This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and selected clinical features.
Testing the association between copy number variation 63 arm-level events and 10 clinical features across 129 patients, 11 significant findings detected with Q value < 0.25.
-
1Q GAIN MUTATION ANALYSIS cnv correlated to 'Time to Death', 'NEOPLASM.DISEASESTAGE', and 'PATHOLOGY.T.STAGE'.
-
6Q GAIN MUTATION ANALYSIS cnv correlated to 'Time to Death'.
-
8Q GAIN MUTATION ANALYSIS cnv correlated to 'Time to Death'.
-
17P GAIN MUTATION ANALYSIS cnv correlated to 'PATHOLOGY.M.STAGE'.
-
3Q LOSS MUTATION ANALYSIS cnv correlated to 'Time to Death'.
-
5P LOSS MUTATION ANALYSIS cnv correlated to 'Time to Death'.
-
16P LOSS MUTATION ANALYSIS cnv correlated to 'Time to Death'.
-
16Q LOSS MUTATION ANALYSIS cnv correlated to 'Time to Death'.
-
17P LOSS MUTATION ANALYSIS cnv correlated to 'Time to Death'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 63 arm-level events and 10 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 11 significant findings detected.
|
Clinical Features |
Time to Death |
AGE |
NEOPLASM DISEASESTAGE |
PATHOLOGY T STAGE |
PATHOLOGY N STAGE |
PATHOLOGY M STAGE |
GENDER |
KARNOFSKY PERFORMANCE SCORE |
NUMBERPACKYEARSSMOKED | YEAROFTOBACCOSMOKINGONSET | ||
| nCNV (%) | nWild-Type | logrank test | t-test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | t-test | t-test | t-test | |
| 1Q GAIN MUTATION ANALYSIS | 11 (9%) | 118 |
1.28e-05 (0.00586) |
0.386 (1.00) |
0.000272 (0.123) |
9.11e-05 (0.0415) |
0.379 (1.00) |
0.156 (1.00) |
0.0944 (1.00) |
|||
| 6Q GAIN MUTATION ANALYSIS | 3 (2%) | 126 |
8.53e-06 (0.00393) |
0.257 (1.00) |
0.0477 (1.00) |
0.513 (1.00) |
0.00616 (1.00) |
0.552 (1.00) |
||||
| 8Q GAIN MUTATION ANALYSIS | 13 (10%) | 116 |
3.61e-05 (0.0166) |
0.838 (1.00) |
0.0576 (1.00) |
0.0442 (1.00) |
0.0305 (1.00) |
0.381 (1.00) |
0.221 (1.00) |
|||
| 17P GAIN MUTATION ANALYSIS | 71 (55%) | 58 |
0.114 (1.00) |
0.0209 (1.00) |
0.00213 (0.946) |
0.0051 (1.00) |
0.227 (1.00) |
