This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and subtypes.
Testing the association between copy number variation 77 arm-level results and 10 molecular subtypes across 182 patients, 22 significant findings detected with Q value < 0.25.
-
1p gain cnv correlated to 'CN_CNMF'.
-
2p gain cnv correlated to 'CN_CNMF'.
-
3q gain cnv correlated to 'CN_CNMF'.
-
5p gain cnv correlated to 'CN_CNMF'.
-
8q gain cnv correlated to 'CN_CNMF'.
-
10p gain cnv correlated to 'CN_CNMF'.
-
17q gain cnv correlated to 'MIRSEQ_MATURE_CNMF'.
-
18p gain cnv correlated to 'CN_CNMF'.
-
5q loss cnv correlated to 'CN_CNMF'.
-
6q loss cnv correlated to 'CN_CNMF'.
-
8p loss cnv correlated to 'CN_CNMF'.
-
9p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
9q loss cnv correlated to 'CN_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
10q loss cnv correlated to 'CN_CNMF'.
-
11p loss cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
16p loss cnv correlated to 'CN_CNMF'.
-
16q loss cnv correlated to 'CN_CNMF'.
-
22q loss cnv correlated to 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 77 arm-level results and 10 molecular subtypes. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 22 significant findings detected.
|
Molecular subtypes |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 9p loss | 0 (0%) | 128 |
2.97e-06 (0.00226) |
0.000178 (0.133) |
0.15 (1.00) |
0.00152 (1.00) |
0.000486 (0.36) |
1.38e-05 (0.0105) |
0.344 (1.00) |
0.548 (1.00) |
0.0741 (1.00) |
0.015 (1.00) |
| 9q loss | 0 (0%) | 130 |
7.61e-05 (0.057) |
0.000459 (0.341) |
0.0166 (1.00) |
0.00117 (0.861) |
0.000535 (0.396) |
3.05e-06 (0.00231) |
0.581 (1.00) |
0.702 (1.00) |
0.085 (1.00) |
0.0414 (1.00) |
| 11p loss | 0 (0%) | 130 |
9.99e-05 (0.0746) |
9.48e-05 (0.0709) |
0.0259 (1.00) |
0.464 (1.00) |
0.00875 (1.00) |
0.00828 (1.00) |
0.172 (1.00) |
0.816 (1.00) |
0.193 (1.00) |
0.14 (1.00) |
| 1p gain | 0 (0%) | 160 |
3.04e-05 (0.023) |
0.28 (1.00) |
0.387 (1.00) |
0.0238 (1.00) |
0.0723 (1.00) |
0.8 (1.00) |
0.957 (1.00) |
0.356 (1.00) |
0.833 (1.00) |
1 (1.00) |
| 2p gain | 0 (0%) | 152 |
5.37e-05 (0.0404) |
0.013 (1.00) |
0.223 (1.00) |
0.627 (1.00) |
0.0186 (1.00) |
0.478 (1.00) |
0.0912 (1.00) |
0.0841 (1.00) |
0.163 (1.00) |
0.0129 (1.00) |
| 3q gain | 0 (0%) | 133 |
1.18e-06 (0.000897) |
0.0967 (1.00) |
0.102 (1.00) |
0.2 (1.00) |
0.0357 (1.00) |
0.0561 (1.00) |
0.216 (1.00) |
0.187 (1.00) |
0.0941 (1.00) |
0.0533 (1.00) |
| 5p gain | 0 (0%) | 130 |
7.4e-05 (0.0555) |
0.832 (1.00) |
0.0227 (1.00) |
0.445 (1.00) |
0.0864 (1.00) |
0.019 (1.00) |
0.609 (1.00) |
0.255 (1.00) |
0.519 (1.00) |
0.26 (1.00) |
| 8q gain | 0 (0%) | 126 |
2.53e-05 (0.0192) |
0.0966 (1.00) |
0.367 (1.00) |
0.14 (1.00) |
0.15 (1.00) |
0.296 (1.00) |
0.934 (1.00) |
0.865 (1.00) |
0.791 (1.00) |
0.273 (1.00) |
| 10p gain | 0 (0%) | 146 |
