This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and subtypes.
Testing the association between copy number variation 71 arm-level results and 6 molecular subtypes across 97 patients, 4 significant findings detected with Q value < 0.25.
-
8q gain cnv correlated to 'CN_CNMF'.
-
7p loss cnv correlated to 'MRNASEQ_CNMF'.
-
7q loss cnv correlated to 'MRNASEQ_CNMF'.
-
16q loss cnv correlated to 'METHLYATION_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 71 arm-level results and 6 molecular subtypes. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 4 significant findings detected.
|
Molecular subtypes |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 8q gain | 0 (0%) | 52 |
4.7e-09 (1.97e-06) |
0.75 (1.00) |
0.199 (1.00) |
0.631 (1.00) |
0.182 (1.00) |
0.218 (1.00) |
| 7p loss | 0 (0%) | 92 |
0.0508 (1.00) |
0.85 (1.00) |
0.000514 (0.214) |
0.634 (1.00) |
0.845 (1.00) |
1 (1.00) |
| 7q loss | 0 (0%) | 90 |
0.0337 (1.00) |
1 (1.00) |
2.41e-05 (0.0101) |
1 (1.00) |
0.773 (1.00) |
0.189 (1.00) |
| 16q loss | 0 (0%) | 70 |
0.00429 (1.00) |
0.000233 (0.0973) |
0.071 (1.00) |
0.068 (1.00) |
0.0366 (1.00) |
0.0312 (1.00) |
| 1p gain | 0 (0%) | 87 |
0.54 (1.00) |
0.392 (1.00) |
0.146 (1.00) |
1 (1.00) |
0.624 (1.00) |
0.331 (1.00) |
| 1q gain | 0 (0%) | 46 |
0.00386 (1.00) |
0.0464 (1.00) |
0.0575 (1.00) |
0.469 (1.00) |
0.741 (1.00) |
1 (1.00) |
| 2p gain | 0 (0%) | 89 |
0.586 (1.00) |
0.137 (1.00) |
0.788 (1.00) |
0.443 (1.00) |
0.00574 (1.00) |
0.59 (1.00) |
| 2q gain | 0 (0%) | 90 |
0.618 (1.00) |
0.0561 (1.00) |
0.748 (1.00) |
0.23 (1.00) |
0.021 (1.00) |
1 (1.00) |
| 3p gain | 0 (0%) | 90 |
0.377 (1.00) |
0.758 (1.00) |
0.109 (1.00) |
0.69 (1.00) |
0.248 (1.00) |
1 (1.00) |
| 3q gain | 0 (0%) | 90 |
0.377 (1.00) |
0.758 (1.00) |
0.109 (1.00) |
0.69 (1.00) |
0.248 (1.00) |
