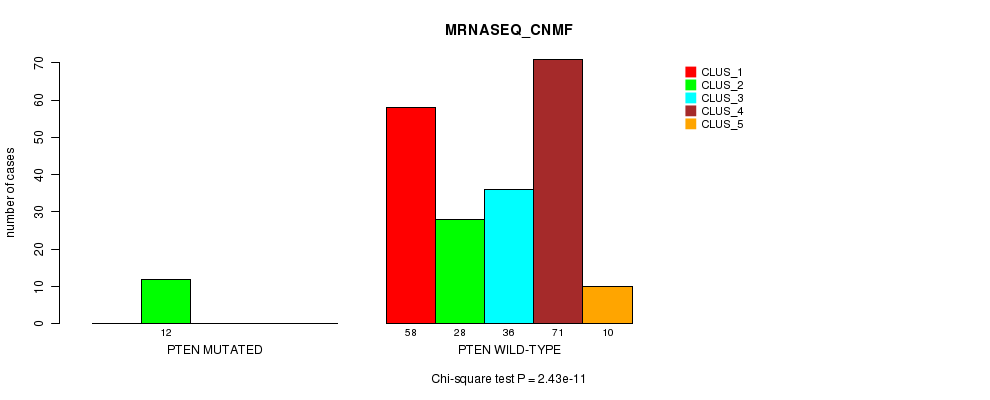
P value = 0.000294 (Fisher's exact test), Q value = 0.06
Table S32. Gene #11: 'PTEN MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 55 | 33 | 82 | 46 |
| PTEN MUTATED | 8 | 0 | 0 | 4 |
| PTEN WILD-TYPE | 47 | 33 | 82 | 42 |
Figure S32. Get High-res Image Gene #11: 'PTEN MUTATION STATUS' versus Clinical Feature #7: 'MIRSEQ_CNMF'
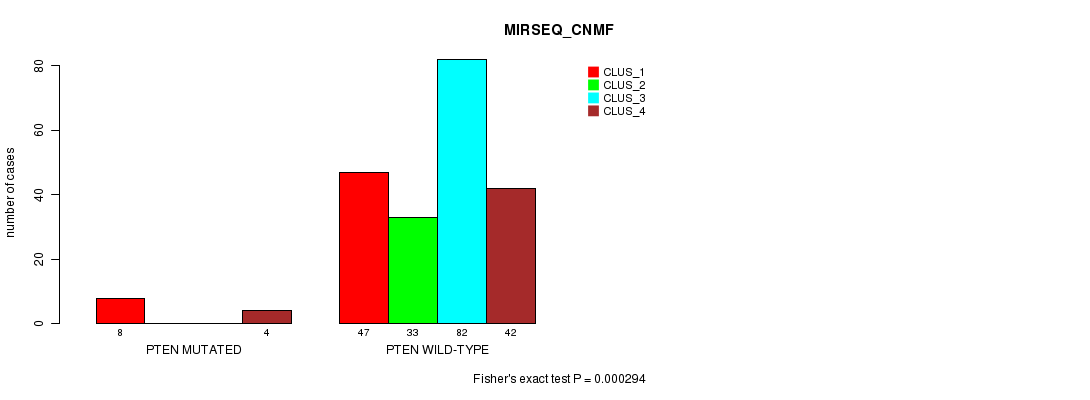
P value = 1.94e-06 (Fisher's exact test), Q value = 0.00041
Table S33. Gene #12: 'NF1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 98 | 32 | 59 | 23 |
| NF1 MUTATED | 1 | 8 | 0 | 3 |
| NF1 WILD-TYPE | 97 | 24 | 59 | 20 |
Figure S33. Get High-res Image Gene #12: 'NF1 MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
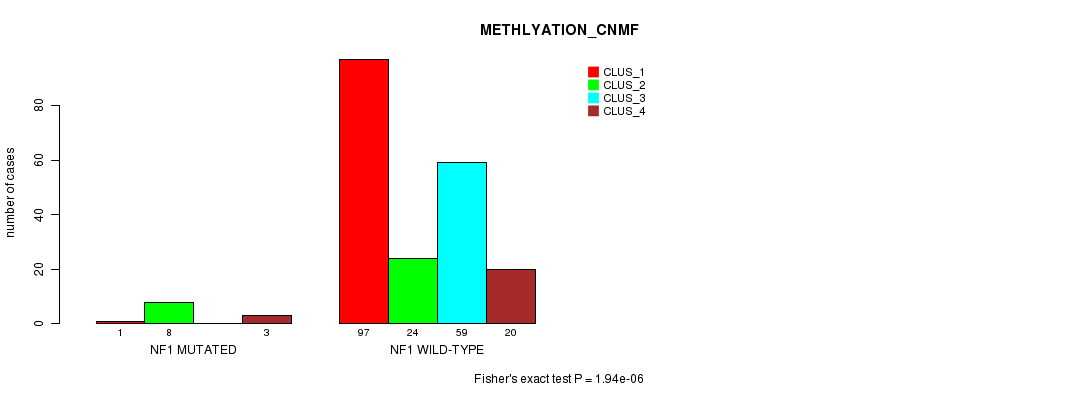
P value = 0.00101 (Chi-square test), Q value = 0.2