4.56e-05 (0.0344) |
0.398 (1.00) |
0.157 (1.00) |
0.342 (1.00) |
0.576 (1.00) |
0.102 (1.00) |
0.344 (1.00) |
0.189 (1.00) |
0.108 (1.00) |
0.578 (1.00) |
| 17q gain | 0 (0%) | 153 |
0.0094 (1.00) |
0.00335 (1.00) |
0.00685 (1.00) |
0.00487 (1.00) |
0.00552 (1.00) |
0.0491 (1.00) |
0.0268 (1.00) |
0.0318 (1.00) |
0.000256 (0.191) |
0.00269 (1.00) |
| 18p gain | 0 (0%) | 150 |
4.52e-05 (0.0341) |
0.00364 (1.00) |
0.812 (1.00) |
0.943 (1.00) |
0.0583 (1.00) |
0.14 (1.00) |
0.569 (1.00) |
0.0899 (1.00) |
0.523 (1.00) |
0.505 (1.00) |
| 5q loss | 0 (0%) | 140 |
3.26e-05 (0.0246) |
0.118 (1.00) |
0.484 (1.00) |
0.194 (1.00) |
0.355 (1.00) |
0.552 (1.00) |
0.16 (1.00) |
0.49 (1.00) |
0.604 (1.00) |
1 (1.00) |
| 6q loss | 0 (0%) | 147 |
0.000115 (0.0857) |
0.0101 (1.00) |
0.105 (1.00) |
0.835 (1.00) |
0.474 (1.00) |
0.214 (1.00) |
1 (1.00) |
0.935 (1.00) |
0.776 (1.00) |
0.495 (1.00) |
| 8p loss | 0 (0%) | 124 |
2.17e-09 (1.66e-06) |
0.535 (1.00) |
0.432 (1.00) |
0.0542 (1.00) |
0.139 (1.00) |
0.0825 (1.00) |
0.035 (1.00) |
0.0664 (1.00) |
0.104 (1.00) |
0.0376 (1.00) |
| 10q loss | 0 (0%) | 148 |
0.000263 (0.196) |
0.0026 (1.00) |
0.601 (1.00) |
0.382 (1.00) |
0.235 (1.00) |
0.207 (1.00) |
0.064 (1.00) |
0.0584 (1.00) |
0.0636 (1.00) |
0.0135 (1.00) |
| 16p loss | 0 (0%) | 159 |
1.92e-07 (0.000146) |
0.305 (1.00) |
0.336 (1.00) |
0.362 (1.00) |
0.207 (1.00) |
0.771 (1.00) |
0.0238 (1.00) |
0.0221 (1.00) |
0.128 (1.00) |
0.606 (1.00) |
| 16q loss | 0 (0%) | 159 |
4.1e-07 (0.000312) |
0.0832 (1.00) |
0.103 (1.00) |
0.976 (1.00) |
0.118 (1.00) |
0.74 (1.00) |
0.112 (1.00) |
0.0574 (1.00) |
0.0184 (1.00) |
0.288 (1.00) |
| 22q loss | 0 (0%) | 146 |
6.22e-05 (0.0467) |
0.219 (1.00) |
0.511 (1.00) |
0.0753 (1.00) |
0.0788 (1.00) |
0.583 (1.00) |
0.578 (1.00) |
0.0129 (1.00) |
0.446 (1.00) |
0.966 (1.00) |
| 1q gain | 0 (0%) | 142 |
0.0372 (1.00) |
0.396 (1.00) |
0.676 (1.00) |
0.885 (1.00) |
0.00478 (1.00) |
1 (1.00) |
0.714 (1.00) |
0.688 (1.00) |
0.363 (1.00) |
0.335 (1.00) |
| 2q gain | 0 (0%) | 169 |
0.00448 (1.00) |
0.00793 (1.00) |
0.369 (1.00) |
0.563 (1.00) |
0.0042 (1.00) |
0.2 (1.00) |
0.127 (1.00) |
0.0576 (1.00) |
0.166 (1.00) |
0.00978 (1.00) |
| 3p gain | 0 (0%) | 149 |
0.0011 (0.806) |
0.00337 (1.00) |
0.0354 (1.00) |
0.185 (1.00) |
0.0791 (1.00) |
0.00396 (1.00) |
0.0335 (1.00) |
0.0224 (1.00) |
0.0177 (1.00) |
0.193 (1.00) |
| 4p gain | 0 (0%) | 171 |
0.224 (1.00) |
0.671 (1.00) |
0.86 (1.00) |
0.521 (1.00) |
0.756 (1.00) |
0.919 (1.00) |
0.198 (1.00) |
0.544 (1.00) |
0.918 (1.00) |
0.682 (1.00) |
| 4q gain | 0 (0%) | 175 |
0.0377 (1.00) |
1 (1.00) |
0.542 (1.00) |
0.287 (1.00) |
0.763 (1.00) |
0.69 (1.00) |
0.201 (1.00) |
0.387 (1.00) |
0.609 (1.00) |
0.761 (1.00) |
| 5q gain | 0 (0%) | 162 |
0.262 (1.00) |
0.131 (1.00) |
0.0845 (1.00) |
0.956 (1.00) |
0.528 (1.00) |
0.583 (1.00) |