1 (1.00) |
| 4p gain | 0 (0%) | 91 |
0.766 (1.00) |
0.101 (1.00) |
0.00914 (1.00) |
0.0106 (1.00) |
0.288 (1.00) |
1 (1.00) |
| 5p gain | 0 (0%) | 69 |
0.0381 (1.00) |
0.247 (1.00) |
0.526 (1.00) |
0.157 (1.00) |
0.797 (1.00) |
0.503 (1.00) |
| 5q gain | 0 (0%) | 77 |
0.129 (1.00) |
0.523 (1.00) |
0.912 (1.00) |
0.335 (1.00) |
0.23 (1.00) |
0.452 (1.00) |
| 6p gain | 0 (0%) | 79 |
0.00474 (1.00) |
0.00692 (1.00) |
0.402 (1.00) |
0.777 (1.00) |
0.322 (1.00) |
1 (1.00) |
| 6q gain | 0 (0%) | 86 |
0.0806 (1.00) |
0.0922 (1.00) |
0.213 (1.00) |
0.732 (1.00) |
0.491 (1.00) |
1 (1.00) |
| 7p gain | 0 (0%) | 71 |
0.0589 (1.00) |
0.195 (1.00) |
0.267 (1.00) |
0.112 (1.00) |
0.107 (1.00) |
1 (1.00) |
| 7q gain | 0 (0%) | 71 |
0.103 (1.00) |
0.00982 (1.00) |
0.16 (1.00) |
0.188 (1.00) |
0.311 (1.00) |
1 (1.00) |
| 8p gain | 0 (0%) | 83 |
0.338 (1.00) |
0.458 (1.00) |
0.0728 (1.00) |
0.302 (1.00) |
0.586 (1.00) |
1 (1.00) |
| 9p gain | 0 (0%) | 93 |
0.201 (1.00) |
0.571 (1.00) |
0.849 (1.00) |
0.568 (1.00) |
0.136 (1.00) |
1 (1.00) |
| 9q gain | 0 (0%) | 94 |
0.633 (1.00) |
0.797 (1.00) |
0.849 (1.00) |
0.568 (1.00) |
0.302 (1.00) |
1 (1.00) |
| 10p gain | 0 (0%) | 90 |
0.433 (1.00) |
0.553 (1.00) |
0.052 (1.00) |
0.0363 (1.00) |
0.469 (1.00) |
1 (1.00) |
| 10q gain | 0 (0%) | 93 |
0.258 (1.00) |
0.444 (1.00) |
0.0672 (1.00) |
0.568 (1.00) |
0.136 (1.00) |
1 (1.00) |
| 11q gain | 0 (0%) | 94 |
0.192 (1.00) |
0.325 (1.00) |
0.42 (1.00) |
0.784 (1.00) |
1 (1.00) |
|
| 12p gain | 0 (0%) | 93 |
0.201 (1.00) |
0.688 (1.00) |
0.173 (1.00) |
0.82 (1.00) |
1 (1.00) |
|
| 12q gain | 0 (0%) | 92 |
0.0736 (1.00) |
0.85 (1.00) |
0.8 (1.00) |
0.568 (1.00) |
0.708 (1.00) |
1 (1.00) |
| 13q gain | 0 (0%) | 92 |
0.0508 (1.00) |
0.595 (1.00) |
0.666 (1.00) |
0.302 (1.00) |
0.213 (1.00) |
1 (1.00) |
| 14q gain | 0 (0%) | 92 |
