This pipeline computes the correlation between significant copy number variation (cnv focal) genes and molecular subtypes.
Testing the association between copy number variation 40 focal events and 12 molecular subtypes across 504 patients, 187 significant findings detected with P value < 0.05 and Q value < 0.25.
-
1p cnv correlated to 'CN_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
1q cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
2p cnv correlated to 'CN_CNMF', 'RPPA_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
2q cnv correlated to 'CN_CNMF' and 'RPPA_CNMF'.
-
3p cnv correlated to 'CN_CNMF', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
3q cnv correlated to 'CN_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_MATURE_CNMF'.
-
4p cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
4q cnv correlated to 'CN_CNMF'.
-
5p cnv correlated to 'CN_CNMF'.
-
6p cnv correlated to 'CN_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
6q cnv correlated to 'CN_CNMF', 'RPPA_CNMF', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
7p cnv correlated to 'CN_CNMF', 'RPPA_CNMF', and 'MRNASEQ_CNMF'.
-
7q cnv correlated to 'CN_CNMF', 'RPPA_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
8p cnv correlated to 'CN_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
8q cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
9p cnv correlated to 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
9q cnv correlated to 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
10p cnv correlated to 'CN_CNMF' and 'MIRSEQ_CHIERARCHICAL'.
-
10q cnv correlated to 'CN_CNMF'.
-
11p cnv correlated to 'CN_CNMF'.
-
11q cnv correlated to 'CN_CNMF'.
-
12p cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_MATURE_CNMF'.
-
12q cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
13q cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
14q cnv correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
15q cnv correlated to 'CN_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
16p cnv correlated to 'CN_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
16q cnv correlated to 'CN_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
17p cnv correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