0.556 (1.00) |
0.654 (1.00) |
0.402 (1.00) |
0.679 (1.00) |
| 6p gain | 0 (0%) | 165 |
0.0433 (1.00) |
0.235 (1.00) |
0.684 (1.00) |
0.533 (1.00) |
0.415 (1.00) |
1 (1.00) |
0.284 (1.00) |
0.134 (1.00) |
0.311 (1.00) |
0.879 (1.00) |
| 6q gain | 0 (0%) | 176 |
0.0125 (1.00) |
1 (1.00) |
0.549 (1.00) |
0.0079 (1.00) |
0.112 (1.00) |
1 (1.00) |
0.0449 (1.00) |
0.0495 (1.00) |
0.155 (1.00) |
0.347 (1.00) |
| 7p gain | 0 (0%) | 130 |
0.00214 (1.00) |
0.00218 (1.00) |
0.0502 (1.00) |
0.38 (1.00) |
0.0493 (1.00) |
0.77 (1.00) |
0.0947 (1.00) |
0.269 (1.00) |
0.785 (1.00) |
0.484 (1.00) |
| 7q gain | 0 (0%) | 137 |
0.000987 (0.727) |
0.00731 (1.00) |
0.392 (1.00) |
0.763 (1.00) |
0.145 (1.00) |
0.66 (1.00) |
0.974 (1.00) |
0.271 (1.00) |
0.765 (1.00) |
0.943 (1.00) |
| 8p gain | 0 (0%) | 163 |
0.0138 (1.00) |
0.0272 (1.00) |
0.165 (1.00) |
0.0283 (1.00) |
0.0272 (1.00) |
0.0389 (1.00) |
0.395 (1.00) |
0.722 (1.00) |
0.409 (1.00) |
0.256 (1.00) |
| 9p gain | 0 (0%) | 168 |
0.123 (1.00) |
0.43 (1.00) |
0.0721 (1.00) |
0.571 (1.00) |
0.00176 (1.00) |
0.163 (1.00) |
0.367 (1.00) |
1 (1.00) |
0.936 (1.00) |
0.74 (1.00) |
| 9q gain | 0 (0%) | 169 |
0.819 (1.00) |
0.221 (1.00) |
0.755 (1.00) |
0.41 (1.00) |
0.0212 (1.00) |
0.135 (1.00) |
1 (1.00) |
0.713 (1.00) |
0.806 (1.00) |
0.405 (1.00) |
| 10q gain | 0 (0%) | 173 |
0.0574 (1.00) |
0.825 (1.00) |
0.684 (1.00) |
0.356 (1.00) |
0.481 (1.00) |
0.551 (1.00) |
0.616 (1.00) |
1 (1.00) |
0.596 (1.00) |
1 (1.00) |
| 11p gain | 0 (0%) | 173 |
0.0416 (1.00) |
0.751 (1.00) |
0.75 (1.00) |
0.596 (1.00) |
0.168 (1.00) |
0.67 (1.00) |
0.22 (1.00) |
0.476 (1.00) |
0.66 (1.00) |
0.801 (1.00) |
| 11q gain | 0 (0%) | 168 |
0.151 (1.00) |
0.878 (1.00) |
0.649 (1.00) |
0.637 (1.00) |
0.292 (1.00) |
0.103 (1.00) |
0.72 (1.00) |
0.639 (1.00) |
0.936 (1.00) |
1 (1.00) |
| 12p gain | 0 (0%) | 153 |
0.00191 (1.00) |
0.785 (1.00) |
0.138 (1.00) |
0.162 (1.00) |
0.0362 (1.00) |
0.0527 (1.00) |
0.0524 (1.00) |
1 (1.00) |
0.143 (1.00) |
0.073 (1.00) |
| 12q gain | 0 (0%) | 162 |
0.0992 (1.00) |
0.354 (1.00) |
0.0221 (1.00) |
0.0226 (1.00) |
0.0119 (1.00) |
0.038 (1.00) |
0.0641 (1.00) |
0.186 (1.00) |
0.0306 (1.00) |
0.0644 (1.00) |
| 13q gain | 0 (0%) | 156 |
0.00995 (1.00) |
0.766 (1.00) |
0.76 (1.00) |
0.66 (1.00) |
0.0804 (1.00) |
0.291 (1.00) |
0.732 (1.00) |
0.561 (1.00) |
0.923 (1.00) |
0.876 (1.00) |
| 14q gain | 0 (0%) | 171 |
0.724 (1.00) |
0.927 (1.00) |
0.395 (1.00) |
0.733 (1.00) |
0.102 (1.00) |
0.0595 (1.00) |
1 (1.00) |
0.677 (1.00) |
0.1 (1.00) |
0.354 (1.00) |
| 15q gain | 0 (0%) | 176 |
1 (1.00) |
0.188 (1.00) |
0.522 (1.00) |
1 (1.00) |
0.3 (1.00) |
0.206 (1.00) |
0.571 (1.00) |
1 (1.00) |
0.489 (1.00) |
0.429 (1.00) |
| 16p gain | 0 (0%) | 169 |
0.0711 (1.00) |
1 (1.00) |
0.203 (1.00) |
0.889 (1.00) |
0.155 (1.00) |
0.751 (1.00) |
1 (1.00) |