0.374 (1.00) |
0.36 (1.00) |
0.506 (1.00) |
1 (1.00) |
0.471 (1.00) |
|
| 15q gain | 0 (0%) | 92 |
0.315 (1.00) |
0.264 (1.00) |
0.48 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| 16p gain | 0 (0%) | 93 |
0.201 (1.00) |
0.571 (1.00) |
0.173 (1.00) |
0.673 (1.00) |
1 (1.00) |
|
| 17p gain | 0 (0%) | 92 |
0.315 (1.00) |
0.0992 (1.00) |
0.849 (1.00) |
0.568 (1.00) |
0.708 (1.00) |
1 (1.00) |
| 17q gain | 0 (0%) | 77 |
0.0406 (1.00) |
0.947 (1.00) |
0.713 (1.00) |
0.566 (1.00) |
0.762 (1.00) |
0.447 (1.00) |
| 18p gain | 0 (0%) | 94 |
0.192 (1.00) |
0.444 (1.00) |
0.173 (1.00) |
0.784 (1.00) |
1 (1.00) |
|
| 18q gain | 0 (0%) | 93 |
0.258 (1.00) |
0.144 (1.00) |
0.661 (1.00) |
0.0697 (1.00) |
0.673 (1.00) |
1 (1.00) |
| 19p gain | 0 (0%) | 88 |
0.223 (1.00) |
0.0144 (1.00) |
0.0482 (1.00) |
0.0363 (1.00) |
0.00214 (0.884) |
0.592 (1.00) |
| 19q gain | 0 (0%) | 87 |
0.489 (1.00) |
0.00543 (1.00) |
0.048 (1.00) |
0.00321 (1.00) |
0.0712 (1.00) |
0.6 (1.00) |
| 20p gain | 0 (0%) | 77 |
0.376 (1.00) |
0.427 (1.00) |
0.076 (1.00) |
0.144 (1.00) |
0.472 (1.00) |
1 (1.00) |
| 20q gain | 0 (0%) | 76 |
0.263 (1.00) |
0.399 (1.00) |
0.0505 (1.00) |
0.25 (1.00) |
0.308 (1.00) |
1 (1.00) |
| 21q gain | 0 (0%) | 91 |
0.316 (1.00) |
0.254 (1.00) |
0.895 (1.00) |
0.302 (1.00) |
1 (1.00) |
1 (1.00) |
| 22q gain | 0 (0%) | 89 |
0.238 (1.00) |
0.35 (1.00) |
0.074 (1.00) |
0.122 (1.00) |
1 (1.00) |
0.59 (1.00) |
| Xq gain | 0 (0%) | 92 |
0.126 (1.00) |
0.08 (1.00) |
0.502 (1.00) |
0.258 (1.00) |
0.213 (1.00) |
0.471 (1.00) |
| 1p loss | 0 (0%) | 82 |
0.496 (1.00) |
0.0422 (1.00) |
0.103 (1.00) |
1 (1.00) |
0.247 (1.00) |
1 (1.00) |
| 1q loss | 0 (0%) | 92 |
0.315 (1.00) |
0.188 (1.00) |
0.0166 (1.00) |
1 (1.00) |
0.351 (1.00) |
1 (1.00) |
| 2p loss | 0 (0%) | 94 |
0.385 (1.00) |