17q cnv correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
18p cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
18q cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
19p cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', and 'MIRSEQ_CNMF'.
-
19q cnv correlated to 'CN_CNMF' and 'MIRSEQ_CNMF'.
-
20p cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
20q cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
21q cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
22q cnv correlated to 'MRNA_CHIERARCHICAL', 'CN_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_MATURE_CNMF'.
-
xq cnv correlated to 'CN_CNMF' and 'RPPA_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 40 focal events and 12 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 187 significant findings detected.
|
Clinical Features |
MRNA CNMF |
MRNA CHIERARCHICAL |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 9q | 159 (32%) | 345 |
0.00628 (1.00) |
0.000826 (0.244) |
1.48e-42 (7.09e-40) |
5.8e-18 (2.71e-15) |
5.31e-07 (0.00021) |
0.000429 (0.133) |
6.51e-22 (3.09e-19) |
7.16e-23 (3.42e-20) |
1.15e-15 (5.31e-13) |
3.6e-05 (0.0127) |
1.47e-08 (5.97e-06) |
3.44e-06 (0.00131) |
| 14q | 227 (45%) | 277 |
4.78e-05 (0.0166) |
4.72e-05 (0.0165) |
1.14e-19 (5.37e-17) |
7.43e-09 (3.03e-06) |
3.64e-13 (1.62e-10) |
1.79e-07 (7.15e-05) |
5.27e-18 (2.48e-15) |
1.69e-15 (7.81e-13) |
1.24e-14 (5.67e-12) |
0.0676 (1.00) |
0.0003 (0.0955) |
0.000568 (0.174) |
| 9p | 158 (31%) | 346 |
0.00378 (0.923) |
0.00044 (0.136) |
6.08e-41 (2.91e-38) |
3.82e-17 (1.78e-14) |
2.54e-05 (0.00908) |
0.0017 (0.453) |
6.25e-20 (2.95e-17) |
5.43e-21 (2.57e-18) |
8.53e-13 (3.76e-10) |
0.00011 (0.0363) |
3.3e-08 (1.33e-05) |
0.000175 (0.0566) |
| 8q | 124 (25%) | 380 |
0.172 (1.00) |
0.218 (1.00) |
4.83e-15 (2.23e-12) |
9.92e-05 (0.0329) |
1.19e-05 (0.00441) |
0.0244 (1.00) |
1.03e-05 (0.00386) |
8.99e-05 (0.0299) |
5.15e-06 (0.00195) |
0.0259 (1.00) |
1.58e-05 (0.00575) |
6.96e-05 (0.0236) |
| 12q | 123 (24%) | 381 |
0.0129 (1.00) |
0.0142 (1.00) |
6.11e-14 (2.78e-11) |
6.02e-05 (0.0206) |
1.87e-13 (8.38e-11) |
0.00162 (0.435) |
3.61e-11 (1.55e-08) |
3.61e-11 (1.55e-08) |
1.21e-09 (5e-07) |
0.05 (1.00) |
2.52e-07 (1e-04) |
0.000654 (0.198) |