Table S34. Gene #12: 'NF1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 58 | 40 | 36 | 71 | 10 |
| NF1 MUTATED | 0 | 9 | 2 | 4 | 1 |
| NF1 WILD-TYPE | 58 | 31 | 34 | 67 | 9 |
Figure S34. Get High-res Image Gene #12: 'NF1 MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
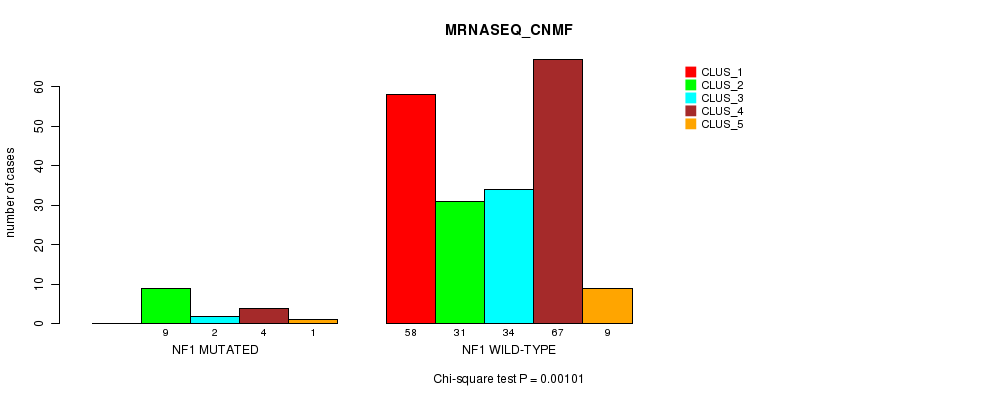
P value = 7.43e-06 (Fisher's exact test), Q value = 0.0016
Table S35. Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 90 | 49 | 76 |
| EGFR MUTATED | 1 | 9 | 0 |
| EGFR WILD-TYPE | 89 | 40 | 76 |
Figure S35. Get High-res Image Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #3: 'CN_CNMF'
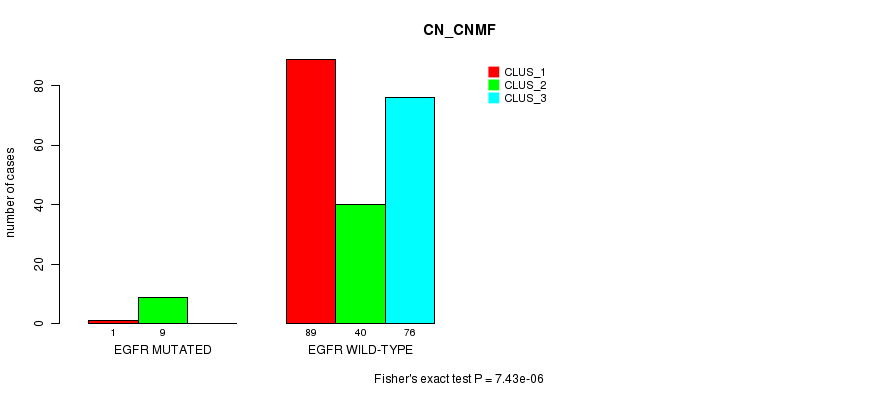
P value = 3.01e-07 (Fisher's exact test), Q value = 6.4e-05
Table S36. Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 98 | 32 | 59 | 23 |
| EGFR MUTATED | 0 | 8 | 0 | 2 |