0.603 (1.00) |
0.283 (1.00) |
0.258 (1.00) |
0.43 (1.00) |
1 (1.00) |
| 2q loss | 0 (0%) | 93 |
0.144 (1.00) |
0.458 (1.00) |
0.568 (1.00) |
0.634 (1.00) |
0.334 (1.00) |
1 (1.00) |
| 3p loss | 0 (0%) | 89 |
0.181 (1.00) |
0.147 (1.00) |
0.0128 (1.00) |
0.00396 (1.00) |
0.21 (1.00) |
0.59 (1.00) |
| 3q loss | 0 (0%) | 94 |
0.192 (1.00) |
0.0353 (1.00) |
0.239 (1.00) |
0.0697 (1.00) |
1 (1.00) |
1 (1.00) |
| 4p loss | 0 (0%) | 88 |
0.021 (1.00) |
0.492 (1.00) |
0.625 (1.00) |
0.477 (1.00) |
0.589 (1.00) |
0.283 (1.00) |
| 4q loss | 0 (0%) | 79 |
0.00105 (0.437) |
0.591 (1.00) |
0.914 (1.00) |
1 (1.00) |
0.28 (1.00) |
0.213 (1.00) |
| 5q loss | 0 (0%) | 92 |
0.0736 (1.00) |
0.571 (1.00) |
0.58 (1.00) |
1 (1.00) |
0.279 (1.00) |
0.471 (1.00) |
| 6q loss | 0 (0%) | 77 |
0.513 (1.00) |
1 (1.00) |
0.73 (1.00) |
0.131 (1.00) |
0.841 (1.00) |
1 (1.00) |
| 8p loss | 0 (0%) | 56 |
0.00184 (0.761) |
0.641 (1.00) |
0.196 (1.00) |
0.626 (1.00) |
0.708 (1.00) |
0.519 (1.00) |
| 8q loss | 0 (0%) | 91 |
0.0015 (0.623) |
0.472 (1.00) |
0.00389 (1.00) |
0.634 (1.00) |
0.288 (1.00) |
1 (1.00) |
| 9p loss | 0 (0%) | 77 |
0.465 (1.00) |
0.85 (1.00) |
0.674 (1.00) |
0.566 (1.00) |
0.573 (1.00) |
0.696 (1.00) |
| 9q loss | 0 (0%) | 78 |
0.499 (1.00) |
0.427 (1.00) |
0.555 (1.00) |
0.771 (1.00) |
0.787 (1.00) |
1 (1.00) |
| 10p loss | 0 (0%) | 91 |
0.204 (1.00) |
1 (1.00) |
0.696 (1.00) |
1 (1.00) |
0.857 (1.00) |
0.536 (1.00) |
| 10q loss | 0 (0%) | 80 |
0.0534 (1.00) |
0.255 (1.00) |
0.16 (1.00) |
0.131 (1.00) |
0.358 (1.00) |
0.108 (1.00) |
| 11p loss | 0 (0%) | 91 |
0.316 (1.00) |
0.553 (1.00) |
0.624 (1.00) |
0.643 (1.00) |
0.857 (1.00) |
0.536 (1.00) |
| 11q loss | 0 (0%) | 89 |
0.238 (1.00) |
1 (1.00) |
0.00885 (1.00) |
0.0615 (1.00) |
0.55 (1.00) |
0.235 (1.00) |
| 12p loss | 0 (0%) | 89 |
0.337 (1.00) |