| 17p | 70 (14%) | 434 |
8.7e-06 (0.00327) |
3.14e-05 (0.0111) |
5.44e-15 (2.5e-12) |
0.145 (1.00) |
0.0018 (0.476) |
0.00033 (0.104) |
1.02e-06 (0.000396) |
1.58e-08 (6.39e-06) |
3.71e-08 (1.5e-05) |
6.44e-11 (2.74e-08) |
0.0127 (1.00) |
0.0428 (1.00) |
| 18p | 116 (23%) | 388 |
0.0129 (1.00) |
0.0376 (1.00) |
1.32e-13 (5.92e-11) |
0.000198 (0.0636) |
1.12e-05 (0.00417) |
0.0471 (1.00) |
1.82e-14 (8.32e-12) |
4.32e-12 (1.88e-09) |
1.84e-09 (7.6e-07) |
9.79e-07 (0.000383) |
3.35e-05 (0.0118) |
0.0173 (1.00) |
| 18q | 118 (23%) | 386 |
0.0136 (1.00) |
0.033 (1.00) |
1.19e-13 (5.36e-11) |
0.000451 (0.139) |
9.47e-06 (0.00354) |
0.061 (1.00) |
8.15e-14 (3.68e-11) |
6.66e-12 (2.88e-09) |
2.43e-09 (1e-06) |
4.95e-07 (0.000196) |
1.59e-05 (0.00578) |
0.00538 (1.00) |
| 20p | 118 (23%) | 386 |
0.0457 (1.00) |
0.0675 (1.00) |
6.46e-22 (3.07e-19) |
6.56e-05 (0.0224) |
6.31e-09 (2.58e-06) |
0.002 (0.517) |
9.78e-15 (4.49e-12) |
8.68e-13 (3.82e-10) |
1.7e-12 (7.48e-10) |
3.4e-05 (0.012) |
1.29e-07 (5.16e-05) |
0.00112 (0.314) |
| 20q | 116 (23%) | 388 |
0.024 (1.00) |
0.033 (1.00) |
5.26e-24 (2.51e-21) |
5.62e-05 (0.0194) |
1.75e-09 (7.23e-07) |
0.00134 (0.367) |
5.17e-16 (2.4e-13) |
2.04e-13 (9.1e-11) |
1.77e-12 (7.73e-10) |
3.04e-05 (0.0108) |
1.29e-07 (5.16e-05) |
0.00112 (0.314) |
| 6p | 110 (22%) | 394 |
0.00526 (1.00) |
0.00159 (0.427) |
5.28e-18 (2.48e-15) |
0.00157 (0.423) |
1.07e-06 (0.000417) |
0.000946 (0.273) |
1.78e-06 (0.000688) |
3.47e-06 (0.00132) |
9.15e-10 (3.81e-07) |
2.38e-05 (0.00853) |
7.8e-07 (0.000306) |
0.00329 (0.809) |
| 12p | 121 (24%) | 383 |
0.0129 (1.00) |
0.0142 (1.00) |
1.66e-13 (7.44e-11) |
0.000101 (0.0336) |
3.83e-12 (1.67e-09) |
0.0031 (0.766) |
5.61e-11 (2.4e-08) |
1.54e-10 (6.52e-08) |
5.93e-09 (2.43e-06) |
0.0455 (1.00) |
6.65e-07 (0.000262) |
0.00128 (0.352) |
| 13q | 91 (18%) | 413 |
0.0129 (1.00) |
0.0213 (1.00) |
4.88e-10 (2.04e-07) |
0.000741 (0.221) |
5.85e-05 (0.0201) |
0.047 (1.00) |
2.41e-05 (0.00863) |
3.13e-06 (0.0012) |
1.1e-05 (0.00408) |
0.000303 (0.096) |
0.0409 (1.00) |
0.0431 (1.00) |
| 17q | 58 (12%) | 446 |
8.36e-05 (0.028) |
0.000175 (0.0566) |
1.32e-10 (5.6e-08) |
0.241 (1.00) |
0.13 (1.00) |
0.00614 (1.00) |
8.28e-05 (0.0278) |
9.69e-07 (0.00038) |
1.28e-05 (0.00471) |
