This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and selected clinical features.
Testing the association between copy number variation 71 arm-level events and 6 clinical features across 229 patients, 19 significant findings detected with Q value < 0.25.
-
3Q GAIN MUTATION ANALYSIS cnv correlated to 'Time to Death'.
-
7Q GAIN MUTATION ANALYSIS cnv correlated to 'AGE'.
-
19Q GAIN MUTATION ANALYSIS cnv correlated to 'Time to Death' and 'AGE'.
-
20P GAIN MUTATION ANALYSIS cnv correlated to 'Time to Death' and 'AGE'.
-
20Q GAIN MUTATION ANALYSIS cnv correlated to 'Time to Death' and 'AGE'.
-
1P LOSS MUTATION ANALYSIS cnv correlated to 'HISTOLOGICAL.TYPE'.
-
10P LOSS MUTATION ANALYSIS cnv correlated to 'Time to Death', 'AGE', and 'HISTOLOGICAL.TYPE'.
-
10Q LOSS MUTATION ANALYSIS cnv correlated to 'Time to Death', 'AGE', and 'HISTOLOGICAL.TYPE'.
-
11Q LOSS MUTATION ANALYSIS cnv correlated to 'Time to Death'.
-
17P LOSS MUTATION ANALYSIS cnv correlated to 'Time to Death' and 'AGE'.
-
19Q LOSS MUTATION ANALYSIS cnv correlated to 'HISTOLOGICAL.TYPE'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 71 arm-level events and 6 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 19 significant findings detected.
|
Clinical Features |
Time to Death |
AGE | GENDER |
KARNOFSKY PERFORMANCE SCORE |
HISTOLOGICAL TYPE |
RADIATIONS RADIATION REGIMENINDICATION |
||
| nCNV (%) | nWild-Type | logrank test | t-test | Fisher's exact test | t-test | Fisher's exact test | Fisher's exact test | |
| 10P LOSS MUTATION ANALYSIS | 39 (17%) | 190 |
1.15e-12 (4.84e-10) |
1.43e-08 (5.97e-06) |
0.0219 (1.00) |
0.305 (1.00) |
0.000156 (0.0639) |
0.473 (1.00) |
| 10Q LOSS MUTATION ANALYSIS | 47 (21%) | 182 |
5.03e-06 (0.00209) |
2.25e-07 (9.36e-05) |
0.022 (1.00) |
0.644 (1.00) |
0.000297 (0.12) |
0.738 (1.00) |
| 19Q GAIN MUTATION ANALYSIS | 11 (5%) | 218 |
5.1e-05 (0.021) |
0.00058 (0.234) |
0.758 (1.00) |
0.243 (1.00) |
0.455 (1.00) |
0.349 (1.00) |
| 20P GAIN MUTATION ANALYSIS | 22 (10%) | 207 |
9.19e-07 (0.000382) |
8.28e-05 (0.034) |
0.178 (1.00) |
0.746 (1.00) |
0.354 (1.00) |
0.5 (1.00) |
| 20Q GAIN MUTATION ANALYSIS | 22 (10%) | 207 |
2.34e-05 (0.00969) |
0.00025 (0.102) |
0.0717 (1.00) |
0.841 (1.00) |
0.515 (1.00) |
0.5 (1.00) |
| 17P LOSS MUTATION ANALYSIS | 10 (4%) | 219 |
0.000276 (0.112) |
0.000572 (0.231) |
0.114 (1.00) |
0.142 (1.00) |
0.849 (1.00) |
0.323 (1.00) |