0.854 (1.00) |
0.75 (1.00) |
1 (1.00) |
| 16q gain | 0 (0%) | 166 |
0.0137 (1.00) |
0.562 (1.00) |
0.0782 (1.00) |
0.395 (1.00) |
0.0472 (1.00) |
0.17 (1.00) |
0.839 (1.00) |
0.778 (1.00) |
0.194 (1.00) |
0.441 (1.00) |
| 17p gain | 0 (0%) | 174 |
0.419 (1.00) |
0.814 (1.00) |
0.841 (1.00) |
1 (1.00) |
0.0705 (1.00) |
0.329 (1.00) |
0.576 (1.00) |
0.251 (1.00) |
0.565 (1.00) |
0.616 (1.00) |
| 18q gain | 0 (0%) | 170 |
0.0549 (1.00) |
0.0145 (1.00) |
0.583 (1.00) |
0.221 (1.00) |
0.464 (1.00) |
0.545 (1.00) |
0.548 (1.00) |
0.321 (1.00) |
0.629 (1.00) |
0.772 (1.00) |
| 19p gain | 0 (0%) | 168 |
0.56 (1.00) |
0.0993 (1.00) |
0.751 (1.00) |
0.89 (1.00) |
0.0496 (1.00) |
0.0347 (1.00) |
0.392 (1.00) |
0.378 (1.00) |
0.197 (1.00) |
0.112 (1.00) |
| 19q gain | 0 (0%) | 149 |
0.00131 (0.961) |
0.301 (1.00) |
0.447 (1.00) |
0.61 (1.00) |
0.137 (1.00) |
0.175 (1.00) |
0.473 (1.00) |
0.866 (1.00) |
0.609 (1.00) |
0.411 (1.00) |
| 20p gain | 0 (0%) | 122 |
0.0812 (1.00) |
0.0829 (1.00) |
0.326 (1.00) |
0.474 (1.00) |
0.188 (1.00) |
0.133 (1.00) |
0.786 (1.00) |
0.43 (1.00) |
0.698 (1.00) |
0.465 (1.00) |
| 20q gain | 0 (0%) | 114 |
0.0138 (1.00) |
0.00237 (1.00) |
0.0598 (1.00) |
0.68 (1.00) |
0.0133 (1.00) |
0.0451 (1.00) |
0.0391 (1.00) |
0.235 (1.00) |
0.11 (1.00) |
0.0275 (1.00) |
| 21q gain | 0 (0%) | 152 |
0.00131 (0.961) |
0.689 (1.00) |
0.228 (1.00) |
0.671 (1.00) |
0.113 (1.00) |
0.0328 (1.00) |
0.0845 (1.00) |
0.392 (1.00) |
0.196 (1.00) |
0.3 (1.00) |
| 22q gain | 0 (0%) | 169 |
0.195 (1.00) |
0.205 (1.00) |
0.147 (1.00) |
0.52 (1.00) |
0.226 (1.00) |
0.61 (1.00) |
0.0133 (1.00) |
0.854 (1.00) |
0.296 (1.00) |
0.476 (1.00) |
| Xq gain | 0 (0%) | 175 |
0.00882 (1.00) |
0.615 (1.00) |
0.964 (1.00) |
0.86 (1.00) |
0.214 (1.00) |
0.69 (1.00) |
0.421 (1.00) |
0.292 (1.00) |
0.261 (1.00) |
0.0827 (1.00) |
| 1p loss | 0 (0%) | 178 |
0.203 (1.00) |
0.124 (1.00) |
0.265 (1.00) |
0.0446 (1.00) |
0.438 (1.00) |
1 (1.00) |
0.18 (1.00) |
0.0515 (1.00) |
||
| 2p loss | 0 (0%) | 173 |
0.827 (1.00) |
0.0335 (1.00) |
0.23 (1.00) |
0.269 (1.00) |
0.0783 (1.00) |
0.0434 (1.00) |
0.441 (1.00) |
1 (1.00) |
0.0808 (1.00) |
0.00774 (1.00) |
| 2q loss | 0 (0%) | 161 |
0.00147 (1.00) |
0.17 (1.00) |
0.757 (1.00) |
0.612 (1.00) |
0.00476 (1.00) |
0.0582 (1.00) |
0.828 (1.00) |
0.526 (1.00) |
0.326 (1.00) |
0.0432 (1.00) |
| 3p loss | 0 (0%) | 170 |
1 (1.00) |
1 (1.00) |
0.0337 (1.00) |
0.367 (1.00) |
0.019 (1.00) |
0.0113 (1.00) |
0.148 (1.00) |
0.117 (1.00) |
0.139 (1.00) |
0.261 (1.00) |
| 3q loss | 0 (0%) | 179 |
0.795 (1.00) |
1 (1.00) |
0.669 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.782 (1.00) |
1 (1.00) |
||
| 4p loss | 0 (0%) | 148 |
0.0143 (1.00) |
0.298 (1.00) |
0.43 (1.00) |
0.933 (1.00) |
0.264 (1.00) |
0.748 (1.00) |
0.372 (1.00) |
0.658 (1.00) |
0.199 (1.00) |
0.561 (1.00) |