| EGFR WILD-TYPE | 98 | 24 | 59 | 21 |
Figure S36. Get High-res Image Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #4: 'METHLYATION_CNMF'
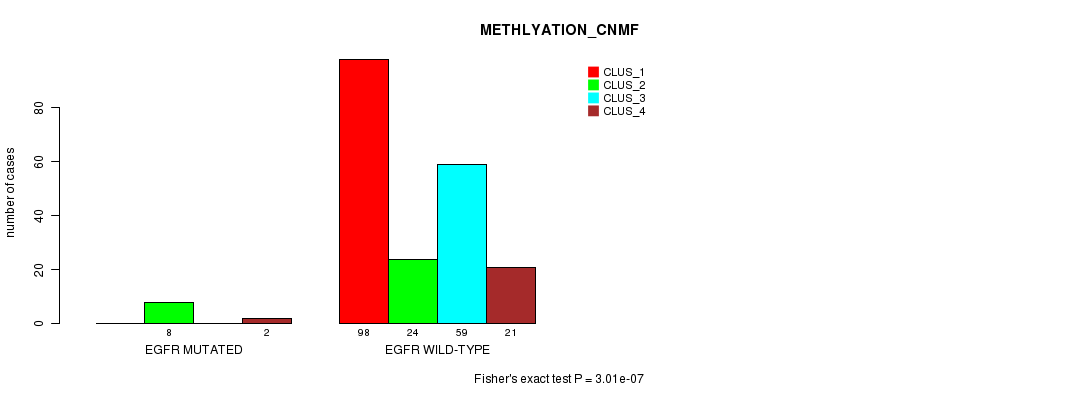
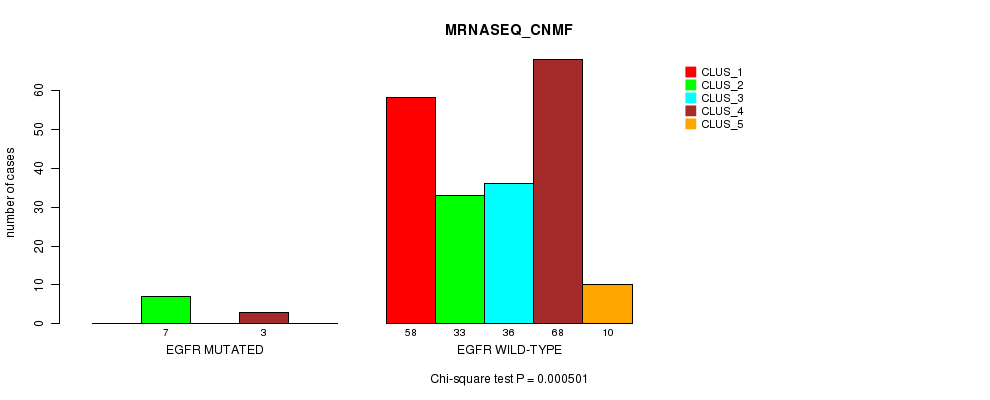
P value = 0.000501 (Chi-square test), Q value = 0.1
Table S37. Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 58 | 40 | 36 | 71 | 10 |
| EGFR MUTATED | 0 | 7 | 0 | 3 | 0 |
| EGFR WILD-TYPE | 58 | 33 | 36 | 68 | 10 |
Figure S37. Get High-res Image Gene #20: 'EGFR MUTATION STATUS' versus Clinical Feature #5: 'MRNASEQ_CNMF'
-
Mutation data file = LGG-TP.mutsig.cluster.txt
-
Molecular subtypes file = LGG-TP.transferedmergedcluster.txt
-
Number of patients = 217
-
Number of significantly mutated genes = 37
-
Number of Molecular subtypes = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.