| 3Q GAIN MUTATION ANALYSIS | 7 (3%) | 222 |
0.000245 (0.0998) |
0.0857 (1.00) |
0.703 (1.00) |
0.273 (1.00) |
0.0708 (1.00) |
0.438 (1.00) |
| 7Q GAIN MUTATION ANALYSIS | 68 (30%) | 161 |
0.00349 (1.00) |
8.53e-05 (0.035) |
0.192 (1.00) |
0.605 (1.00) |
0.188 (1.00) |
0.768 (1.00) |
| 1P LOSS MUTATION ANALYSIS | 76 (33%) | 153 |
0.0678 (1.00) |
0.0285 (1.00) |
1 (1.00) |
0.647 (1.00) |
9.42e-18 (3.97e-15) |
0.0616 (1.00) |
| 11Q LOSS MUTATION ANALYSIS | 9 (4%) | 220 |
4.94e-05 (0.0204) |
0.159 (1.00) |
0.192 (1.00) |
0.439 (1.00) |
0.14 (1.00) |
0.488 (1.00) |
| 19Q LOSS MUTATION ANALYSIS | 87 (38%) | 142 |
0.161 (1.00) |
0.186 (1.00) |
0.495 (1.00) |
0.882 (1.00) |
6.56e-14 (2.75e-11) |
0.405 (1.00) |
| 1P GAIN MUTATION ANALYSIS | 8 (3%) | 221 |
0.00145 (0.58) |
0.0897 (1.00) |
0.144 (1.00) |
0.0763 (1.00) |
0.00267 (1.00) |
1 (1.00) |
| 1Q GAIN MUTATION ANALYSIS | 12 (5%) | 217 |
0.828 (1.00) |
0.00363 (1.00) |
0.14 (1.00) |
0.44 (1.00) |
0.134 (1.00) |
1 (1.00) |
| 2P GAIN MUTATION ANALYSIS | 5 (2%) | 224 |
0.865 (1.00) |
0.259 (1.00) |
0.658 (1.00) |
0.353 (1.00) |
0.522 (1.00) |
1 (1.00) |
| 2Q GAIN MUTATION ANALYSIS | 5 (2%) | 224 |
0.865 (1.00) |
0.259 (1.00) |
0.658 (1.00) |
0.353 (1.00) |
0.522 (1.00) |
1 (1.00) |
| 3P GAIN MUTATION ANALYSIS | 7 (3%) | 222 |
0.0109 (1.00) |
0.0901 (1.00) |
0.247 (1.00) |
0.704 (1.00) |
0.159 (1.00) |
1 (1.00) |
| 6P GAIN MUTATION ANALYSIS | 4 (2%) | 225 |
0.751 (1.00) |
0.341 (1.00) |
1 (1.00) |
0.0763 (1.00) |
0.487 (1.00) |
0.647 (1.00) |
| 6Q GAIN MUTATION ANALYSIS | 3 (1%) | 226 |
0.712 (1.00) |
0.666 (1.00) |
0.587 (1.00) |
0.0763 (1.00) |
0.487 (1.00) |
0.563 (1.00) |
| 7P GAIN MUTATION ANALYSIS | 54 (24%) | 175 |
0.00683 (1.00) |
0.00193 (0.772) |
0.0844 (1.00) |
0.876 (1.00) |
0.17 (1.00) |
0.874 (1.00) |
| 8P GAIN MUTATION ANALYSIS | 22 (10%) | 207 |
0.609 (1.00) |
0.447 (1.00) |
0.261 (1.00) |
0.978 (1.00) |
0.138 (1.00) |
0.822 (1.00) |
| 8Q GAIN MUTATION ANALYSIS | 27 (12%) | 202 |
0.884 (1.00) |
0.106 (1.00) |
0.225 (1.00) |
0.554 (1.00) |
0.0278 (1.00) |
0.837 (1.00) |
| 9P GAIN MUTATION ANALYSIS | 8 (3%) | 221 |
0.021 (1.00) |
0.102 (1.00) |
0.144 (1.00) |
0.5 (1.00) |
0.547 (1.00) |
1 (1.00) |
| 9Q GAIN MUTATION ANALYSIS | 9 (4%) | 220 |
0.00693 (1.00) |
0.0708 (1.00) |
0.0118 (1.00) |
0.788 (1.00) |
0.479 (1.00) |
1 (1.00) |
| 10P GAIN MUTATION ANALYSIS | 25 (11%) | 204 |
0.645 (1.00) |