| 4q loss | 0 (0%) | 154 |
0.524 (1.00) |
0.19 (1.00) |
0.106 (1.00) |
0.861 (1.00) |
0.106 (1.00) |
0.565 (1.00) |
0.774 (1.00) |
0.178 (1.00) |
0.254 (1.00) |
0.746 (1.00) |
| 5p loss | 0 (0%) | 167 |
0.0527 (1.00) |
1 (1.00) |
0.326 (1.00) |
0.17 (1.00) |
0.868 (1.00) |
0.527 (1.00) |
0.122 (1.00) |
0.562 (1.00) |
0.217 (1.00) |
0.236 (1.00) |
| 6p loss | 0 (0%) | 161 |
0.148 (1.00) |
0.17 (1.00) |
0.196 (1.00) |
0.541 (1.00) |
0.344 (1.00) |
0.484 (1.00) |
0.227 (1.00) |
1 (1.00) |
0.254 (1.00) |
1 (1.00) |
| 7q loss | 0 (0%) | 176 |
0.0259 (1.00) |
0.0938 (1.00) |
0.844 (1.00) |
0.363 (1.00) |
0.352 (1.00) |
0.7 (1.00) |
0.13 (1.00) |
1 (1.00) |
||
| 8q loss | 0 (0%) | 174 |
0.293 (1.00) |
0.369 (1.00) |
0.643 (1.00) |
0.0826 (1.00) |
0.255 (1.00) |
0.511 (1.00) |
0.33 (1.00) |
0.0901 (1.00) |
0.357 (1.00) |
0.0458 (1.00) |
| 10p loss | 0 (0%) | 162 |
0.128 (1.00) |
0.487 (1.00) |
0.759 (1.00) |
0.343 (1.00) |
0.111 (1.00) |
0.273 (1.00) |
0.864 (1.00) |
0.241 (1.00) |
0.707 (1.00) |
0.345 (1.00) |
| 11q loss | 0 (0%) | 142 |
0.0027 (1.00) |
0.0431 (1.00) |
0.0638 (1.00) |
0.0328 (1.00) |
0.0124 (1.00) |
0.0604 (1.00) |
0.674 (1.00) |
0.785 (1.00) |
0.409 (1.00) |
0.437 (1.00) |
| 12p loss | 0 (0%) | 175 |
0.0487 (1.00) |
0.433 (1.00) |
0.964 (1.00) |
0.86 (1.00) |
0.243 (1.00) |
0.243 (1.00) |
0.888 (1.00) |
1 (1.00) |
0.689 (1.00) |
0.868 (1.00) |
| 12q loss | 0 (0%) | 174 |
0.419 (1.00) |
0.814 (1.00) |
0.356 (1.00) |
0.442 (1.00) |
0.224 (1.00) |
0.417 (1.00) |
0.576 (1.00) |
0.251 (1.00) |
0.405 (1.00) |
0.616 (1.00) |
| 13q loss | 0 (0%) | 159 |
0.000902 (0.666) |
0.0547 (1.00) |
0.101 (1.00) |
0.0628 (1.00) |
0.206 (1.00) |
0.383 (1.00) |
0.771 (1.00) |
0.413 (1.00) |
0.877 (1.00) |
1 (1.00) |
| 14q loss | 0 (0%) | 154 |
0.000588 (0.434) |
0.112 (1.00) |
0.126 (1.00) |
0.0601 (1.00) |
0.117 (1.00) |
0.1 (1.00) |
0.0228 (1.00) |
0.195 (1.00) |
0.0455 (1.00) |
0.452 (1.00) |
| 15q loss | 0 (0%) | 156 |
0.0374 (1.00) |
0.565 (1.00) |
0.027 (1.00) |
0.0979 (1.00) |
0.459 (1.00) |
0.23 (1.00) |
0.376 (1.00) |
0.0263 (1.00) |
0.57 (1.00) |
0.8 (1.00) |
| 17p loss | 0 (0%) | 131 |
0.252 (1.00) |
0.377 (1.00) |
0.974 (1.00) |
0.446 (1.00) |
0.176 (1.00) |
0.301 (1.00) |
0.019 (1.00) |
0.0135 (1.00) |
0.0586 (1.00) |
0.0705 (1.00) |
| 17q loss | 0 (0%) | 176 |
1 (1.00) |
0.0405 (1.00) |
0.735 (1.00) |
0.426 (1.00) |
0.452 (1.00) |
0.308 (1.00) |
0.484 (1.00) |
0.503 (1.00) |
0.351 (1.00) |
0.23 (1.00) |
| 18p loss | 0 (0%) | 158 |
0.146 (1.00) |
0.28 (1.00) |
0.731 (1.00) |
0.785 (1.00) |
0.895 (1.00) |
0.555 (1.00) |
0.281 (1.00) |
0.0114 (1.00) |
0.00852 (1.00) |
0.0108 (1.00) |
| 18q loss | 0 (0%) | 140 |
0.00508 (1.00) |
0.238 (1.00) |
0.758 (1.00) |
0.669 (1.00) |
0.152 (1.00) |
0.0929 (1.00) |
0.24 (1.00) |
0.224 (1.00) |
0.658 (1.00) |
0.691 (1.00) |
| 19p loss | 0 (0%) | 168 |