0.000167 (0.0757) |
0.0235 (1.00) |
0.182 (1.00) |
||
| 3Q LOSS MUTATION ANALYSIS | 3 (2%) | 126 |
2.37e-06 (0.00109) |
0.323 (1.00) |
0.0943 (1.00) |
0.0665 (1.00) |
0.434 (1.00) |
0.144 (1.00) |
1 (1.00) |
|||
| 5P LOSS MUTATION ANALYSIS | 6 (5%) | 123 |
0.000124 (0.0564) |
0.114 (1.00) |
0.0558 (1.00) |
0.0262 (1.00) |
0.0172 (1.00) |
0.193 (1.00) |
0.0739 (1.00) |
|||
| 16P LOSS MUTATION ANALYSIS | 4 (3%) | 125 |
4.28e-05 (0.0195) |
0.653 (1.00) |
0.255 (1.00) |
0.157 (1.00) |
1 (1.00) |
0.0882 (1.00) |
||||
| 16Q LOSS MUTATION ANALYSIS | 4 (3%) | 125 |
4.28e-05 (0.0195) |
0.653 (1.00) |
0.255 (1.00) |
0.157 (1.00) |
1 (1.00) |
0.0882 (1.00) |
||||
| 17P LOSS MUTATION ANALYSIS | 7 (5%) | 122 |
4.06e-09 (1.88e-06) |
0.863 (1.00) |
0.00668 (1.00) |
0.0332 (1.00) |
0.108 (1.00) |
0.0555 (1.00) |
0.202 (1.00) |
|||
| 2P GAIN MUTATION ANALYSIS | 21 (16%) | 108 |
0.092 (1.00) |
0.22 (1.00) |
0.0549 (1.00) |
0.586 (1.00) |
0.203 (1.00) |
0.00141 (0.629) |
0.449 (1.00) |
0.541 (1.00) |
||
| 2Q GAIN MUTATION ANALYSIS | 23 (18%) | 106 |
0.177 (1.00) |
0.62 (1.00) |
0.0138 (1.00) |
0.571 (1.00) |
0.127 (1.00) |
0.00263 (1.00) |
0.456 (1.00) |
0.501 (1.00) |
||
| 3P GAIN MUTATION ANALYSIS | 36 (28%) | 93 |
0.239 (1.00) |
0.792 (1.00) |
0.0926 (1.00) |
0.0322 (1.00) |
1 (1.00) |
0.254 (1.00) |
0.403 (1.00) |
0.13 (1.00) |
||
| 3Q GAIN MUTATION ANALYSIS | 41 (32%) | 88 |
0.105 (1.00) |
0.675 (1.00) |
0.129 (1.00) |
0.0804 (1.00) |
1 (1.00) |
0.143 (1.00) |
0.155 (1.00) |
0.248 (1.00) |
||
| 4P GAIN MUTATION ANALYSIS | 6 (5%) | 123 |
0.0226 (1.00) |
0.00425 (1.00) |
0.092 (1.00) |
0.85 (1.00) |
0.53 (1.00) |
0.193 (1.00) |
0.665 (1.00) |
|||
| 4Q GAIN MUTATION ANALYSIS | 5 (4%) | 124 |
0.36 (1.00) |
0.0141 (1.00) |
0.387 (1.00) |
0.662 (1.00) |
0.284 (1.00) |
0.69 (1.00) |
1 (1.00) |
|||
| 5P GAIN MUTATION ANALYSIS | 20 (16%) | 109 |
0.449 (1.00) |
0.323 (1.00) |
0.233 (1.00) |
0.217 (1.00) |
0.862 (1.00) |
0.291 (1.00) |
0.608 (1.00) |
0.397 (1.00) |
||
| 5Q GAIN MUTATION ANALYSIS | 19 (15%) | 110 |
0.149 (1.00) |