0.0345 (1.00) |
0.0904 (1.00) |
0.0594 (1.00) |
0.0641 (1.00) |
0.00393 (1.00) |
| 10Q GAIN MUTATION ANALYSIS | 3 (1%) | 226 |
0.414 (1.00) |
0.113 (1.00) |
0.256 (1.00) |
0.0507 (1.00) |
0.0595 (1.00) |
|
| 11P GAIN MUTATION ANALYSIS | 17 (7%) | 212 |
0.892 (1.00) |
0.0255 (1.00) |
0.613 (1.00) |
0.884 (1.00) |
0.409 (1.00) |
0.801 (1.00) |
| 11Q GAIN MUTATION ANALYSIS | 25 (11%) | 204 |
0.489 (1.00) |
0.0185 (1.00) |
0.832 (1.00) |
0.973 (1.00) |
0.492 (1.00) |
1 (1.00) |
| 12P GAIN MUTATION ANALYSIS | 16 (7%) | 213 |
0.53 (1.00) |
0.147 (1.00) |
0.307 (1.00) |
0.0763 (1.00) |
1 (1.00) |
1 (1.00) |
| 12Q GAIN MUTATION ANALYSIS | 8 (3%) | 221 |
0.0586 (1.00) |
0.721 (1.00) |
1 (1.00) |
0.0763 (1.00) |
0.815 (1.00) |
0.485 (1.00) |
| 15Q GAIN MUTATION ANALYSIS | 5 (2%) | 224 |
0.48 (1.00) |
0.14 (1.00) |
0.385 (1.00) |
0.666 (1.00) |
0.617 (1.00) |
1 (1.00) |
| 16P GAIN MUTATION ANALYSIS | 7 (3%) | 222 |
0.433 (1.00) |
0.136 (1.00) |
0.247 (1.00) |
0.425 (1.00) |
0.707 (1.00) |
|
| 16Q GAIN MUTATION ANALYSIS | 9 (4%) | 220 |
0.149 (1.00) |
0.157 (1.00) |
0.517 (1.00) |
0.771 (1.00) |
0.0946 (1.00) |
0.741 (1.00) |
| 17P GAIN MUTATION ANALYSIS | 5 (2%) | 224 |
0.198 (1.00) |
0.495 (1.00) |
0.385 (1.00) |
0.704 (1.00) |
0.278 (1.00) |
0.0795 (1.00) |
| 17Q GAIN MUTATION ANALYSIS | 5 (2%) | 224 |
0.198 (1.00) |
0.495 (1.00) |
0.385 (1.00) |
0.704 (1.00) |
0.278 (1.00) |
0.0795 (1.00) |
| 18P GAIN MUTATION ANALYSIS | 9 (4%) | 220 |
0.315 (1.00) |
0.9 (1.00) |
1 (1.00) |
0.0763 (1.00) |
0.291 (1.00) |
0.488 (1.00) |
| 18Q GAIN MUTATION ANALYSIS | 6 (3%) | 223 |
0.0455 (1.00) |
0.502 (1.00) |
1 (1.00) |
0.44 (1.00) |
0.407 (1.00) |
|
| 19P GAIN MUTATION ANALYSIS | 36 (16%) | 193 |
0.416 (1.00) |
0.00344 (1.00) |
1 (1.00) |
0.796 (1.00) |
0.127 (1.00) |
1 (1.00) |
| 21Q GAIN MUTATION ANALYSIS | 10 (4%) | 219 |
0.966 (1.00) |
0.484 (1.00) |
0.755 (1.00) |
0.105 (1.00) |
1 (1.00) |
|
| 22Q GAIN MUTATION ANALYSIS | 7 (3%) | 222 |
0.295 (1.00) |
0.212 (1.00) |
0.703 (1.00) |
0.704 (1.00) |
0.0611 (1.00) |
1 (1.00) |
| XQ GAIN MUTATION ANALYSIS | 18 (8%) | 211 |
0.559 (1.00) |
0.165 (1.00) |
0.0813 (1.00) |
0.836 (1.00) |
0.234 (1.00) |
0.451 (1.00) |
| 1Q LOSS MUTATION ANALYSIS | 14 (6%) | 215 |
0.726 (1.00) |
0.152 (1.00) |
0.409 (1.00) |
0.289 (1.00) |
0.0463 (1.00) |
0.784 (1.00) |
| 2P LOSS MUTATION ANALYSIS | 9 (4%) | 220 |
0.138 (1.00) |
0.701 (1.00) |
0.305 (1.00) |