0.0375 (1.00) |
0.599 (1.00) |
0.739 (1.00) |
0.151 (1.00) |
0.241 (1.00) |
0.514 (1.00) |
0.367 (1.00) |
0.378 (1.00) |
0.437 (1.00) |
0.59 (1.00) |
| 19q loss | 0 (0%) | 175 |
0.089 (1.00) |
1 (1.00) |
0.714 (1.00) |
0.109 (1.00) |
0.214 (1.00) |
0.42 (1.00) |
0.0223 (1.00) |
0.387 (1.00) |
0.062 (1.00) |
0.14 (1.00) |
| 20p loss | 0 (0%) | 174 |
0.371 (1.00) |
0.126 (1.00) |
0.301 (1.00) |
0.0272 (1.00) |
0.737 (1.00) |
0.151 (1.00) |
0.458 (1.00) |
0.251 (1.00) |
0.1 (1.00) |
0.691 (1.00) |
| 21q loss | 0 (0%) | 160 |
0.202 (1.00) |
0.916 (1.00) |
0.361 (1.00) |
0.419 (1.00) |
0.811 (1.00) |
0.531 (1.00) |
0.638 (1.00) |
0.169 (1.00) |
0.158 (1.00) |
0.86 (1.00) |
| Xq loss | 0 (0%) | 176 |
0.0259 (1.00) |
0.758 (1.00) |
0.78 (1.00) |
0.636 (1.00) |
0.162 (1.00) |
0.308 (1.00) |
0.309 (1.00) |
0.503 (1.00) |
0.351 (1.00) |
0.23 (1.00) |
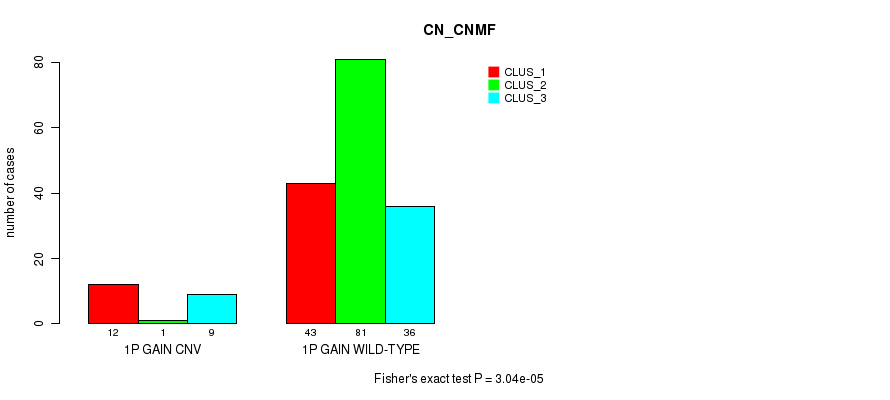
P value = 3.04e-05 (Fisher's exact test), Q value = 0.023
Table S1. Gene #1: '1p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 1P GAIN CNV | 12 | 1 | 9 |
| 1P GAIN WILD-TYPE | 43 | 81 | 36 |
Figure S1. Get High-res Image Gene #1: '1p gain' versus Molecular Subtype #1: 'CN_CNMF'
P value = 5.37e-05 (Fisher's exact test), Q value = 0.04
Table S2. Gene #3: '2p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 2P GAIN CNV | 18 | 4 | 8 |
| 2P GAIN WILD-TYPE | 37 | 78 | 37 |
Figure S2. Get High-res Image Gene #3: '2p gain' versus Molecular Subtype #1: 'CN_CNMF'

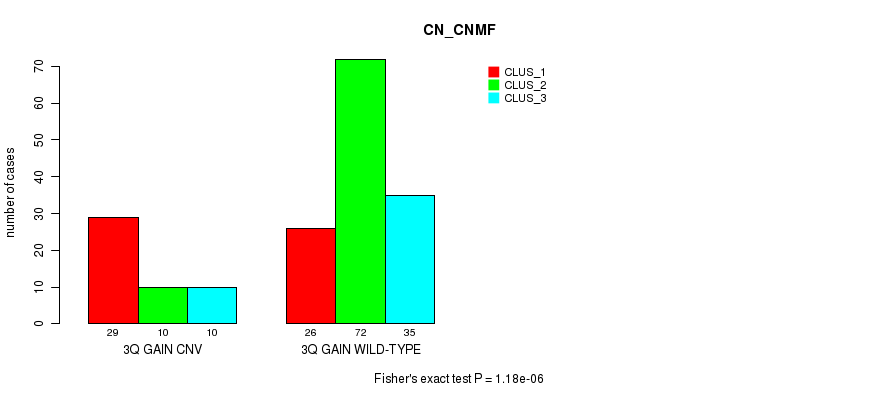
P value = 1.18e-06 (Fisher's exact test), Q value = 9e-04
Table S3. Gene #6: '3q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 3Q GAIN CNV | 29 | 10 | 10 |
| 3Q GAIN WILD-TYPE | 26 | 72 | 35 |
Figure S3. Get High-res Image Gene #6: '3q gain' versus Molecular Subtype #1: 'CN_CNMF'
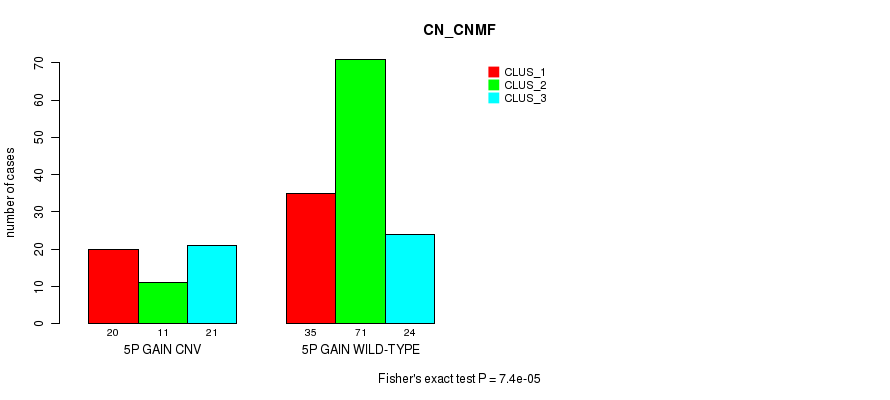
P value = 7.4e-05 (Fisher's exact test), Q value = 0.055
Table S4. Gene #9: '5p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 5P GAIN CNV | 20 | 11 | 21 |
| 5P GAIN WILD-TYPE | 35 | 71 | 24 |
Figure S4. Get High-res Image Gene #9: '5p gain' versus Molecular Subtype #1: 'CN_CNMF'
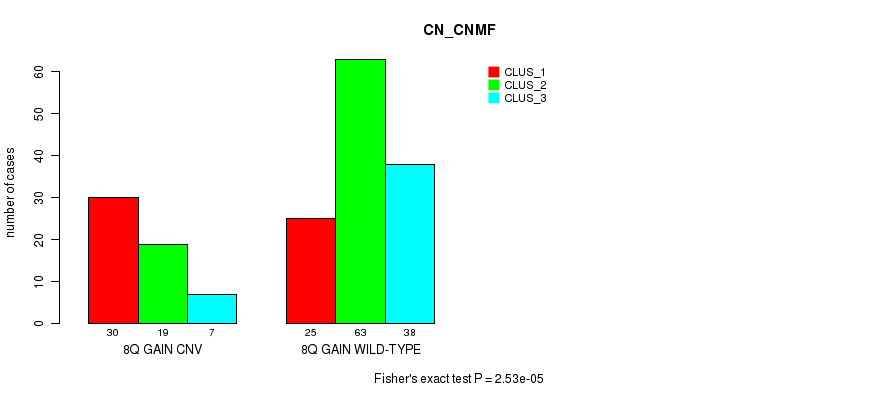
P value = 2.53e-05 (Fisher's exact test), Q value = 0.019
Table S5. Gene #16: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 8Q GAIN CNV | 30 | 19 | 7 |
| 8Q GAIN WILD-TYPE | 25 | 63 | 38 |
Figure S5. Get High-res Image Gene #16: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'
P value = 4.56e-05 (Fisher's exact test), Q value = 0.034
Table S6. Gene #19: '10p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 10P GAIN CNV | 19 | 5 | 12 |
| 10P GAIN WILD-TYPE | 36 | 77 | 33 |
Figure S6. Get High-res Image Gene #19: '10p gain' versus Molecular Subtype #1: 'CN_CNMF'
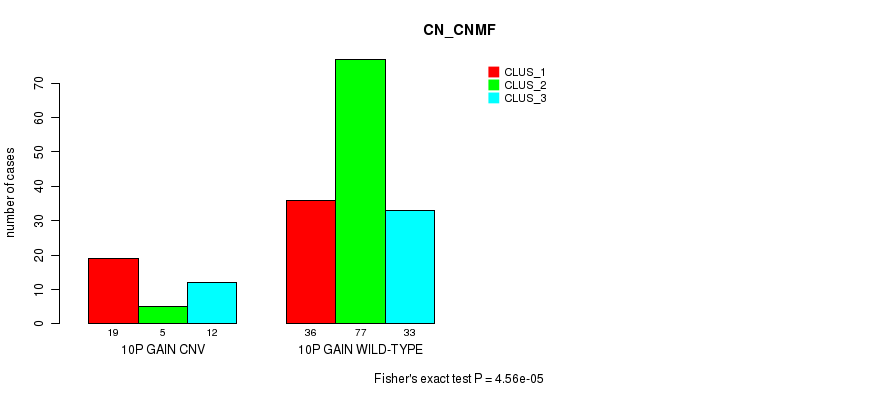
P value = 0.000256 (Fisher's exact test), Q value = 0.19