1.9e-10 (8.04e-08) |
0.018 (1.00) |
0.0304 (1.00) |
| 21q | 105 (21%) | 399 |
0.116 (1.00) |
0.163 (1.00) |
2.08e-11 (8.93e-09) |
8.91e-05 (0.0298) |
0.000125 (0.041) |
0.00632 (1.00) |
0.000154 (0.0501) |
2.21e-05 (0.00798) |
0.000114 (0.0374) |
0.00108 (0.306) |
0.0213 (1.00) |
0.0596 (1.00) |
| 22q | 79 (16%) | 425 |
0.00432 (1.00) |
0.000485 (0.149) |
5.67e-14 (2.58e-11) |
0.0176 (1.00) |
0.0306 (1.00) |
0.158 (1.00) |
9.26e-06 (0.00347) |
1.52e-05 (0.00555) |
2.35e-06 (0.000906) |
0.0125 (1.00) |
0.000659 (0.198) |
0.141 (1.00) |
| 3q | 158 (31%) | 346 |
0.236 (1.00) |
0.251 (1.00) |
5.98e-10 (2.49e-07) |
0.0104 (1.00) |
0.0348 (1.00) |
0.0967 (1.00) |
1.22e-05 (0.0045) |
8.44e-06 (0.00318) |
1.59e-05 (0.00578) |
0.0294 (1.00) |
0.000598 (0.182) |
0.0965 (1.00) |
| 6q | 135 (27%) | 369 |
0.0194 (1.00) |
0.0107 (1.00) |
1.01e-17 (4.69e-15) |
0.0476 (1.00) |
2.1e-05 (0.00762) |
0.00917 (1.00) |
0.00101 (0.288) |
0.00306 (0.76) |
4.41e-07 (0.000175) |
0.000418 (0.131) |
5.88e-05 (0.0202) |
0.0294 (1.00) |
| 7q | 172 (34%) | 332 |
0.416 (1.00) |
0.577 (1.00) |
2.11e-13 (9.4e-11) |
0.00729 (1.00) |
4.52e-06 (0.00172) |
0.000615 (0.186) |
0.000108 (0.0357) |
0.000597 (0.182) |
0.00209 (0.537) |
0.0463 (1.00) |
0.0358 (1.00) |
0.163 (1.00) |
| 8p | 165 (33%) | 339 |
0.144 (1.00) |
0.224 (1.00) |
8.91e-12 (3.84e-09) |
0.00124 (0.344) |
0.00183 (0.482) |
0.0763 (1.00) |
0.000424 (0.132) |
0.000826 (0.244) |
1.36e-05 (0.00498) |
0.0406 (1.00) |
0.00185 (0.484) |
0.0078 (1.00) |
| 19p | 61 (12%) | 443 |
0.433 (1.00) |
0.486 (1.00) |
3.73e-13 (1.65e-10) |
0.000771 (0.229) |
0.00594 (1.00) |
0.00168 (0.449) |
0.000682 (0.204) |
0.00135 (0.37) |
4.75e-05 (0.0165) |
0.00305 (0.76) |
0.00151 (0.41) |
0.353 (1.00) |
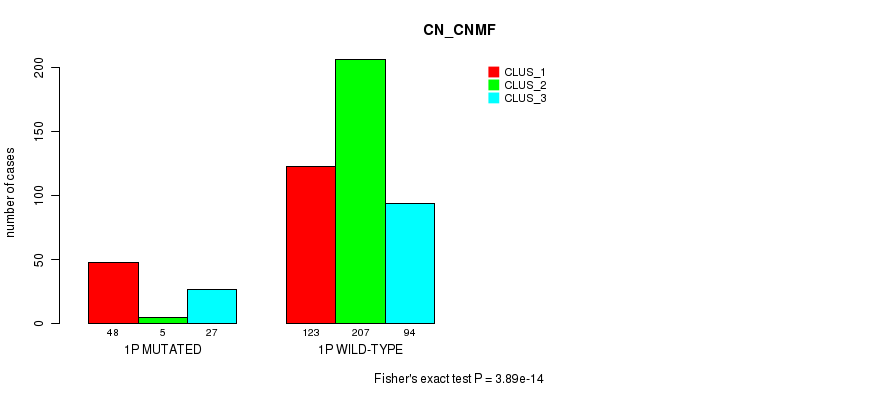
| 1p | 80 (16%) | 424 |
0.0145 (1.00) |
0.00641 (1.00) |
3.89e-14 (1.78e-11) |
0.125 (1.00) |
0.00878 (1.00) |
0.00814 (1.00) |
0.0515 (1.00) |
0.00142 (0.385) |
0.000929 (0.269) |
1.08e-09 (4.48e-07) |
0.00931 (1.00) |
6.92e-05 (0.0235) |
| 1q | 87 (17%) | 417 |
0.0837 (1.00) |
0.126 (1.00) |
9.8e-16 (4.55e-13) |
0.0846 (1.00) |
0.0349 (1.00) |
0.00939 (1.00) |
0.0194 (1.00) |
0.00937 (1.00) |
4.08e-05 (0.0143) |
1.32e-06 (0.000513) |
0.0184 (1.00) |
0.0166 (1.00) |