0.631 (1.00) |
0.305 (1.00) |
0.302 (1.00) |
1 (1.00) |
0.303 (1.00) |
0.79 (1.00) |
0.397 (1.00) |
||
| 6P GAIN MUTATION ANALYSIS | 4 (3%) | 125 |
0.0639 (1.00) |
0.356 (1.00) |
0.0624 (1.00) |
0.157 (1.00) |
0.434 (1.00) |
0.00611 (1.00) |
0.31 (1.00) |
|||
| 7P GAIN MUTATION ANALYSIS | 71 (55%) | 58 |
0.11 (1.00) |
0.809 (1.00) |
0.0027 (1.00) |
0.0132 (1.00) |
0.805 (1.00) |
0.16 (1.00) |
0.0593 (1.00) |
0.184 (1.00) |
||
| 7Q GAIN MUTATION ANALYSIS | 73 (57%) | 56 |
0.0991 (1.00) |
0.716 (1.00) |
0.00387 (1.00) |
0.0186 (1.00) |
0.726 (1.00) |
0.276 (1.00) |
0.0863 (1.00) |
0.184 (1.00) |
||
| 8P GAIN MUTATION ANALYSIS | 10 (8%) | 119 |
0.0466 (1.00) |
0.894 (1.00) |
0.487 (1.00) |
0.358 (1.00) |
0.441 (1.00) |
0.849 (1.00) |
0.499 (1.00) |
|||
| 10P GAIN MUTATION ANALYSIS | 5 (4%) | 124 |
0.54 (1.00) |
0.932 (1.00) |
0.255 (1.00) |
0.515 (1.00) |
0.51 (1.00) |
1 (1.00) |
||||
| 10Q GAIN MUTATION ANALYSIS | 4 (3%) | 125 |
0.501 (1.00) |
0.811 (1.00) |
0.271 (1.00) |
0.589 (1.00) |
0.69 (1.00) |
1 (1.00) |
||||
| 11P GAIN MUTATION ANALYSIS | 6 (5%) | 123 |
0.0357 (1.00) |
0.303 (1.00) |
0.0558 (1.00) |
0.0508 (1.00) |
0.721 (1.00) |
0.012 (1.00) |
1 (1.00) |
|||
| 11Q GAIN MUTATION ANALYSIS | 4 (3%) | 125 |
0.755 (1.00) |
0.127 (1.00) |
0.271 (1.00) |
0.0956 (1.00) |
1 (1.00) |
0.144 (1.00) |
0.587 (1.00) |
|||
| 12P GAIN MUTATION ANALYSIS | 49 (38%) | 80 |
0.348 (1.00) |
0.776 (1.00) |
0.582 (1.00) |
0.463 (1.00) |
0.493 (1.00) |
0.109 (1.00) |
0.119 (1.00) |
0.248 (1.00) |
||
| 12Q GAIN MUTATION ANALYSIS | 49 (38%) | 80 |
0.348 (1.00) |
0.776 (1.00) |
0.582 (1.00) |
0.463 (1.00) |
0.493 (1.00) |
0.109 (1.00) |
0.119 (1.00) |
0.248 (1.00) |
||
| 13Q GAIN MUTATION ANALYSIS | 15 (12%) | 114 |
0.485 (1.00) |
0.155 (1.00) |
0.481 (1.00) |
0.527 (1.00) |
1 (1.00) |
1 (1.00) |
0.197 (1.00) |
|||
| 16P GAIN MUTATION ANALYSIS | 66 (51%) | 63 |
0.984 (1.00) |
0.45 (1.00) |
0.599 (1.00) |
0.967 (1.00) |
1 (1.00) |
0.263 (1.00) |
0.00728 (1.00) |
0.529 (1.00) |
||
| 16Q GAIN MUTATION ANALYSIS | 64 (50%) | 65 |