Table S7. Gene #31: '17q gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 79 | 70 |
| 17Q GAIN CNV | 11 | 4 | 14 |
| 17Q GAIN WILD-TYPE | 22 | 75 | 56 |
Figure S7. Get High-res Image Gene #31: '17q gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
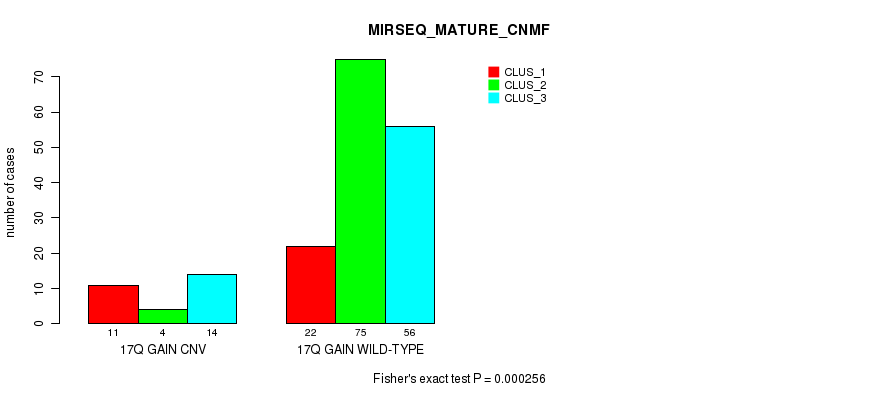
P value = 4.52e-05 (Fisher's exact test), Q value = 0.034
Table S8. Gene #32: '18p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 18P GAIN CNV | 13 | 4 | 15 |
| 18P GAIN WILD-TYPE | 42 | 78 | 30 |
Figure S8. Get High-res Image Gene #32: '18p gain' versus Molecular Subtype #1: 'CN_CNMF'
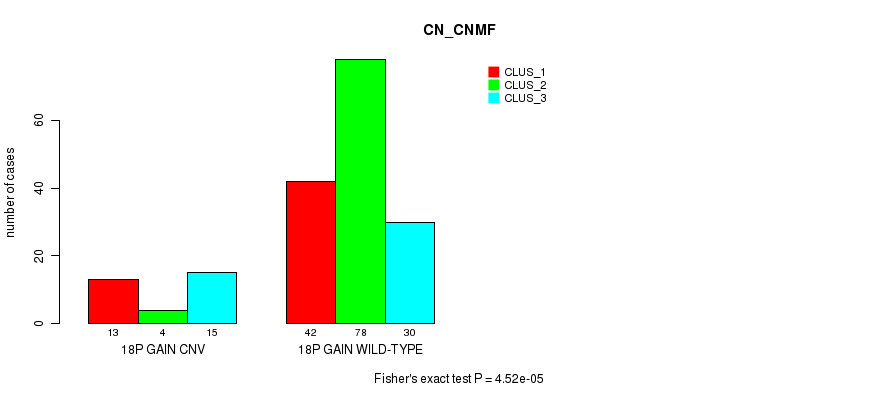
P value = 3.26e-05 (Fisher's exact test), Q value = 0.025
Table S9. Gene #49: '5q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 5Q LOSS CNV | 22 | 7 | 13 |
| 5Q LOSS WILD-TYPE | 33 | 75 | 32 |
Figure S9. Get High-res Image Gene #49: '5q loss' versus Molecular Subtype #1: 'CN_CNMF'
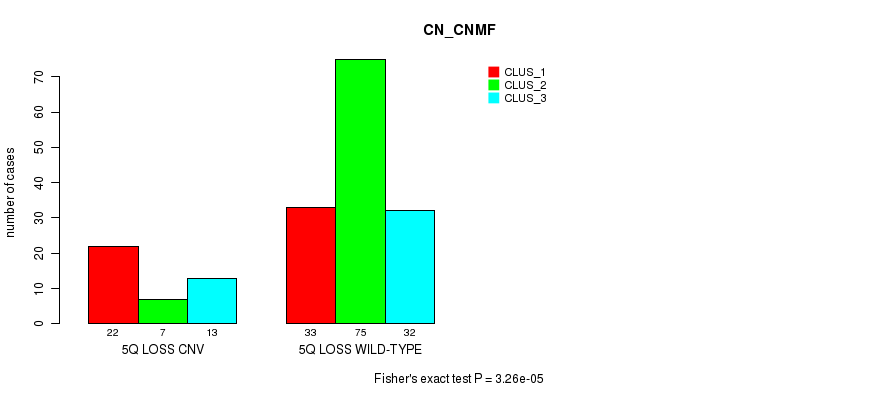
P value = 0.000115 (Fisher's exact test), Q value = 0.086
Table S10. Gene #51: '6q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 6Q LOSS CNV | 21 | 7 | 7 |
| 6Q LOSS WILD-TYPE | 34 | 75 | 38 |
Figure S10. Get High-res Image Gene #51: '6q loss' versus Molecular Subtype #1: 'CN_CNMF'
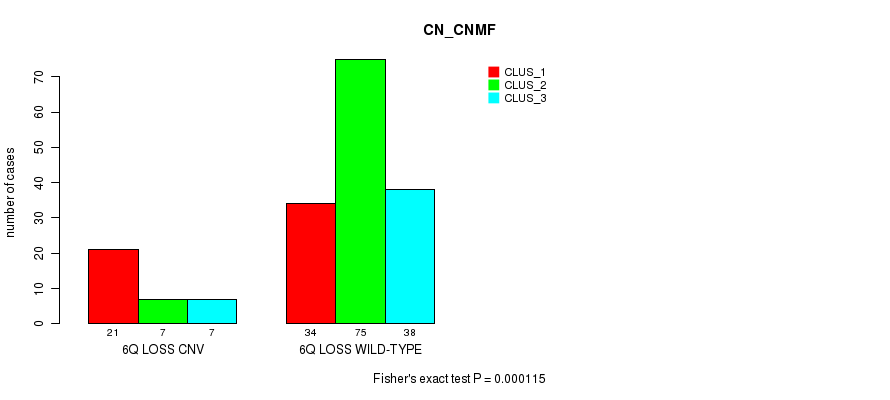
P value = 2.17e-09 (Fisher's exact test), Q value = 1.7e-06
Table S11. Gene #53: '8p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 8P LOSS CNV | 18 | 10 | 30 |
| 8P LOSS WILD-TYPE | 37 | 72 | 15 |
Figure S11. Get High-res Image Gene #53: '8p loss' versus Molecular Subtype #1: 'CN_CNMF'
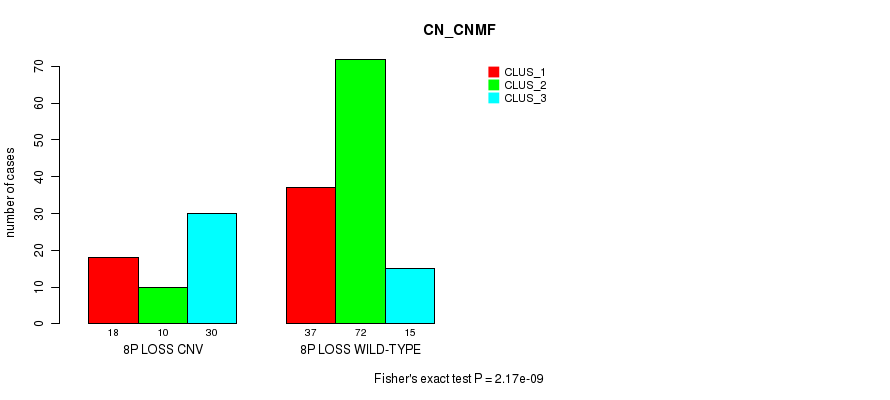
P value = 2.97e-06 (Fisher's exact test), Q value = 0.0023
Table S12. Gene #55: '9p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 9P LOSS CNV | 26 | 26 | 2 |
| 9P LOSS WILD-TYPE | 29 | 56 | 43 |
Figure S12. Get High-res Image Gene #55: '9p loss' versus Molecular Subtype #1: 'CN_CNMF'
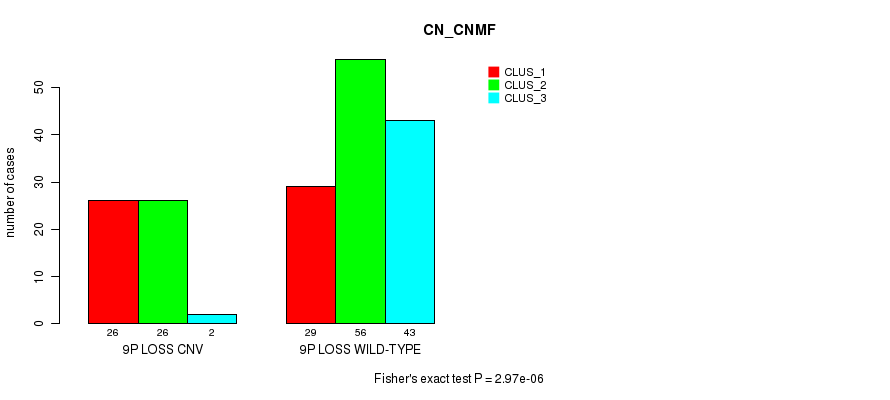
P value = 0.000178 (Fisher's exact test), Q value = 0.13
Table S13. Gene #55: '9p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 71 | 41 |
| 9P LOSS CNV | 28 | 9 | 17 |
| 9P LOSS WILD-TYPE | 42 | 62 | 24 |
Figure S13. Get High-res Image Gene #55: '9p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
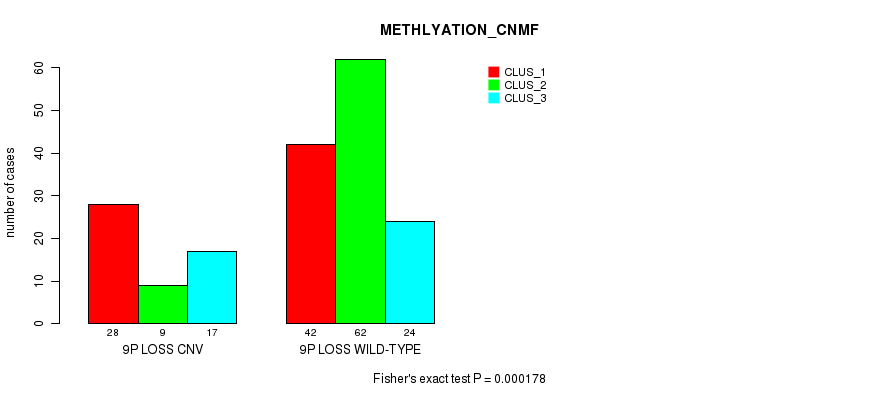
P value = 1.38e-05 (Fisher's exact test), Q value = 0.01
Table S14. Gene #55: '9p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 36 | 67 | 76 |
| 9P LOSS CNV | 13 | 31 | 9 |
| 9P LOSS WILD-TYPE | 23 | 36 | 67 |
Figure S14. Get High-res Image Gene #55: '9p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
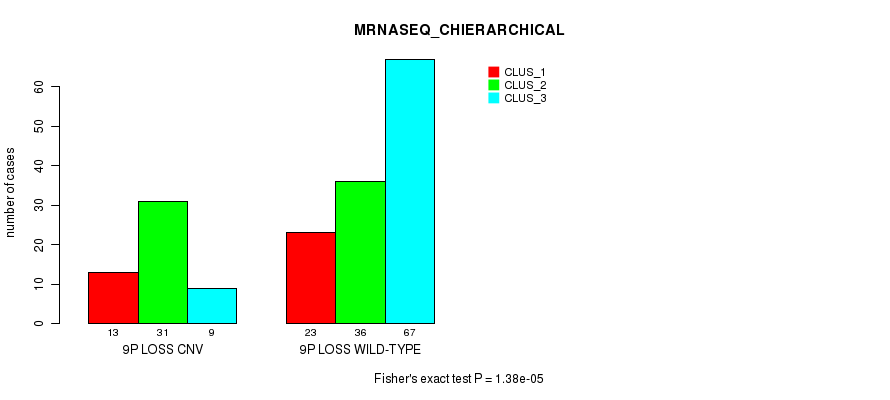
P value = 7.61e-05 (Fisher's exact test), Q value = 0.057
Table S15. Gene #56: '9q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 9Q LOSS CNV | 24 | 25 | 3 |