| 2p | 89 (18%) | 415 |
0.0326 (1.00) |
0.0513 (1.00) |
4.65e-10 (1.95e-07) |
0.369 (1.00) |
0.000178 (0.0573) |
0.0549 (1.00) |
0.473 (1.00) |
0.378 (1.00) |
0.06 (1.00) |
0.000369 (0.116) |
0.00531 (1.00) |
0.231 (1.00) |
| 3p | 382 (76%) | 122 |
0.00933 (1.00) |
0.00108 (0.306) |
8.43e-22 (3.99e-19) |
0.0382 (1.00) |
0.116 (1.00) |
0.0593 (1.00) |
5.96e-06 (0.00225) |
1.16e-08 (4.73e-06) |
0.302 (1.00) |
0.0372 (1.00) |
0.028 (1.00) |
0.00434 (1.00) |
| 7p | 170 (34%) | 334 |
0.416 (1.00) |
0.577 (1.00) |
7.13e-14 (3.23e-11) |
0.00453 (1.00) |
3.22e-06 (0.00124) |
0.00189 (0.491) |
0.000147 (0.0479) |
0.000893 (0.262) |
0.00249 (0.629) |
0.0902 (1.00) |
0.00914 (1.00) |
0.244 (1.00) |
| 2q | 90 (18%) | 414 |
0.106 (1.00) |
0.167 (1.00) |
4.86e-08 (1.95e-05) |
0.413 (1.00) |
0.000326 (0.103) |
0.0792 (1.00) |
0.65 (1.00) |
0.604 (1.00) |
0.0939 (1.00) |
0.00133 (0.367) |
0.0169 (1.00) |
0.161 (1.00) |
| 4p | 84 (17%) | 420 |
0.82 (1.00) |
0.819 (1.00) |
2.41e-10 (1.01e-07) |
0.000658 (0.198) |
0.0251 (1.00) |
0.0773 (1.00) |
0.0149 (1.00) |
0.0908 (1.00) |
0.0957 (1.00) |
0.0493 (1.00) |
0.0447 (1.00) |
0.453 (1.00) |
| 10p | 82 (16%) | 422 |
0.0649 (1.00) |
0.1 (1.00) |
2.02e-12 (8.82e-10) |
0.0389 (1.00) |
0.583 (1.00) |
0.161 (1.00) |
0.00208 (0.537) |
0.0169 (1.00) |
0.00288 (0.722) |
0.000829 (0.244) |
0.021 (1.00) |
0.173 (1.00) |
| 15q | 61 (12%) | 443 |
0.133 (1.00) |
0.143 (1.00) |
5.02e-12 (2.17e-09) |
0.0386 (1.00) |
0.0153 (1.00) |
0.0855 (1.00) |
0.00111 (0.312) |
0.000472 (0.145) |
0.00712 (1.00) |
0.0122 (1.00) |
0.053 (1.00) |
0.37 (1.00) |
| 16p | 115 (23%) | 389 |
0.149 (1.00) |
0.216 (1.00) |
7.59e-05 (0.0257) |
0.299 (1.00) |
0.00785 (1.00) |
0.0377 (1.00) |
0.00601 (1.00) |
0.000225 (0.0719) |
0.0129 (1.00) |
0.193 (1.00) |
0.113 (1.00) |
0.253 (1.00) |
| 16q | 112 (22%) | 392 |
0.282 (1.00) |
0.12 (1.00) |
2.99e-06 (0.00115) |
0.0884 (1.00) |
0.0471 (1.00) |
0.0907 (1.00) |
0.00371 (0.908) |
0.000244 (0.0779) |
0.00105 (0.299) |
0.134 (1.00) |
0.0341 (1.00) |
0.0975 (1.00) |
| 19q | 60 (12%) | 444 |
0.18 (1.00) |
0.302 (1.00) |
2.75e-13 (1.22e-10) |
0.000916 (0.267) |
0.00228 (0.579) |
0.00254 (0.64) |
0.00105 (0.299) |
0.00301 (0.752) |
4.89e-05 (0.0169) |
0.000965 (0.277) |
0.00177 (0.468) |
0.471 (1.00) |