0.0625 (1.00) |
0.479 (1.00) |
0.741 (1.00) |
| 2Q LOSS MUTATION ANALYSIS | 7 (3%) | 222 |
0.501 (1.00) |
0.00747 (1.00) |
0.466 (1.00) |
0.166 (1.00) |
0.267 (1.00) |
1 (1.00) |
| 3P LOSS MUTATION ANALYSIS | 9 (4%) | 220 |
0.122 (1.00) |
0.0584 (1.00) |
0.305 (1.00) |
0.958 (1.00) |
0.914 (1.00) |
0.741 (1.00) |
| 3Q LOSS MUTATION ANALYSIS | 15 (7%) | 214 |
0.107 (1.00) |
0.836 (1.00) |
0.185 (1.00) |
0.271 (1.00) |
0.748 (1.00) |
1 (1.00) |
| 4P LOSS MUTATION ANALYSIS | 37 (16%) | 192 |
0.735 (1.00) |
0.0494 (1.00) |
0.278 (1.00) |
0.554 (1.00) |
0.0913 (1.00) |
0.587 (1.00) |
| 4Q LOSS MUTATION ANALYSIS | 46 (20%) | 183 |
0.306 (1.00) |
0.424 (1.00) |
0.507 (1.00) |
0.61 (1.00) |
0.0566 (1.00) |
0.866 (1.00) |
| 5P LOSS MUTATION ANALYSIS | 19 (8%) | 210 |
0.497 (1.00) |
0.0974 (1.00) |
0.147 (1.00) |
0.362 (1.00) |
0.038 (1.00) |
0.047 (1.00) |
| 5Q LOSS MUTATION ANALYSIS | 17 (7%) | 212 |
0.137 (1.00) |
0.819 (1.00) |
0.806 (1.00) |
0.0687 (1.00) |
0.565 (1.00) |
0.0372 (1.00) |
| 6P LOSS MUTATION ANALYSIS | 14 (6%) | 215 |
0.0214 (1.00) |
0.586 (1.00) |
0.784 (1.00) |
0.319 (1.00) |
0.0252 (1.00) |
0.574 (1.00) |
| 6Q LOSS MUTATION ANALYSIS | 29 (13%) | 200 |
0.00389 (1.00) |
0.124 (1.00) |
0.113 (1.00) |
0.166 (1.00) |
0.00422 (1.00) |
0.314 (1.00) |
| 8P LOSS MUTATION ANALYSIS | 8 (3%) | 221 |
0.124 (1.00) |
0.112 (1.00) |
1 (1.00) |
0.824 (1.00) |
0.243 (1.00) |
0.715 (1.00) |
| 8Q LOSS MUTATION ANALYSIS | 6 (3%) | 223 |
0.00478 (1.00) |
0.0997 (1.00) |
0.411 (1.00) |
0.824 (1.00) |
0.216 (1.00) |
1 (1.00) |
| 9P LOSS MUTATION ANALYSIS | 55 (24%) | 174 |
0.0098 (1.00) |
0.053 (1.00) |
0.534 (1.00) |
0.74 (1.00) |
0.104 (1.00) |
0.113 (1.00) |
| 9Q LOSS MUTATION ANALYSIS | 22 (10%) | 207 |
0.0403 (1.00) |
0.0402 (1.00) |
1 (1.00) |
0.158 (1.00) |
0.47 (1.00) |
0.822 (1.00) |
| 11P LOSS MUTATION ANALYSIS | 28 (12%) | 201 |
0.451 (1.00) |
0.308 (1.00) |
0.162 (1.00) |
0.731 (1.00) |
0.0929 (1.00) |
0.417 (1.00) |
| 12P LOSS MUTATION ANALYSIS | 13 (6%) | 216 |
0.539 (1.00) |
0.2 (1.00) |
0.571 (1.00) |
0.345 (1.00) |
0.333 (1.00) |
1 (1.00) |
| 12Q LOSS MUTATION ANALYSIS | 21 (9%) | 208 |
0.348 (1.00) |
0.307 (1.00) |
1 (1.00) |
0.667 (1.00) |
0.304 (1.00) |
0.484 (1.00) |
| 13Q LOSS MUTATION ANALYSIS | 59 (26%) | 170 |
0.769 (1.00) |
0.248 (1.00) |
0.544 (1.00) |
0.362 (1.00) |
0.2 (1.00) |
0.0888 (1.00) |
| 14Q LOSS MUTATION ANALYSIS | 37 (16%) | 192 |