0.304 (1.00) |
0.663 (1.00) |
0.599 (1.00) |
0.907 (1.00) |
0.804 (1.00) |
0.0373 (1.00) |
0.0359 (1.00) |
0.25 (1.00) |
||
| 17Q GAIN MUTATION ANALYSIS | 84 (65%) | 45 |
0.718 (1.00) |
0.0637 (1.00) |
0.449 (1.00) |
0.377 (1.00) |
0.506 (1.00) |
0.00265 (1.00) |
0.115 (1.00) |
0.561 (1.00) |
||
| 18P GAIN MUTATION ANALYSIS | 7 (5%) | 122 |
0.85 (1.00) |
0.586 (1.00) |
0.485 (1.00) |
0.538 (1.00) |
0.136 (1.00) |
1 (1.00) |
0.541 (1.00) |
|||
| 18Q GAIN MUTATION ANALYSIS | 5 (4%) | 124 |
0.363 (1.00) |
0.954 (1.00) |
0.733 (1.00) |
0.662 (1.00) |
0.69 (1.00) |
1 (1.00) |
||||
| 20P GAIN MUTATION ANALYSIS | 42 (33%) | 87 |
0.815 (1.00) |
0.0189 (1.00) |
0.606 (1.00) |
0.53 (1.00) |
0.876 (1.00) |
0.492 (1.00) |
0.839 (1.00) |
0.636 (1.00) |
||
| 20Q GAIN MUTATION ANALYSIS | 45 (35%) | 84 |
0.483 (1.00) |
0.0144 (1.00) |
0.398 (1.00) |
0.432 (1.00) |
0.771 (1.00) |
0.898 (1.00) |
1 (1.00) |
0.636 (1.00) |
||
| 21Q GAIN MUTATION ANALYSIS | 6 (5%) | 123 |
0.755 (1.00) |
0.0276 (1.00) |
0.561 (1.00) |
0.251 (1.00) |
0.51 (1.00) |
0.665 (1.00) |
||||
| XQ GAIN MUTATION ANALYSIS | 38 (29%) | 91 |
0.673 (1.00) |
0.609 (1.00) |
0.16 (1.00) |
0.146 (1.00) |
1 (1.00) |
0.569 (1.00) |
0.145 (1.00) |
0.136 (1.00) |
||
| 1P LOSS MUTATION ANALYSIS | 17 (13%) | 112 |
0.7 (1.00) |
0.645 (1.00) |
0.446 (1.00) |
0.565 (1.00) |
0.34 (1.00) |
0.912 (1.00) |
0.779 (1.00) |
0.342 (1.00) |
||
| 1Q LOSS MUTATION ANALYSIS | 10 (8%) | 119 |
0.563 (1.00) |
0.398 (1.00) |
0.255 (1.00) |
0.645 (1.00) |
0.481 (1.00) |
1 (1.00) |
0.342 (1.00) |
|||
| 3P LOSS MUTATION ANALYSIS | 9 (7%) | 120 |
0.178 (1.00) |
0.111 (1.00) |
0.00718 (1.00) |
0.00599 (1.00) |
0.724 (1.00) |
0.0885 (1.00) |
0.72 (1.00) |
|||
| 4P LOSS MUTATION ANALYSIS | 11 (9%) | 118 |
0.193 (1.00) |
0.0531 (1.00) |
0.00996 (1.00) |
0.00414 (1.00) |
0.0279 (1.00) |
0.468 (1.00) |
0.00371 (1.00) |
|||
| 4Q LOSS MUTATION ANALYSIS | 12 (9%) | 117 |
0.817 (1.00) |
0.237 (1.00) |
0.0387 (1.00) |
0.0377 (1.00) |
0.267 (1.00) |
0.12 (1.00) |
0.0472 (1.00) |