| xq | 75 (15%) | 429 |
0.0775 (1.00) |
0.0868 (1.00) |
7.68e-05 (0.0259) |
0.0197 (1.00) |
0.00121 (0.337) |
2.25e-05 (0.00808) |
0.015 (1.00) |
0.00713 (1.00) |
0.0178 (1.00) |
0.00093 (0.269) |
0.0155 (1.00) |
0.0021 (0.537) |
| 4q | 78 (15%) | 426 |
0.544 (1.00) |
0.554 (1.00) |
8.35e-13 (3.69e-10) |
0.000915 (0.267) |
0.00924 (1.00) |
0.0439 (1.00) |
0.00218 (0.555) |
0.0106 (1.00) |
0.0167 (1.00) |
0.0515 (1.00) |
0.0163 (1.00) |
0.235 (1.00) |
| 5p | 192 (38%) | 312 |
0.924 (1.00) |
0.878 (1.00) |
3.75e-05 (0.0132) |
0.0957 (1.00) |
0.0677 (1.00) |
0.0967 (1.00) |
0.036 (1.00) |
0.00186 (0.486) |
0.0765 (1.00) |
0.921 (1.00) |
0.172 (1.00) |
0.94 (1.00) |
| 10q | 98 (19%) | 406 |
0.141 (1.00) |
0.201 (1.00) |
4.45e-12 (1.93e-09) |
0.0102 (1.00) |
0.969 (1.00) |
0.0801 (1.00) |
0.00631 (1.00) |
0.032 (1.00) |
0.0082 (1.00) |
0.007 (1.00) |
0.021 (1.00) |
0.0732 (1.00) |
| 11p | 48 (10%) | 456 |
0.086 (1.00) |
0.126 (1.00) |
9.37e-11 (3.98e-08) |
0.0848 (1.00) |
0.0387 (1.00) |
0.15 (1.00) |
0.0464 (1.00) |
0.0507 (1.00) |
0.0785 (1.00) |
0.00713 (1.00) |
0.0419 (1.00) |
0.0379 (1.00) |
| 11q | 51 (10%) | 453 |
0.173 (1.00) |
0.239 (1.00) |
2.9e-10 (1.22e-07) |
0.103 (1.00) |
0.0532 (1.00) |
0.305 (1.00) |
0.0433 (1.00) |
0.045 (1.00) |
0.0699 (1.00) |
0.0141 (1.00) |
0.0419 (1.00) |
0.074 (1.00) |
| 5q | 205 (41%) | 299 |
0.395 (1.00) |
0.291 (1.00) |
0.00421 (1.00) |
0.241 (1.00) |
0.271 (1.00) |
0.0864 (1.00) |
0.264 (1.00) |
0.0157 (1.00) |
0.638 (1.00) |
0.918 (1.00) |
0.587 (1.00) |
0.868 (1.00) |
P value = 3.89e-14 (Fisher's exact test), Q value = 1.8e-11
Table S1. Gene #1: '1p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 1P MUTATED | 48 | 5 | 27 |
| 1P WILD-TYPE | 123 | 207 | 94 |
Figure S1. Get High-res Image Gene #1: '1p' versus Molecular Subtype #3: 'CN_CNMF'
P value = 1.08e-09 (Fisher's exact test), Q value = 4.5e-07
Table S2. Gene #1: '1p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 1P MUTATED | 30 | 23 | 27 |
| 1P WILD-TYPE | 234 | 17 | 155 |
Figure S2. Get High-res Image Gene #1: '1p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
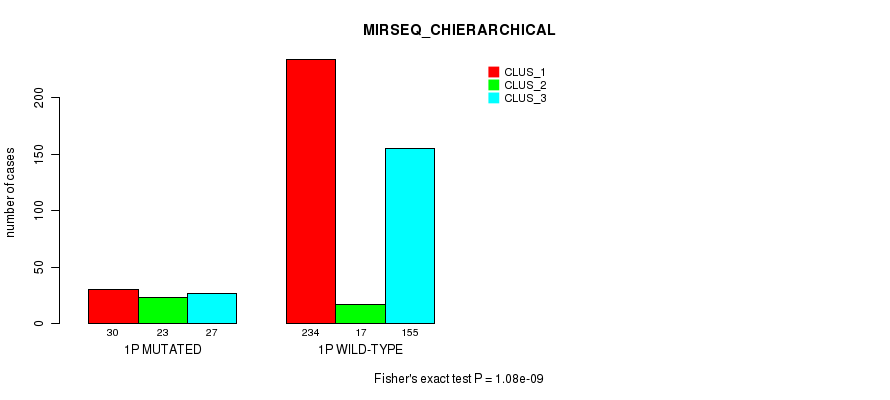
P value = 6.92e-05 (Fisher's exact test), Q value = 0.024