0.00103 (0.414) |
0.0208 (1.00) |
0.373 (1.00) |
0.198 (1.00) |
0.289 (1.00) |
0.714 (1.00) |
| 15Q LOSS MUTATION ANALYSIS | 19 (8%) | 210 |
0.464 (1.00) |
0.931 (1.00) |
0.631 (1.00) |
0.811 (1.00) |
0.83 (1.00) |
0.81 (1.00) |
| 16P LOSS MUTATION ANALYSIS | 7 (3%) | 222 |
0.408 (1.00) |
0.0647 (1.00) |
1 (1.00) |
0.439 (1.00) |
0.425 (1.00) |
0.438 (1.00) |
| 16Q LOSS MUTATION ANALYSIS | 12 (5%) | 217 |
0.275 (1.00) |
0.0195 (1.00) |
0.379 (1.00) |
0.619 (1.00) |
0.361 (1.00) |
0.546 (1.00) |
| 17Q LOSS MUTATION ANALYSIS | 7 (3%) | 222 |
0.0228 (1.00) |
0.0181 (1.00) |
0.703 (1.00) |
0.319 (1.00) |
0.721 (1.00) |
0.707 (1.00) |
| 18P LOSS MUTATION ANALYSIS | 27 (12%) | 202 |
0.563 (1.00) |
0.709 (1.00) |
0.225 (1.00) |
0.548 (1.00) |
0.813 (1.00) |
0.675 (1.00) |
| 18Q LOSS MUTATION ANALYSIS | 28 (12%) | 201 |
0.304 (1.00) |
0.663 (1.00) |
0.686 (1.00) |
0.852 (1.00) |
0.565 (1.00) |
0.685 (1.00) |
| 19P LOSS MUTATION ANALYSIS | 13 (6%) | 216 |
0.88 (1.00) |
0.197 (1.00) |
0.777 (1.00) |
0.352 (1.00) |
0.937 (1.00) |
0.141 (1.00) |
| 20P LOSS MUTATION ANALYSIS | 3 (1%) | 226 |
0.485 (1.00) |
0.545 (1.00) |
0.587 (1.00) |
1 (1.00) |
1 (1.00) |
|
| 21Q LOSS MUTATION ANALYSIS | 19 (8%) | 210 |
0.318 (1.00) |
0.429 (1.00) |
0.238 (1.00) |
0.257 (1.00) |
0.0193 (1.00) |
0.625 (1.00) |
| 22Q LOSS MUTATION ANALYSIS | 25 (11%) | 204 |
0.0462 (1.00) |
0.0478 (1.00) |
0.0179 (1.00) |
0.208 (1.00) |
0.0196 (1.00) |
0.83 (1.00) |
| XQ LOSS MUTATION ANALYSIS | 31 (14%) | 198 |
0.141 (1.00) |
0.411 (1.00) |
1 (1.00) |
0.998 (1.00) |
0.298 (1.00) |
1 (1.00) |
P value = 0.000245 (logrank test), Q value = 0.1
Table S1. Gene #6: '3Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 229 | 55 | 0.0 - 211.2 (14.5) |
| 3Q GAIN MUTATED | 7 | 4 | 0.2 - 41.1 (8.8) |
| 3Q GAIN WILD-TYPE | 222 | 51 | 0.0 - 211.2 (14.5) |
Figure S1. Get High-res Image Gene #6: '3Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
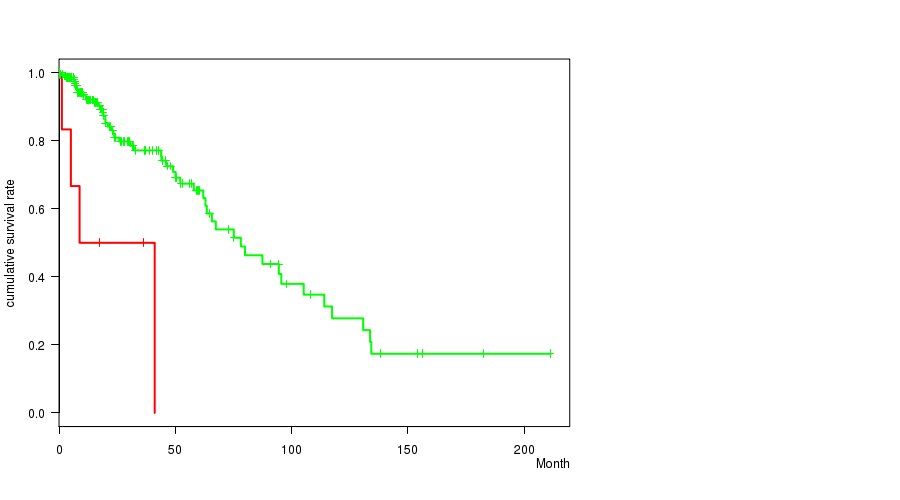
P value = 8.53e-05 (t-test), Q value = 0.035
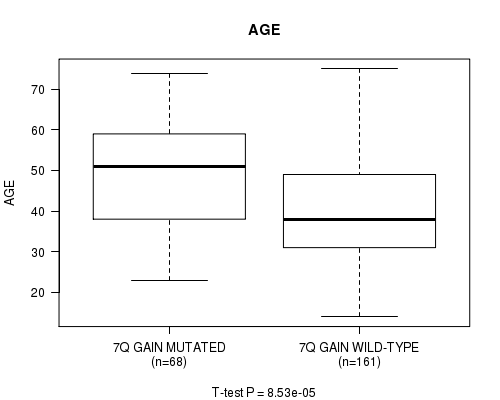
Table S2. Gene #10: '7Q GAIN MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 229 | 42.8 (13.4) |
| 7Q GAIN MUTATED | 68 | 48.2 (13.4) |
| 7Q GAIN WILD-TYPE | 161 | 40.5 (12.7) |
Figure S2. Get High-res Image Gene #10: '7Q GAIN MUTATION STATUS' versus Clinical Feature #2: 'AGE'
P value = 5.1e-05 (logrank test), Q value = 0.021