|||
| 5Q LOSS MUTATION ANALYSIS | 5 (4%) | 124 |
0.0201 (1.00) |
0.114 (1.00) |
0.255 (1.00) |
0.0466 (1.00) |
0.0394 (1.00) |
0.284 (1.00) |
0.173 (1.00) |
|||
| 6P LOSS MUTATION ANALYSIS | 10 (8%) | 119 |
0.0941 (1.00) |
0.501 (1.00) |
0.147 (1.00) |
0.101 (1.00) |
0.53 (1.00) |
1 (1.00) |
0.283 (1.00) |
0.248 (1.00) |
||
| 6Q LOSS MUTATION ANALYSIS | 12 (9%) | 117 |
0.341 (1.00) |
0.82 (1.00) |
0.119 (1.00) |
0.0253 (1.00) |
0.587 (1.00) |
0.431 (1.00) |
0.0472 (1.00) |
0.383 (1.00) |
||
| 8P LOSS MUTATION ANALYSIS | 5 (4%) | 124 |
0.000834 (0.374) |
0.209 (1.00) |
0.041 (1.00) |
0.515 (1.00) |
0.0196 (1.00) |
0.173 (1.00) |
||||
| 8Q LOSS MUTATION ANALYSIS | 3 (2%) | 126 |
0.0145 (1.00) |
0.601 (1.00) |
0.119 (1.00) |
0.356 (1.00) |
0.144 (1.00) |
0.227 (1.00) |
||||
| 9P LOSS MUTATION ANALYSIS | 17 (13%) | 112 |
0.0721 (1.00) |
0.535 (1.00) |
0.00317 (1.00) |
0.00353 (1.00) |
0.374 (1.00) |
0.0495 (1.00) |
0.0116 (1.00) |
0.398 (1.00) |
||
| 9Q LOSS MUTATION ANALYSIS | 18 (14%) | 111 |
0.0954 (1.00) |
0.868 (1.00) |
0.0249 (1.00) |
0.0024 (1.00) |
0.203 (1.00) |
0.259 (1.00) |
0.0963 (1.00) |
0.398 (1.00) |
||
| 10P LOSS MUTATION ANALYSIS | 8 (6%) | 121 |
0.878 (1.00) |
0.736 (1.00) |
0.14 (1.00) |
0.0795 (1.00) |
0.102 (1.00) |
0.136 (1.00) |
0.703 (1.00) |
0.397 (1.00) |
||
| 10Q LOSS MUTATION ANALYSIS | 8 (6%) | 121 |
0.219 (1.00) |
0.473 (1.00) |
0.0723 (1.00) |
0.0237 (1.00) |
0.34 (1.00) |
0.3 (1.00) |
1 (1.00) |
0.397 (1.00) |
||
| 11P LOSS MUTATION ANALYSIS | 9 (7%) | 120 |
0.013 (1.00) |
0.0568 (1.00) |
0.185 (1.00) |
0.231 (1.00) |
0.434 (1.00) |
0.481 (1.00) |
0.458 (1.00) |
|||
| 11Q LOSS MUTATION ANALYSIS | 12 (9%) | 117 |
0.00109 (0.488) |
0.0288 (1.00) |
0.00424 (1.00) |
0.00215 (0.953) |
0.285 (1.00) |
0.156 (1.00) |
1 (1.00) |
|||
| 13Q LOSS MUTATION ANALYSIS | 11 (9%) | 118 |
0.0237 (1.00) |
0.273 (1.00) |
0.0719 (1.00) |
0.0427 (1.00) |
0.0823 (1.00) |
0.468 (1.00) |
0.00371 (1.00) |
|||
| 14Q LOSS MUTATION ANALYSIS | 27 (21%) | 102 |
0.596 (1.00) |
0.73 (1.00) |