Table S3. Gene #1: '1p' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 86 | 69 | 71 |
| 1P MUTATED | 3 | 18 | 15 |
| 1P WILD-TYPE | 83 | 51 | 56 |
Figure S3. Get High-res Image Gene #1: '1p' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
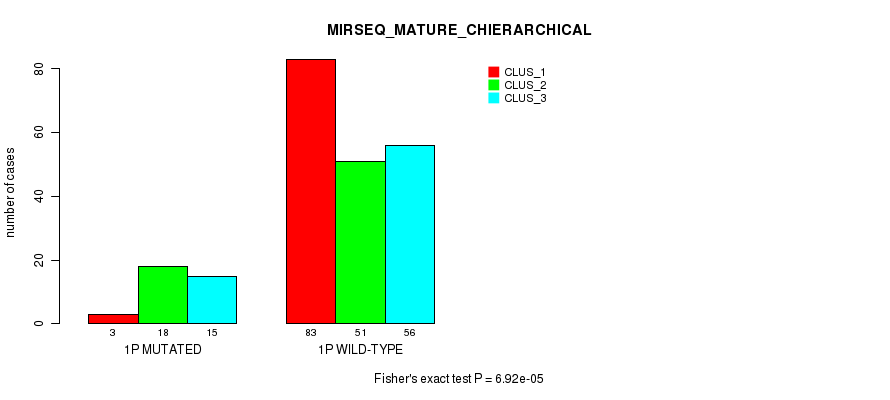
P value = 9.8e-16 (Fisher's exact test), Q value = 4.5e-13
Table S4. Gene #2: '1q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 1Q MUTATED | 56 | 6 | 25 |
| 1Q WILD-TYPE | 115 | 206 | 96 |
Figure S4. Get High-res Image Gene #2: '1q' versus Molecular Subtype #3: 'CN_CNMF'
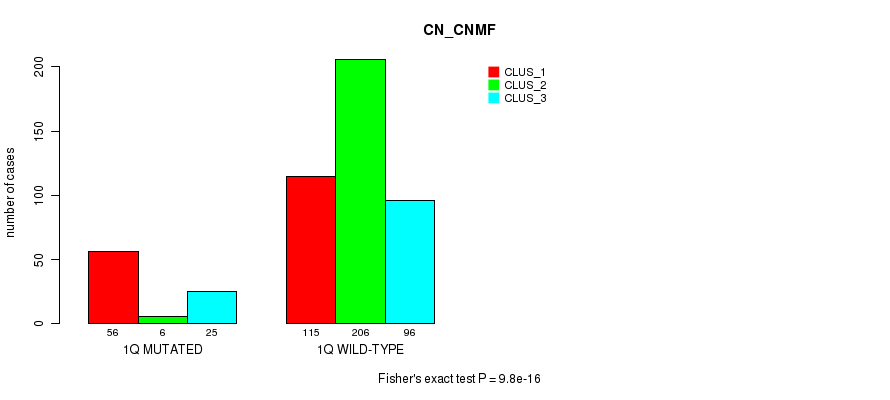
P value = 4.08e-05 (Fisher's exact test), Q value = 0.014
Table S5. Gene #2: '1q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 1Q MUTATED | 12 | 26 | 47 |
| 1Q WILD-TYPE | 106 | 177 | 118 |
Figure S5. Get High-res Image Gene #2: '1q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
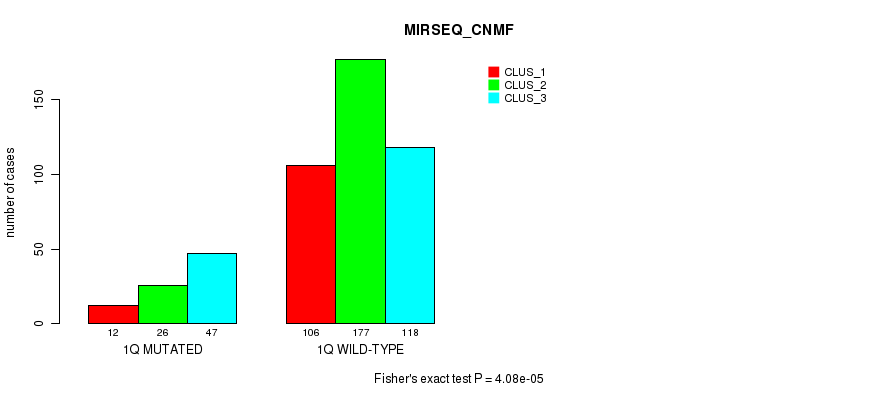
P value = 1.32e-06 (Fisher's exact test), Q value = 0.00051
Table S6. Gene #2: '1q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 1Q MUTATED | 34 | 20 | 31 |
| 1Q WILD-TYPE | 230 | 20 | 151 |
Figure S6. Get High-res Image Gene #2: '1q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
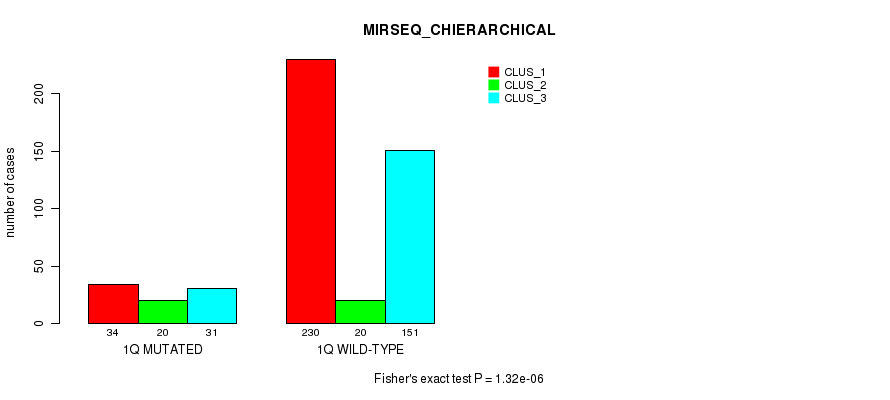
P value = 4.65e-10 (Fisher's exact test), Q value = 1.9e-07
Table S7. Gene #3: '2p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 2P MUTATED | 51 | 12 | 26 |
| 2P WILD-TYPE | 120 | 200 | 95 |
Figure S7. Get High-res Image Gene #3: '2p' versus Molecular Subtype #3: 'CN_CNMF'
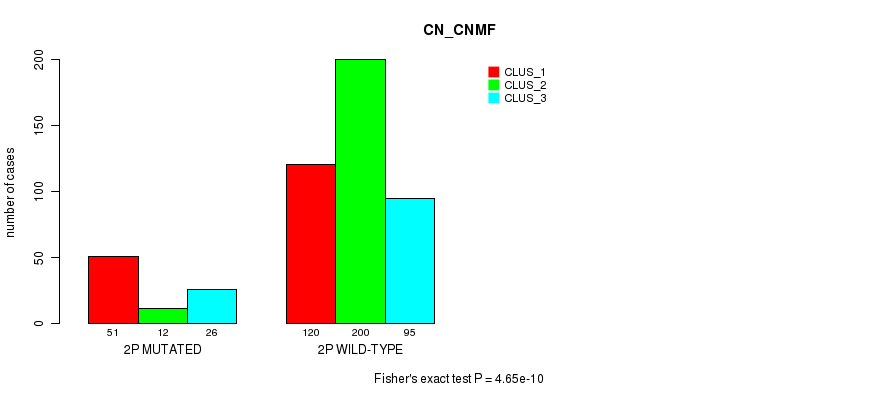
P value = 0.000178 (Chi-square test), Q value = 0.057
Table S8. Gene #3: '2p' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 2P MUTATED | 8 | 7 | 18 | 24 | 9 | 10 |
| 2P WILD-TYPE | 92 | 81 | 66 | 51 | 34 | 47 |
Figure S8. Get High-res Image Gene #3: '2p' versus Molecular Subtype #5: 'RPPA_CNMF'
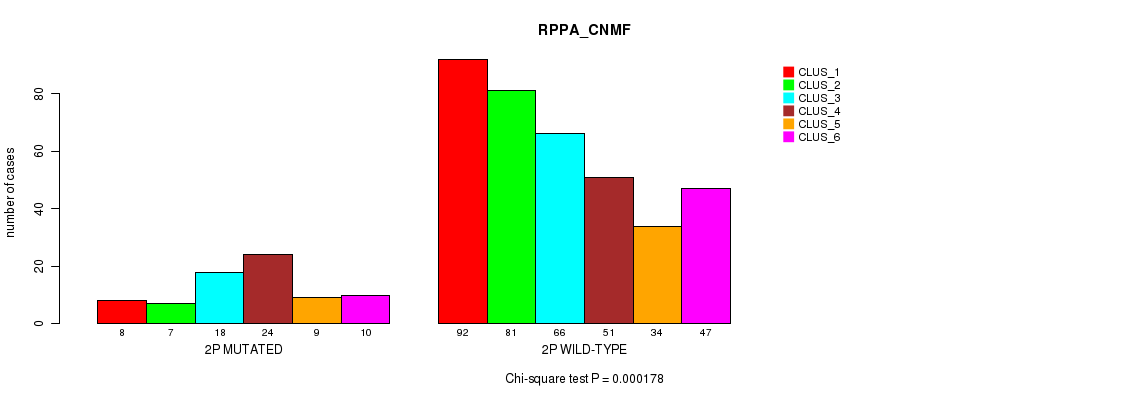
P value = 0.000369 (Fisher's exact test), Q value = 0.12
Table S9. Gene #3: '2p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 2P MUTATED | 47 | 17 | 25 |
| 2P WILD-TYPE | 217 | 23 | 157 |
Figure S9. Get High-res Image Gene #3: '2p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
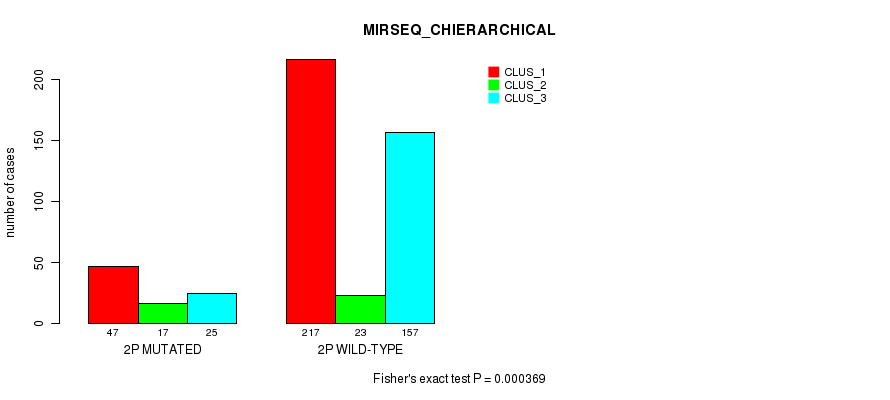