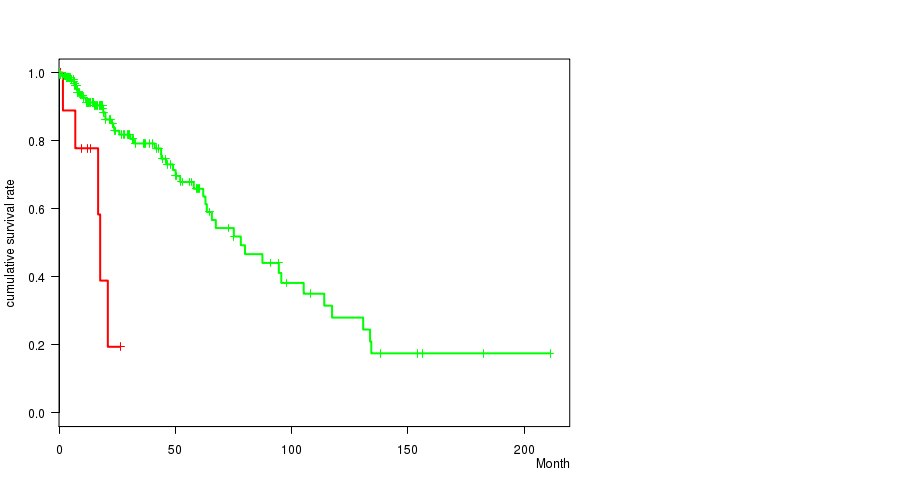
Table S3. Gene #29: '19Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 229 | 55 | 0.0 - 211.2 (14.5) |
| 19Q GAIN MUTATED | 11 | 5 | 0.5 - 26.3 (12.2) |
| 19Q GAIN WILD-TYPE | 218 | 50 | 0.0 - 211.2 (14.7) |
Figure S3. Get High-res Image Gene #29: '19Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
P value = 0.00058 (t-test), Q value = 0.23
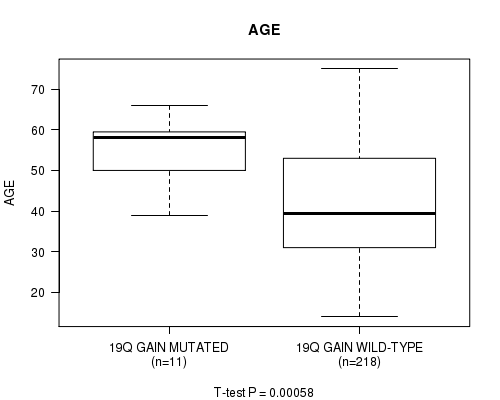
Table S4. Gene #29: '19Q GAIN MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 229 | 42.8 (13.4) |
| 19Q GAIN MUTATED | 11 | 54.4 (8.4) |
| 19Q GAIN WILD-TYPE | 218 | 42.2 (13.3) |
Figure S4. Get High-res Image Gene #29: '19Q GAIN MUTATION STATUS' versus Clinical Feature #2: 'AGE'
P value = 9.19e-07 (logrank test), Q value = 0.00038
Table S5. Gene #30: '20P GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 229 | 55 | 0.0 - 211.2 (14.5) |
| 20P GAIN MUTATED | 22 | 9 | 0.5 - 41.1 (12.2) |
| 20P GAIN WILD-TYPE | 207 | 46 | 0.0 - 211.2 (14.9) |
Figure S5. Get High-res Image Gene #30: '20P GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
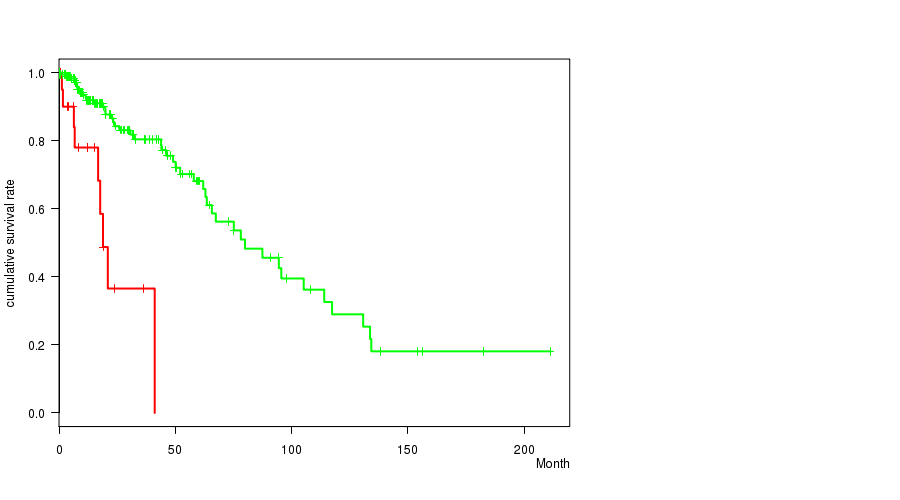
P value = 8.28e-05 (t-test), Q value = 0.034
Table S6. Gene #30: '20P GAIN MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 229 | 42.8 (13.4) |
| 20P GAIN MUTATED | 22 | 52.3 (9.9) |
| 20P GAIN WILD-TYPE | 207 | 41.8 (13.3) |
Figure S6. Get High-res Image Gene #30: '20P GAIN MUTATION STATUS' versus Clinical Feature #2: 'AGE'
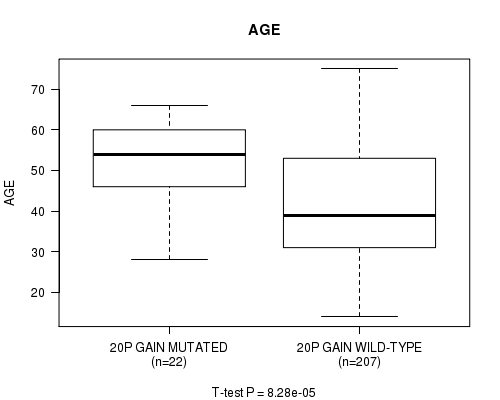
P value = 2.34e-05 (logrank test), Q value = 0.0097
Table S7. Gene #31: '20Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 229 | 55 | 0.0 - 211.2 (14.5) |
| 20Q GAIN MUTATED | 22 | 8 | 0.5 - 41.1 (12.2) |
| 20Q GAIN WILD-TYPE | 207 | 47 | 0.0 - 211.2 (14.9) |
Figure S7. Get High-res Image Gene #31: '20Q GAIN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
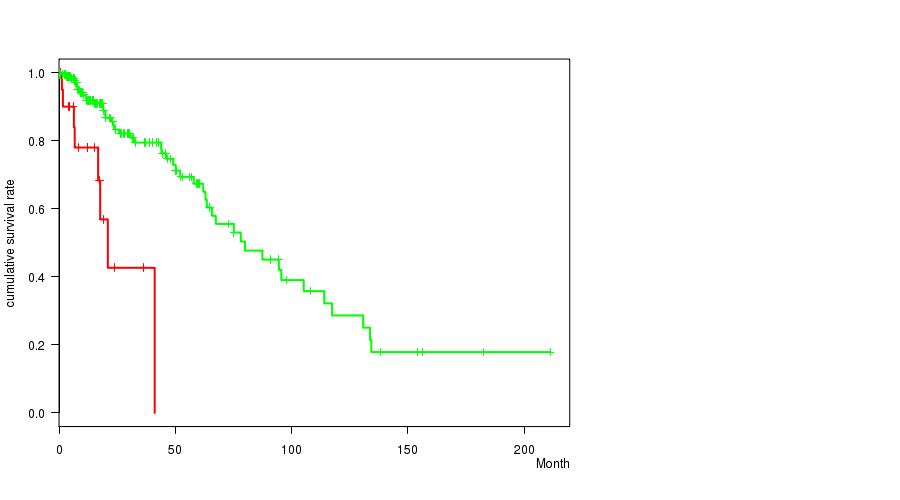
P value = 0.00025 (t-test), Q value = 0.1
Table S8. Gene #31: '20Q GAIN MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 229 | 42.8 (13.4) |
| 20Q GAIN MUTATED | 22 | 51.9 (10.5) |
| 20Q GAIN WILD-TYPE | 207 | 41.8 (13.3) |
Figure S8. Get High-res Image Gene #31: '20Q GAIN MUTATION STATUS' versus Clinical Feature #2: 'AGE'
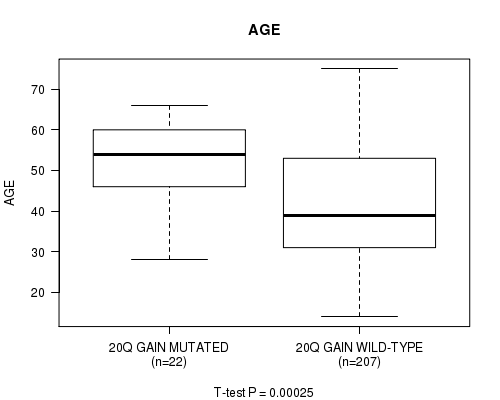
P value = 9.42e-18 (Fisher's exact test), Q value = 4e-15