0.0817 (1.00) |
0.117 (1.00) |
0.274 (1.00) |
0.213 (1.00) |
0.246 (1.00) |
0.377 (1.00) |
||
| 15Q LOSS MUTATION ANALYSIS | 14 (11%) | 115 |
0.00495 (1.00) |
0.178 (1.00) |
0.298 (1.00) |
0.0761 (1.00) |
0.267 (1.00) |
0.0578 (1.00) |
0.762 (1.00) |
0.654 (1.00) |
||
| 18P LOSS MUTATION ANALYSIS | 20 (16%) | 109 |
0.0589 (1.00) |
0.427 (1.00) |
0.0107 (1.00) |
0.00267 (1.00) |
0.203 (1.00) |
0.0575 (1.00) |
1 (1.00) |
0.342 (1.00) |
||
| 18Q LOSS MUTATION ANALYSIS | 21 (16%) | 108 |
0.0589 (1.00) |
0.427 (1.00) |
0.00487 (1.00) |
0.000971 (0.435) |
0.203 (1.00) |
0.042 (1.00) |
0.801 (1.00) |
0.342 (1.00) |
||
| 19P LOSS MUTATION ANALYSIS | 9 (7%) | 120 |
0.647 (1.00) |
0.47 (1.00) |
0.118 (1.00) |
0.0524 (1.00) |
1 (1.00) |
0.3 (1.00) |
1 (1.00) |
|||
| 19Q LOSS MUTATION ANALYSIS | 8 (6%) | 121 |
0.741 (1.00) |
0.747 (1.00) |
0.0474 (1.00) |
0.0179 (1.00) |
1 (1.00) |
0.136 (1.00) |
0.703 (1.00) |
|||
| 21Q LOSS MUTATION ANALYSIS | 23 (18%) | 106 |
0.425 (1.00) |
0.472 (1.00) |
0.0112 (1.00) |
0.00971 (1.00) |
0.33 (1.00) |
0.12 (1.00) |
0.456 (1.00) |
|||
| 22Q LOSS MUTATION ANALYSIS | 31 (24%) | 98 |
0.788 (1.00) |
0.602 (1.00) |
0.000733 (0.33) |
0.000626 (0.282) |
0.14 (1.00) |
0.118 (1.00) |
0.0735 (1.00) |
0.445 (1.00) |
||
| XQ LOSS MUTATION ANALYSIS | 19 (15%) | 110 |
0.00127 (0.564) |
0.0873 (1.00) |
0.0621 (1.00) |
0.016 (1.00) |
0.0392 (1.00) |
0.317 (1.00) |
0.288 (1.00) |
0.458 (1.00) |
P value = 1.28e-05 (logrank test), Q value = 0.0059
Table S1. Gene #1: '1Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 119 | 15 | 0.0 - 194.8 (13.7) |
| 1Q GAIN MUTATED | 10 | 3 | 0.7 - 30.3 (5.8) |
| 1Q GAIN WILD-TYPE | 109 | 12 | 0.0 - 194.8 (14.6) |
Figure S1. Get High-res Image Gene #1: '1Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
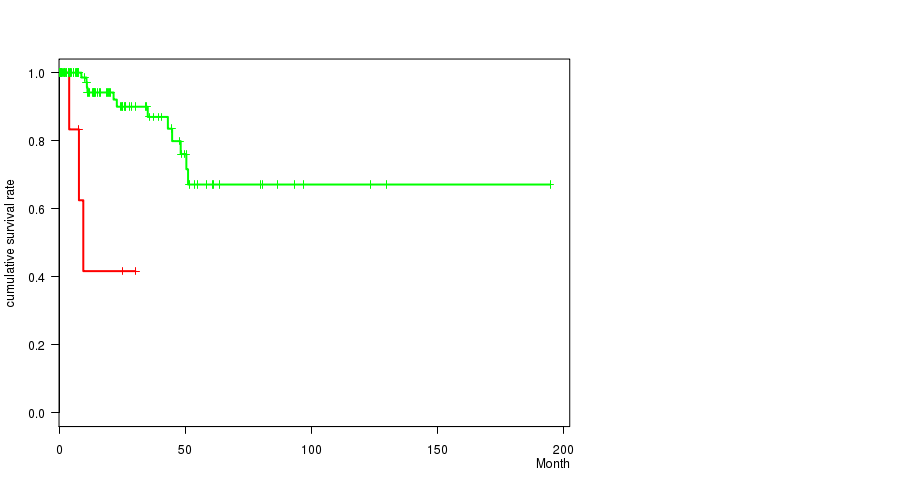
P value = 0.000272 (Fisher's exact test), Q value = 0.12
Table S2. Gene #1: '1Q GAIN MUTATION STATUS' versus Clinical Feature #3: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE I | STAGE II | STAGE III | STAGE IV |