| 9Q LOSS WILD-TYPE | 31 | 57 | 42 |
Figure S15. Get High-res Image Gene #56: '9q loss' versus Molecular Subtype #1: 'CN_CNMF'
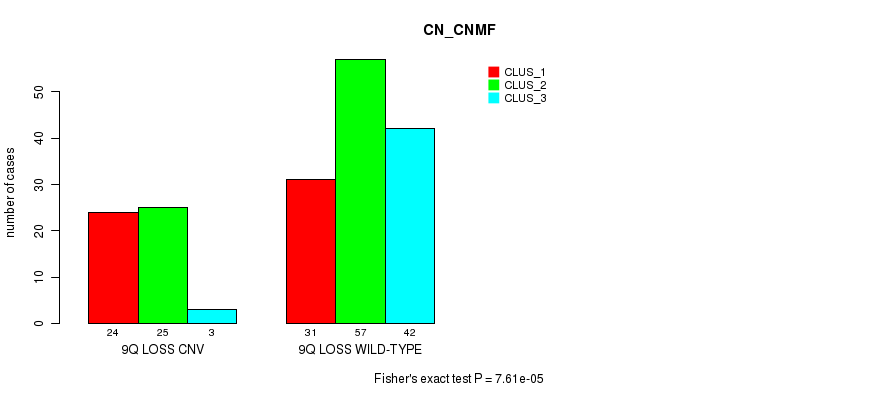
P value = 3.05e-06 (Fisher's exact test), Q value = 0.0023
Table S16. Gene #56: '9q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 36 | 67 | 76 |
| 9Q LOSS CNV | 14 | 29 | 7 |
| 9Q LOSS WILD-TYPE | 22 | 38 | 69 |
Figure S16. Get High-res Image Gene #56: '9q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
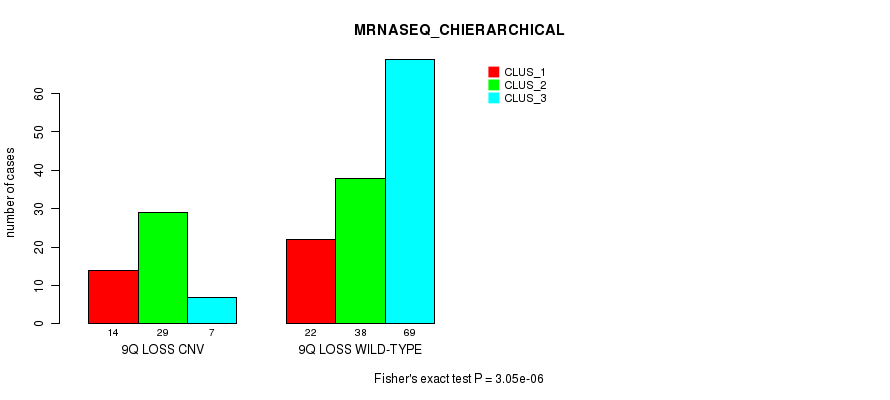
P value = 0.000263 (Fisher's exact test), Q value = 0.2
Table S17. Gene #58: '10q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 10Q LOSS CNV | 19 | 6 | 9 |
| 10Q LOSS WILD-TYPE | 36 | 76 | 36 |
Figure S17. Get High-res Image Gene #58: '10q loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 9.99e-05 (Fisher's exact test), Q value = 0.075
Table S18. Gene #59: '11p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 11P LOSS CNV | 28 | 16 | 8 |
| 11P LOSS WILD-TYPE | 27 | 66 | 37 |
Figure S18. Get High-res Image Gene #59: '11p loss' versus Molecular Subtype #1: 'CN_CNMF'
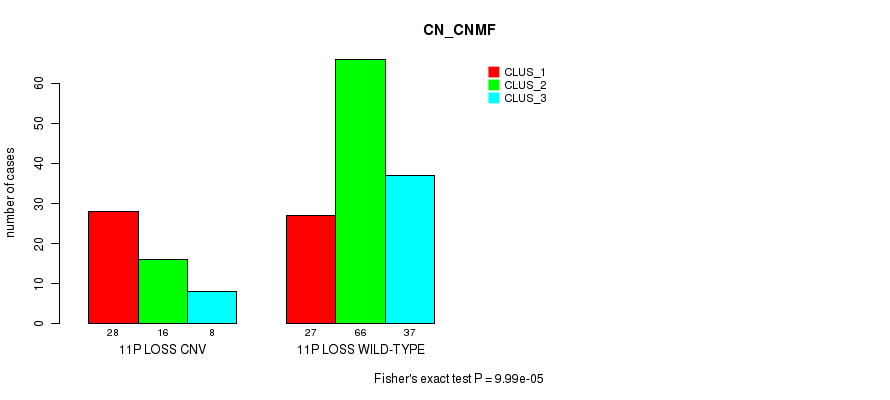
P value = 9.48e-05 (Fisher's exact test), Q value = 0.071
Table S19. Gene #59: '11p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 71 | 41 |
| 11P LOSS CNV | 28 | 8 | 16 |
| 11P LOSS WILD-TYPE | 42 | 63 | 25 |
Figure S19. Get High-res Image Gene #59: '11p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
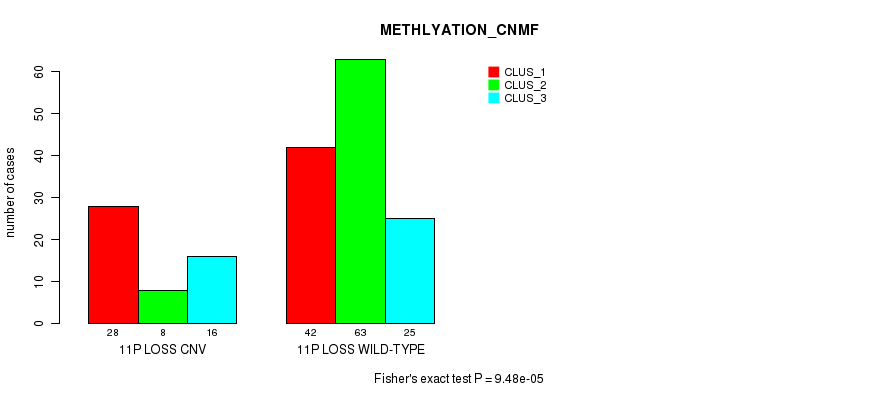
P value = 1.92e-07 (Fisher's exact test), Q value = 0.00015
Table S20. Gene #66: '16p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 16P LOSS CNV | 10 | 0 | 13 |
| 16P LOSS WILD-TYPE | 45 | 82 | 32 |
Figure S20. Get High-res Image Gene #66: '16p loss' versus Molecular Subtype #1: 'CN_CNMF'
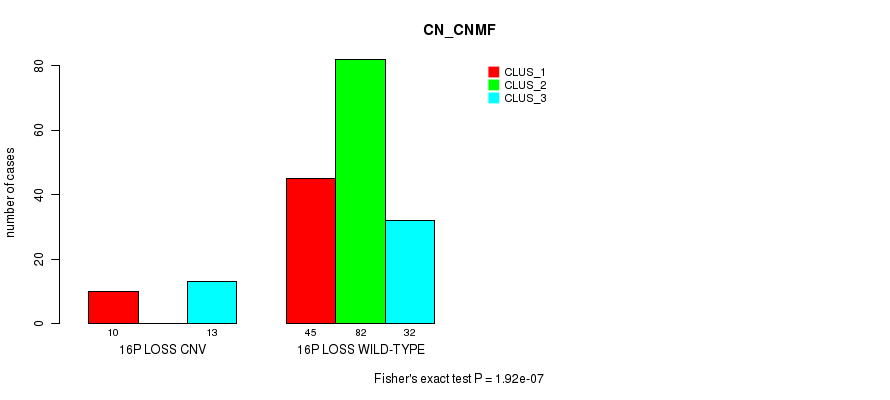
P value = 4.1e-07 (Fisher's exact test), Q value = 0.00031
Table S21. Gene #67: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 16Q LOSS CNV | 11 | 0 | 12 |
| 16Q LOSS WILD-TYPE | 44 | 82 | 33 |
Figure S21. Get High-res Image Gene #67: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
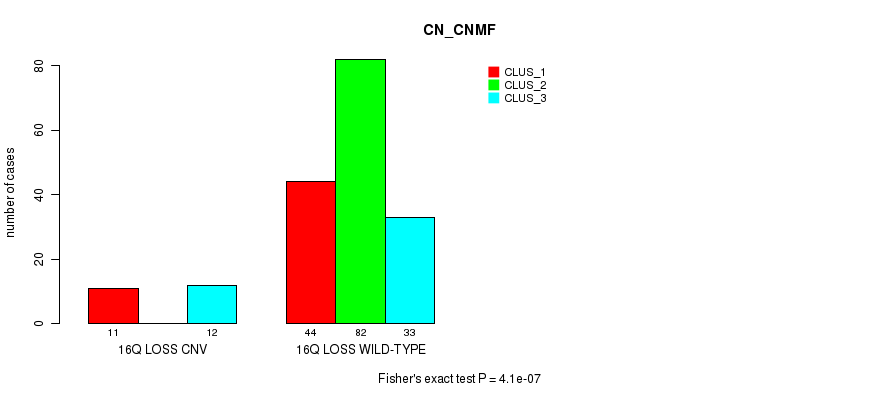
P value = 6.22e-05 (Fisher's exact test), Q value = 0.047
Table S22. Gene #76: '22q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 55 | 82 | 45 |
| 22Q LOSS CNV | 16 | 5 | 15 |
| 22Q LOSS WILD-TYPE | 39 | 77 | 30 |
Figure S22. Get High-res Image Gene #76: '22q loss' versus Molecular Subtype #1: 'CN_CNMF'
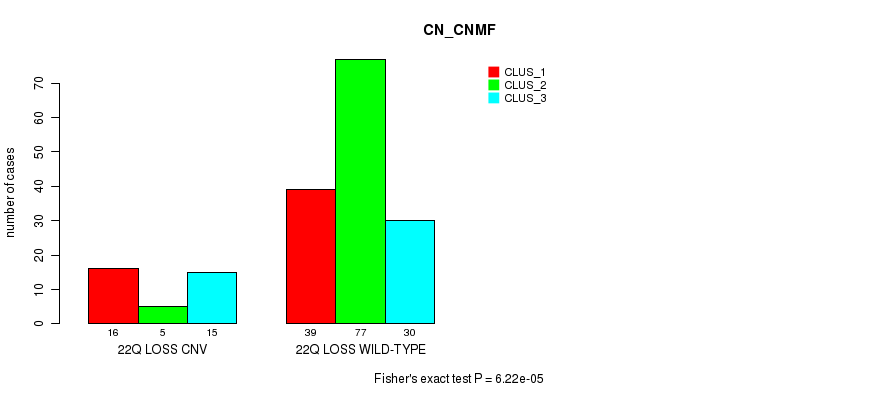
-
Mutation data file = broad_values_by_arm.mutsig.cluster.txt
-
Molecular subtypes file = BLCA-TP.transferedmergedcluster.txt
-
Number of patients = 182
-
Number of significantly arm-level cnvs = 77
-
Number of molecular subtypes = 10
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.