Table S9. Gene #35: '1P LOSS MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL.TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 71 | 64 | 93 |
| 1P LOSS MUTATED | 5 | 10 | 61 |
| 1P LOSS WILD-TYPE | 66 | 54 | 32 |
Figure S9. Get High-res Image Gene #35: '1P LOSS MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL.TYPE'
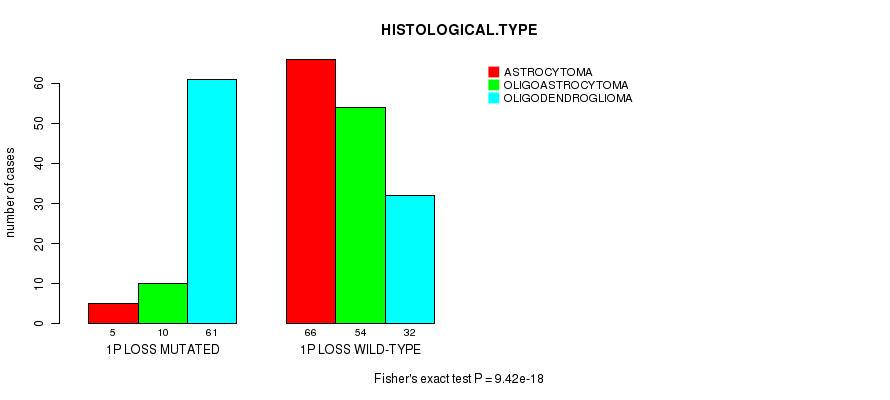
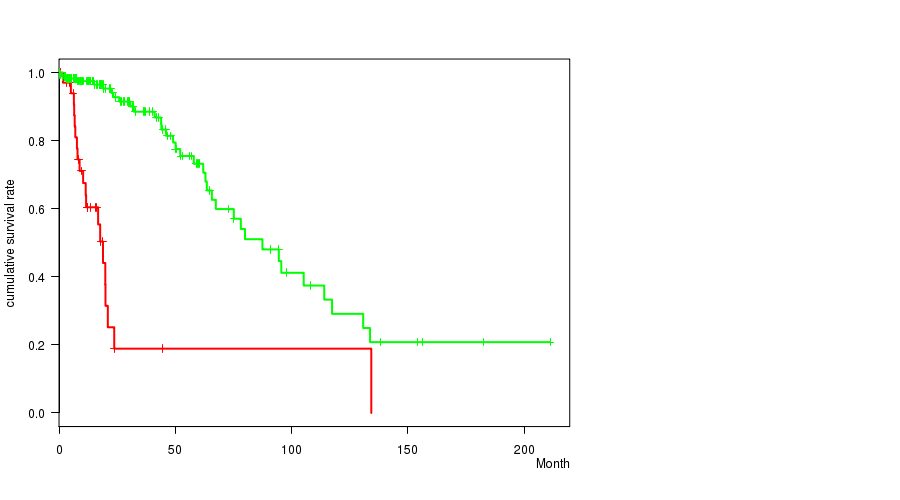
P value = 1.15e-12 (logrank test), Q value = 4.8e-10
Table S10. Gene #51: '10P LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 229 | 55 | 0.0 - 211.2 (14.5) |
| 10P LOSS MUTATED | 39 | 20 | 0.1 - 134.3 (10.4) |
| 10P LOSS WILD-TYPE | 190 | 35 | 0.0 - 211.2 (15.2) |
Figure S10. Get High-res Image Gene #51: '10P LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
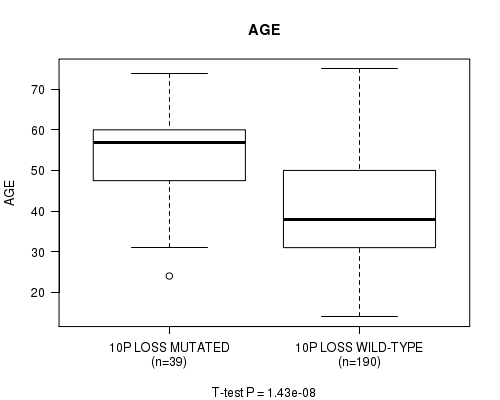
P value = 1.43e-08 (t-test), Q value = 6e-06
Table S11. Gene #51: '10P LOSS MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 229 | 42.8 (13.4) |
| 10P LOSS MUTATED | 39 | 53.7 (11.1) |
| 10P LOSS WILD-TYPE | 190 | 40.5 (12.7) |
Figure S11. Get High-res Image Gene #51: '10P LOSS MUTATION STATUS' versus Clinical Feature #2: 'AGE'
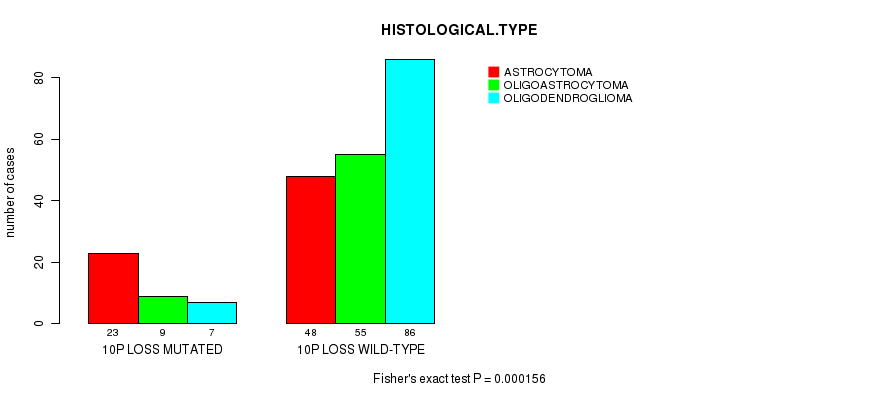
P value = 0.000156 (Fisher's exact test), Q value = 0.064
Table S12. Gene #51: '10P LOSS MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL.TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 71 | 64 | 93 |
| 10P LOSS MUTATED | 23 | 9 | 7 |
| 10P LOSS WILD-TYPE | 48 | 55 | 86 |
Figure S12. Get High-res Image Gene #51: '10P LOSS MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL.TYPE'
P value = 5.03e-06 (logrank test), Q value = 0.0021
Table S13. Gene #52: '10Q LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 229 | 55 | 0.0 - 211.2 (14.5) |
| 10Q LOSS MUTATED | 47 | 25 | 0.1 - 156.2 (12.2) |
| 10Q LOSS WILD-TYPE | 182 | 30 | 0.0 - 211.2 (14.9) |
Figure S13. Get High-res Image Gene #52: '10Q LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
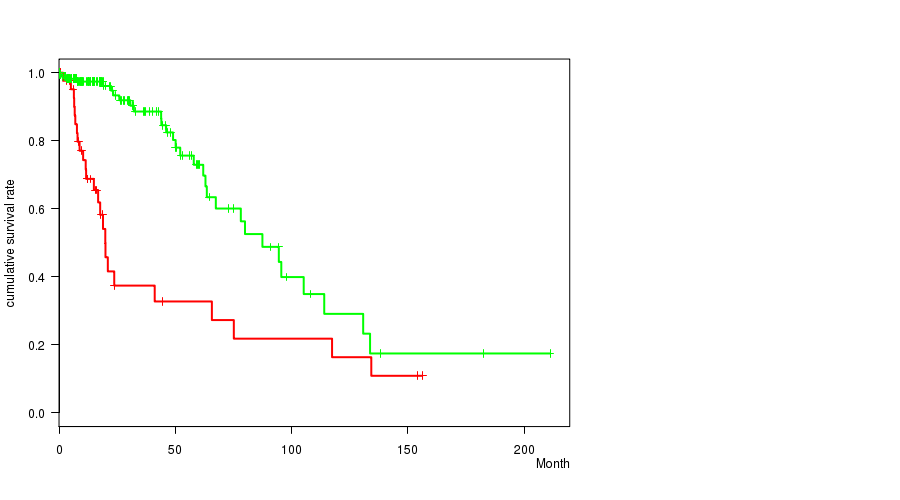
P value = 2.25e-07 (t-test), Q value = 9.4e-05
Table S14. Gene #52: '10Q LOSS MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 229 | 42.8 (13.4) |
| 10Q LOSS MUTATED | 47 | 52.0 (12.3) |
| 10Q LOSS WILD-TYPE | 182 | 40.4 (12.6) |
Figure S14. Get High-res Image Gene #52: '10Q LOSS MUTATION STATUS' versus Clinical Feature #2: 'AGE'
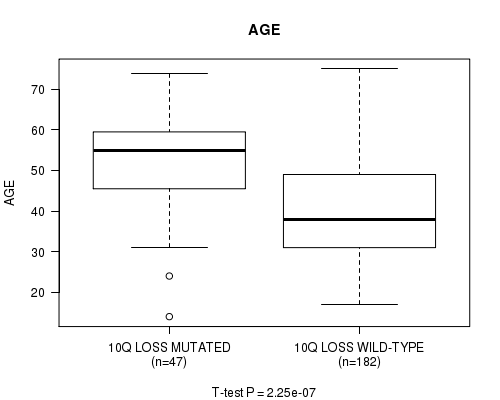
P value = 0.000297 (Fisher's exact test), Q value = 0.12