0.00863 (1.00) |
0.0374 (1.00) |
0.69 (1.00) |
0.41 (1.00) |
1 (1.00) |
| 13q loss | 0 (0%) | 67 |
0.0219 (1.00) |
0.115 (1.00) |
0.0456 (1.00) |
0.436 (1.00) |
0.0552 (1.00) |
0.733 (1.00) |
| 14q loss | 0 (0%) | 71 |
0.0114 (1.00) |
0.508 (1.00) |
0.0101 (1.00) |
0.112 (1.00) |
0.566 (1.00) |
1 (1.00) |
| 15q loss | 0 (0%) | 88 |
0.00523 (1.00) |
0.492 (1.00) |
0.987 (1.00) |
0.443 (1.00) |
0.589 (1.00) |
0.283 (1.00) |
| 16p loss | 0 (0%) | 77 |
0.0776 (1.00) |
0.0149 (1.00) |
0.00753 (1.00) |
0.0397 (1.00) |
0.0807 (1.00) |
0.452 (1.00) |
| 17p loss | 0 (0%) | 56 |
0.00899 (1.00) |
0.018 (1.00) |
0.299 (1.00) |
0.634 (1.00) |
0.135 (1.00) |
0.009 (1.00) |
| 17q loss | 0 (0%) | 91 |
0.766 (1.00) |
0.078 (1.00) |
0.693 (1.00) |
0.634 (1.00) |
0.857 (1.00) |
0.536 (1.00) |
| 18p loss | 0 (0%) | 87 |
0.191 (1.00) |
0.029 (1.00) |
0.00493 (1.00) |
0.712 (1.00) |
0.826 (1.00) |
1 (1.00) |
| 18q loss | 0 (0%) | 86 |
0.0245 (1.00) |
0.0938 (1.00) |
0.0131 (1.00) |
0.477 (1.00) |
0.649 (1.00) |
1 (1.00) |
| 19p loss | 0 (0%) | 92 |
0.52 (1.00) |
0.198 (1.00) |
0.401 (1.00) |
1 (1.00) |
1 (1.00) |
0.471 (1.00) |
| 20p loss | 0 (0%) | 92 |
0.734 (1.00) |
0.595 (1.00) |
1 (1.00) |
0.0618 (1.00) |
0.103 (1.00) |
|
| 21q loss | 0 (0%) | 81 |
0.357 (1.00) |
0.563 (1.00) |
0.13 (1.00) |
0.477 (1.00) |
0.247 (1.00) |
0.0189 (1.00) |
| 22q loss | 0 (0%) | 87 |
0.374 (1.00) |
0.473 (1.00) |
0.0981 (1.00) |
0.152 (1.00) |
0.91 (1.00) |
0.331 (1.00) |
P value = 4.7e-09 (Fisher's exact test), Q value = 2e-06
Table S1. Gene #15: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 28 | 36 | 33 |
| 8Q GAIN CNV | 26 | 8 | 11 |
| 8Q GAIN WILD-TYPE | 2 | 28 | 22 |
Figure S1. Get High-res Image Gene #15: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'
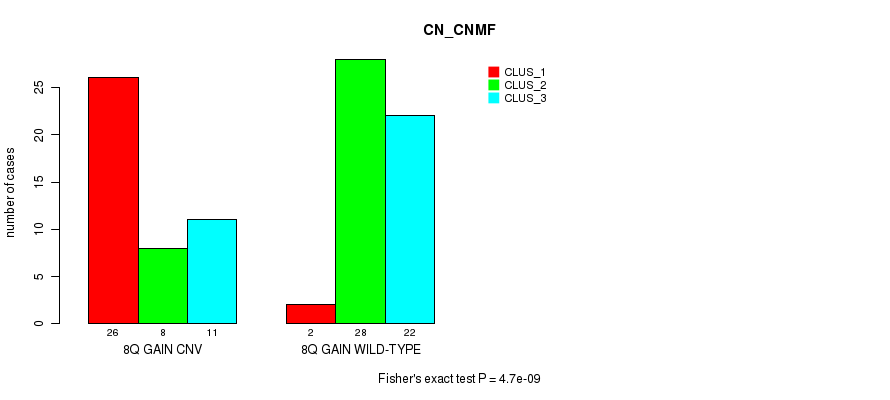