|---|---|---|---|---|
| ALL | 69 | 10 | 30 | 10 |
| 1Q GAIN MUTATED | 1 | 0 | 7 | 3 |
| 1Q GAIN WILD-TYPE | 68 | 10 | 23 | 7 |
Figure S2. Get High-res Image Gene #1: '1Q GAIN MUTATION STATUS' versus Clinical Feature #3: 'NEOPLASM.DISEASESTAGE'

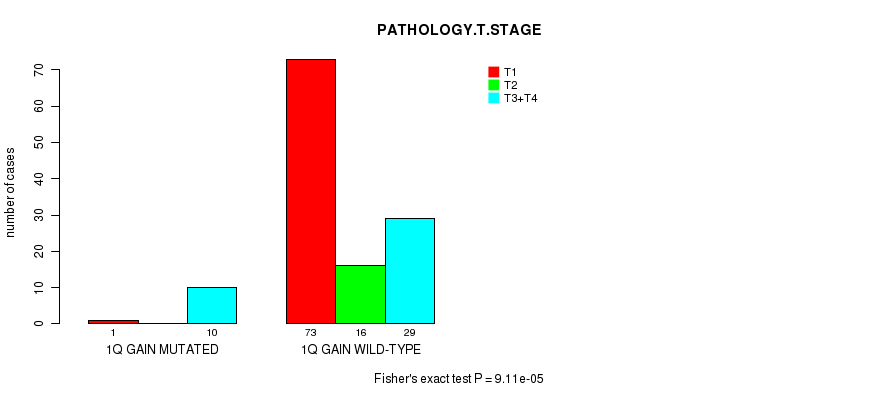
P value = 9.11e-05 (Fisher's exact test), Q value = 0.041
Table S3. Gene #1: '1Q GAIN MUTATION STATUS' versus Clinical Feature #4: 'PATHOLOGY.T.STAGE'
| nPatients | T1 | T2 | T3+T4 |
|---|---|---|---|
| ALL | 74 | 16 | 39 |
| 1Q GAIN MUTATED | 1 | 0 | 10 |
| 1Q GAIN WILD-TYPE | 73 | 16 | 29 |
Figure S3. Get High-res Image Gene #1: '1Q GAIN MUTATION STATUS' versus Clinical Feature #4: 'PATHOLOGY.T.STAGE'
P value = 8.53e-06 (logrank test), Q value = 0.0039
Table S4. Gene #11: '6Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 119 | 15 | 0.0 - 194.8 (13.7) |
| 6Q GAIN MUTATED | 3 | 2 | 7.9 - 13.6 (9.6) |
| 6Q GAIN WILD-TYPE | 116 | 13 | 0.0 - 194.8 (14.1) |
Figure S4. Get High-res Image Gene #11: '6Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'

P value = 3.61e-05 (logrank test), Q value = 0.017
Table S5. Gene #15: '8Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 119 | 15 | 0.0 - 194.8 (13.7) |
| 8Q GAIN MUTATED | 12 | 4 | 0.2 - 43.2 (6.8) |
| 8Q GAIN WILD-TYPE | 107 | 11 | 0.0 - 194.8 (14.6) |
Figure S5. Get High-res Image Gene #15: '8Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'

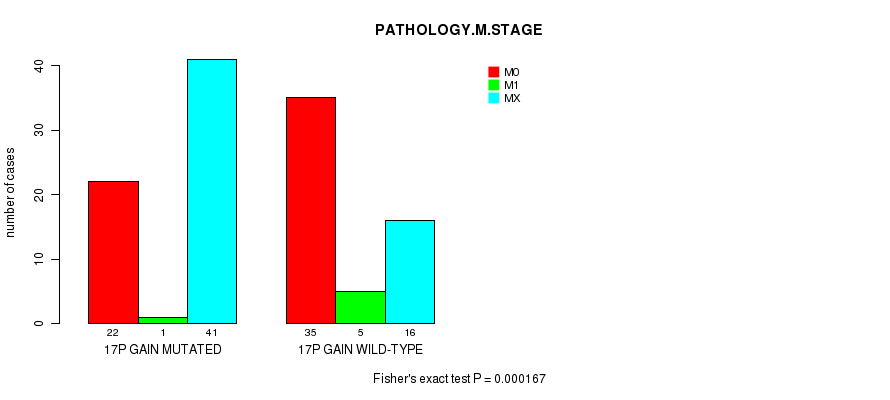
P value = 0.000167 (Fisher's exact test), Q value = 0.076
Table S6. Gene #25: '17P GAIN MUTATION STATUS' versus Clinical Feature #6: 'PATHOLOGY.M.STAGE'
| nPatients | M0 | M1 | MX |
|---|---|---|---|
| ALL | 57 | 6 | 57 |
| 17P GAIN MUTATED | 22 | 1 | 41 |
| 17P GAIN WILD-TYPE | 35 | 5 | 16 |
Figure S6. Get High-res Image Gene #25: '17P GAIN MUTATION STATUS' versus Clinical Feature #6: 'PATHOLOGY.M.STAGE'
P value = 2.37e-06 (logrank test), Q value = 0.0011
Table S7. Gene #36: '3Q LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 119 | 15 | 0.0 - 194.8 (13.7) |