Table S15. Gene #52: '10Q LOSS MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL.TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 71 | 64 | 93 |
| 10Q LOSS MUTATED | 26 | 11 | 10 |
| 10Q LOSS WILD-TYPE | 45 | 53 | 83 |
Figure S15. Get High-res Image Gene #52: '10Q LOSS MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL.TYPE'
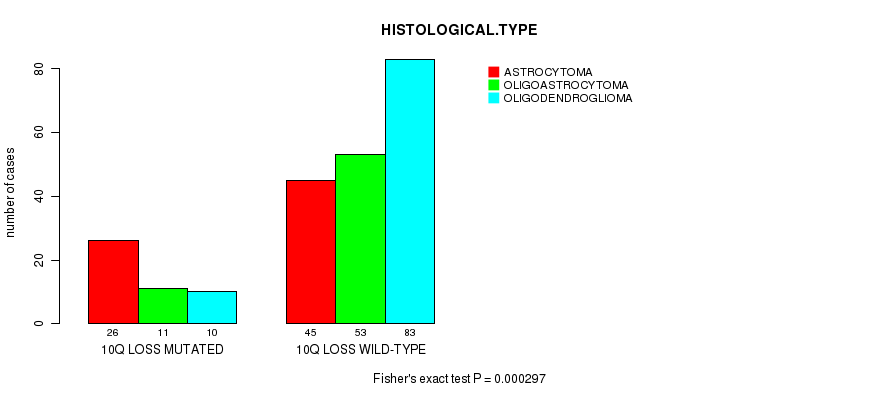
P value = 4.94e-05 (logrank test), Q value = 0.02
Table S16. Gene #54: '11Q LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 229 | 55 | 0.0 - 211.2 (14.5) |
| 11Q LOSS MUTATED | 9 | 5 | 0.2 - 41.1 (15.0) |
| 11Q LOSS WILD-TYPE | 220 | 50 | 0.0 - 211.2 (14.5) |
Figure S16. Get High-res Image Gene #54: '11Q LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
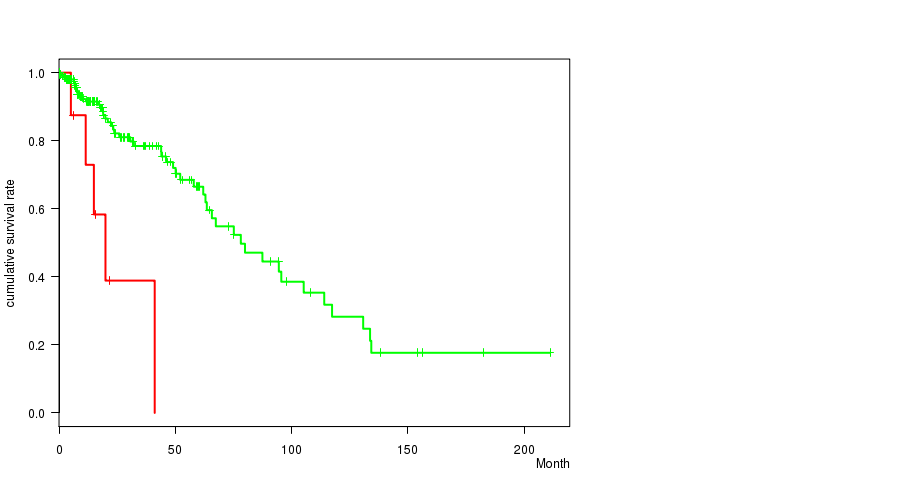
P value = 0.000276 (logrank test), Q value = 0.11
Table S17. Gene #62: '17P LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 229 | 55 | 0.0 - 211.2 (14.5) |
| 17P LOSS MUTATED | 10 | 4 | 0.1 - 23.7 (12.3) |
| 17P LOSS WILD-TYPE | 219 | 51 | 0.0 - 211.2 (14.5) |
Figure S17. Get High-res Image Gene #62: '17P LOSS MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
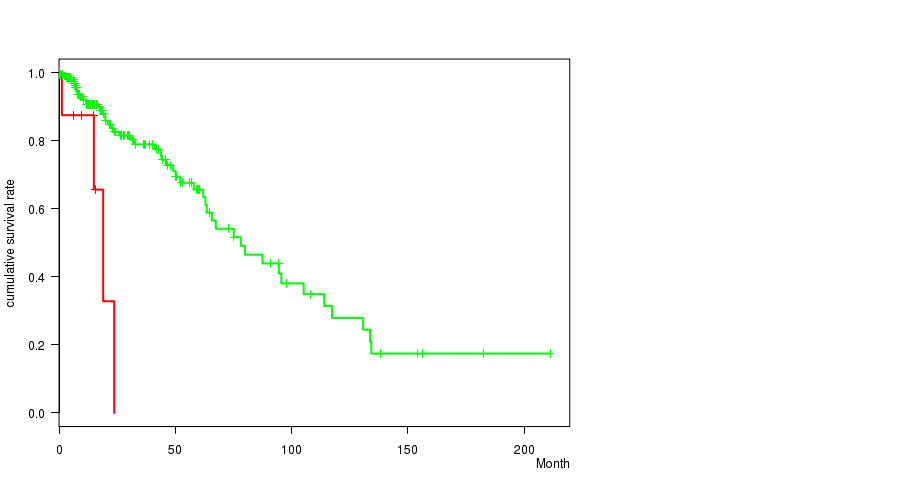
P value = 0.000572 (t-test), Q value = 0.23
Table S18. Gene #62: '17P LOSS MUTATION STATUS' versus Clinical Feature #2: 'AGE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 229 | 42.8 (13.4) |
| 17P LOSS MUTATED | 10 | 54.7 (7.9) |
| 17P LOSS WILD-TYPE | 219 | 42.2 (13.3) |
Figure S18. Get High-res Image Gene #62: '17P LOSS MUTATION STATUS' versus Clinical Feature #2: 'AGE'
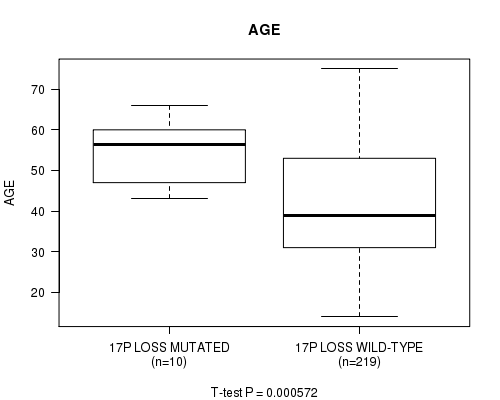
P value = 6.56e-14 (Fisher's exact test), Q value = 2.8e-11
Table S19. Gene #67: '19Q LOSS MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL.TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 71 | 64 | 93 |
| 19Q LOSS MUTATED | 10 | 14 | 63 |
| 19Q LOSS WILD-TYPE | 61 | 50 | 30 |
Figure S19. Get High-res Image Gene #67: '19Q LOSS MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL.TYPE'
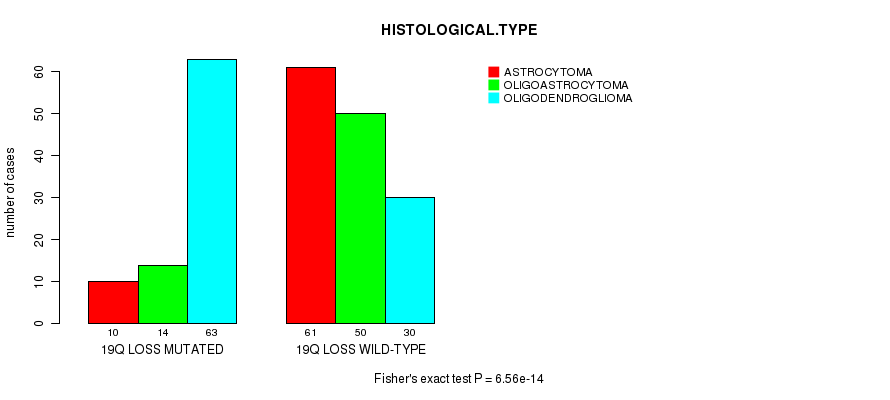
-
Copy number data file = transformed.cor.cli.txt
-
Clinical data file = LGG-TP.clin.merged.picked.txt
-
Number of patients = 229
-
Number of significantly arm-level cnvs = 71
-
Number of selected clinical features = 6
-
Exclude regions that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.