P value = 0.000514 (Chi-square test), Q value = 0.21
Table S2. Gene #48: '7p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 | CLUS_8 | CLUS_9 |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 16 | 8 | 6 | 14 | 11 | 3 | 6 | 1 | 4 |
| 7P LOSS CNV | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 1 |
| 7P LOSS WILD-TYPE | 16 | 8 | 6 | 14 | 11 | 1 | 5 | 1 | 3 |
Figure S2. Get High-res Image Gene #48: '7p loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
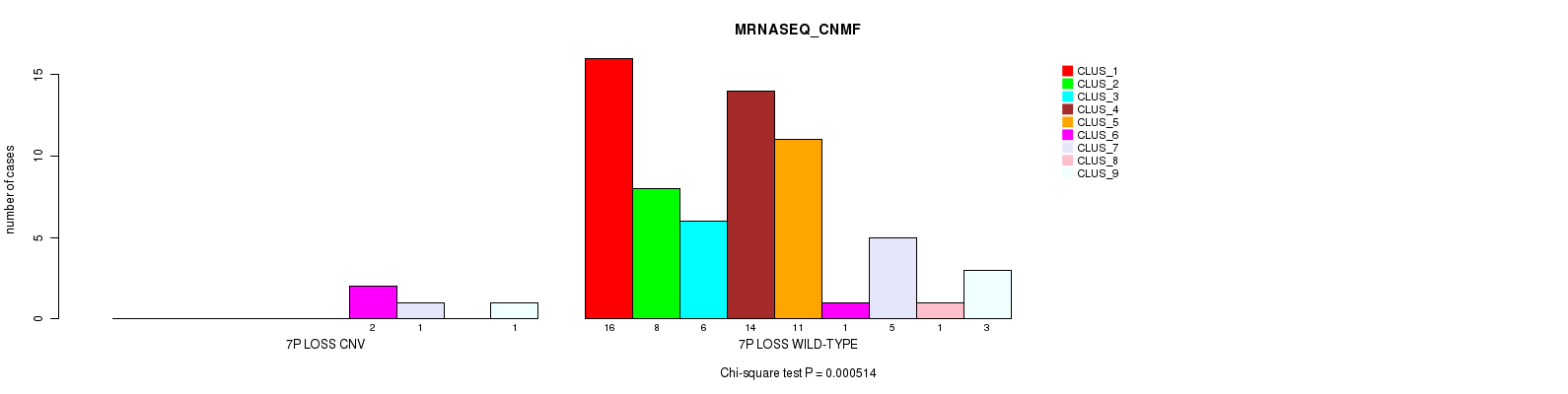
P value = 2.41e-05 (Chi-square test), Q value = 0.01
Table S3. Gene #49: '7q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 | CLUS_8 | CLUS_9 |
|---|---|---|---|---|---|---|---|---|---|
| ALL | 16 | 8 | 6 | 14 | 11 | 3 | 6 | 1 | 4 |
| 7Q LOSS CNV | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 0 | 0 |
| 7Q LOSS WILD-TYPE | 15 | 8 | 6 | 14 | 10 | 0 | 5 | 1 | 4 |
Figure S3. Get High-res Image Gene #49: '7q loss' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
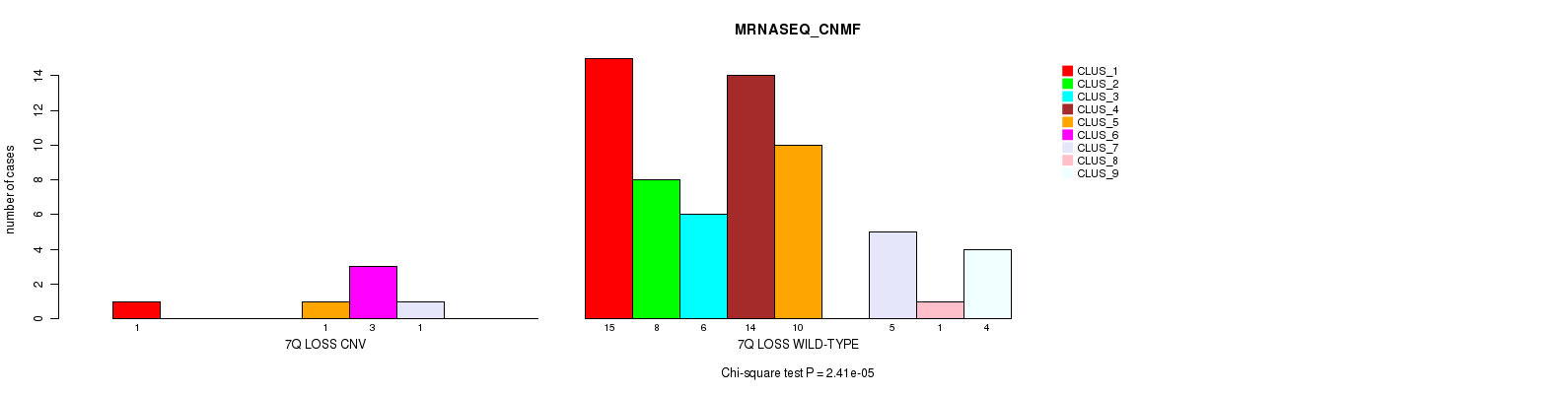
P value = 0.000233 (Fisher's exact test), Q value = 0.097
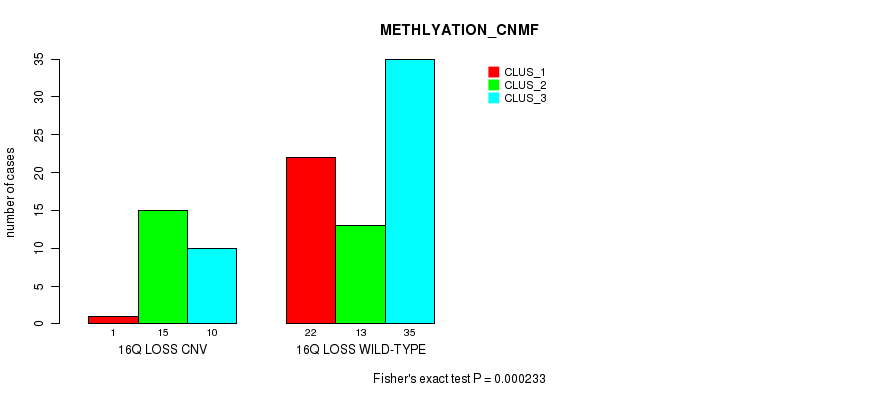
Table S4. Gene #63: '16q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 23 | 28 | 45 |
| 16Q LOSS CNV | 1 | 15 | 10 |
| 16Q LOSS WILD-TYPE | 22 | 13 | 35 |
Figure S4. Get High-res Image Gene #63: '16q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
-
Mutation data file = broad_values_by_arm.mutsig.cluster.txt
-
Molecular subtypes file = LIHC-TP.transferedmergedcluster.txt
-
Number of patients = 97
-
Number of significantly arm-level cnvs = 71
-
Number of molecular subtypes = 6
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
This is an experimental feature. The full results of the analysis summarized in this report can be downloaded from the TCGA Data Coordination Center.