| 3Q LOSS MUTATED | 3 | 2 | 3.7 - 21.6 (8.8) |
| 3Q LOSS WILD-TYPE | 116 | 13 | 0.0 - 194.8 (13.9) |
Figure S7. Get High-res Image Gene #36: '3Q LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'

P value = 0.000124 (logrank test), Q value = 0.056
Table S8. Gene #39: '5P LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 119 | 15 | 0.0 - 194.8 (13.7) |
| 5P LOSS MUTATED | 6 | 2 | 0.0 - 22.9 (3.2) |
| 5P LOSS WILD-TYPE | 113 | 13 | 0.0 - 194.8 (14.1) |
Figure S8. Get High-res Image Gene #39: '5P LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'

P value = 4.28e-05 (logrank test), Q value = 0.02
Table S9. Gene #54: '16P LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 119 | 15 | 0.0 - 194.8 (13.7) |
| 16P LOSS MUTATED | 3 | 2 | 0.7 - 21.6 (11.1) |
| 16P LOSS WILD-TYPE | 116 | 13 | 0.0 - 194.8 (13.9) |
Figure S9. Get High-res Image Gene #54: '16P LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'

P value = 4.28e-05 (logrank test), Q value = 0.02
Table S10. Gene #55: '16Q LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 119 | 15 | 0.0 - 194.8 (13.7) |
| 16Q LOSS MUTATED | 3 | 2 | 0.7 - 21.6 (11.1) |
| 16Q LOSS WILD-TYPE | 116 | 13 | 0.0 - 194.8 (13.9) |
Figure S10. Get High-res Image Gene #55: '16Q LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'

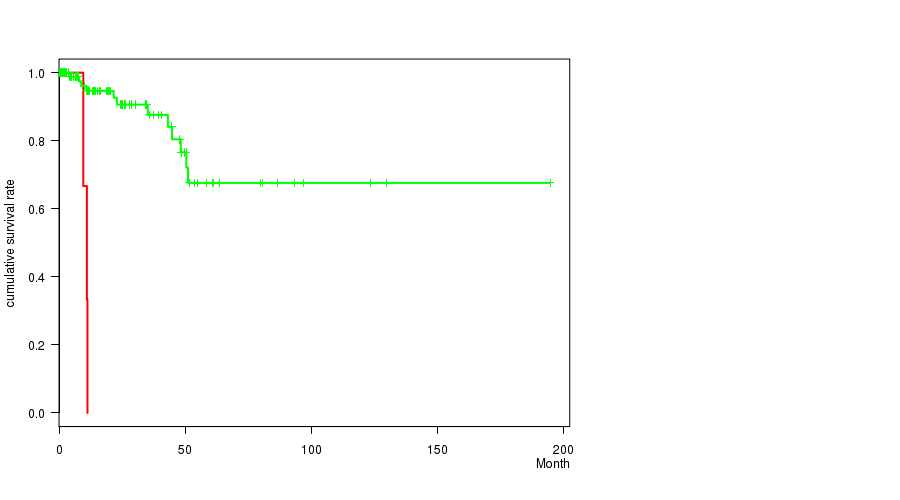
P value = 4.06e-09 (logrank test), Q value = 1.9e-06
Table S11. Gene #56: '17P LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 119 | 15 | 0.0 - 194.8 (13.7) |
| 17P LOSS MUTATED | 5 | 3 | 0.2 - 11.3 (9.6) |
| 17P LOSS WILD-TYPE | 114 | 12 | 0.0 - 194.8 (14.4) |
Figure S11. Get High-res Image Gene #56: '17P LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
-
Copy number data file = transformed.cor.cli.txt
-
Clinical data file = KIRP-TP.clin.merged.picked.txt
-
Number of patients = 129
-
Number of significantly arm-level cnvs = 63
-
Number of selected clinical features = 10
-
Exclude regions that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.