P value = 4.86e-08 (Fisher's exact test), Q value = 2e-05
Table S10. Gene #4: '2q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 2Q MUTATED | 49 | 15 | 26 |
| 2Q WILD-TYPE | 122 | 197 | 95 |
Figure S10. Get High-res Image Gene #4: '2q' versus Molecular Subtype #3: 'CN_CNMF'
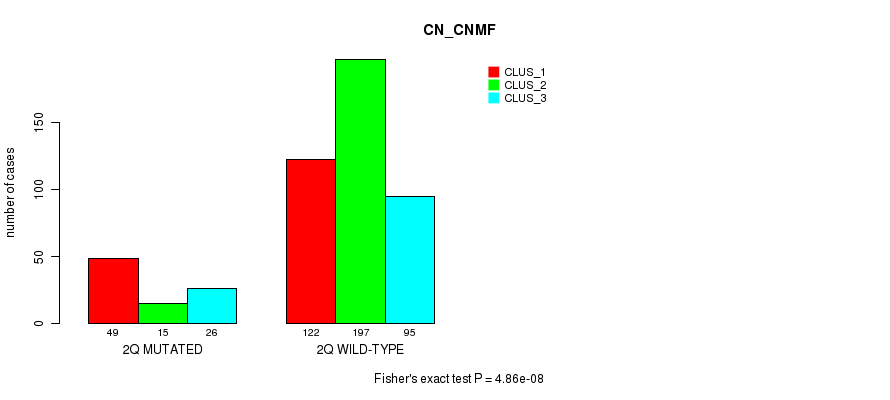
P value = 0.000326 (Chi-square test), Q value = 0.1
Table S11. Gene #4: '2q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 2Q MUTATED | 9 | 7 | 17 | 24 | 10 | 11 |
| 2Q WILD-TYPE | 91 | 81 | 67 | 51 | 33 | 46 |
Figure S11. Get High-res Image Gene #4: '2q' versus Molecular Subtype #5: 'RPPA_CNMF'
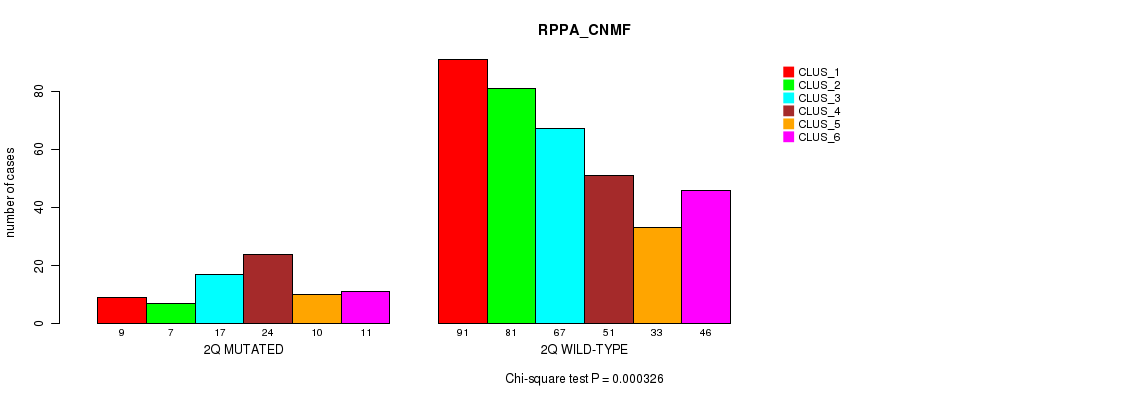
P value = 8.43e-22 (Fisher's exact test), Q value = 4e-19
Table S12. Gene #5: '3p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 3P MUTATED | 159 | 171 | 52 |
| 3P WILD-TYPE | 12 | 41 | 69 |
Figure S12. Get High-res Image Gene #5: '3p' versus Molecular Subtype #3: 'CN_CNMF'
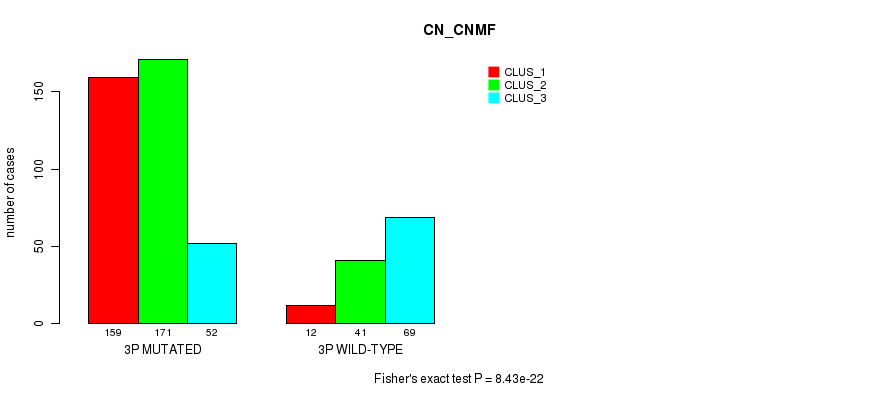
P value = 5.96e-06 (Fisher's exact test), Q value = 0.0023
Table S13. Gene #5: '3p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 3P MUTATED | 168 | 150 | 59 |
| 3P WILD-TYPE | 37 | 39 | 45 |
Figure S13. Get High-res Image Gene #5: '3p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
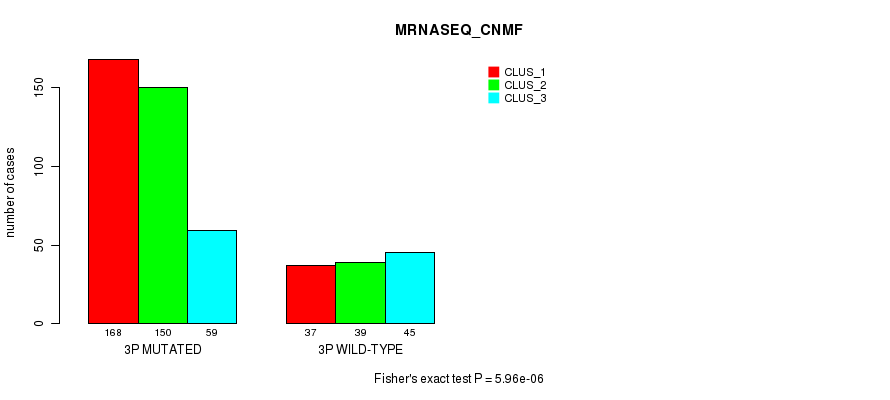
P value = 1.16e-08 (Fisher's exact test), Q value = 4.7e-06
Table S14. Gene #5: '3p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 3P MUTATED | 175 | 166 | 36 |
| 3P WILD-TYPE | 40 | 40 | 41 |
Figure S14. Get High-res Image Gene #5: '3p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
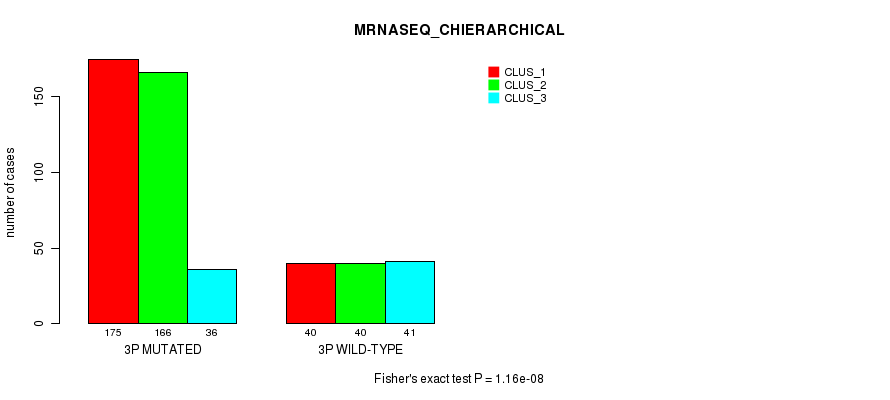
P value = 5.98e-10 (Fisher's exact test), Q value = 2.5e-07
Table S15. Gene #6: '3q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 3Q MUTATED | 86 | 43 | 29 |
| 3Q WILD-TYPE | 85 | 169 | 92 |
Figure S15. Get High-res Image Gene #6: '3q' versus Molecular Subtype #3: 'CN_CNMF'
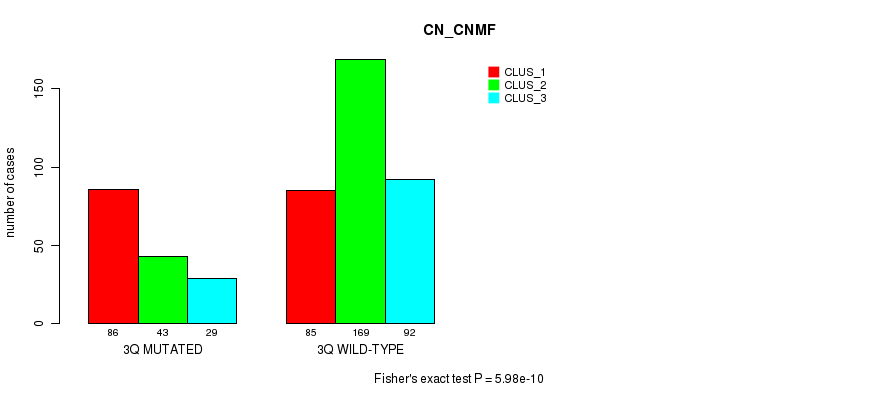
P value = 1.22e-05 (Fisher's exact test), Q value = 0.0045
Table S16. Gene #6: '3q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 3Q MUTATED | 42 | 80 | 36 |
| 3Q WILD-TYPE | 163 | 109 | 68 |
Figure S16. Get High-res Image Gene #6: '3q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
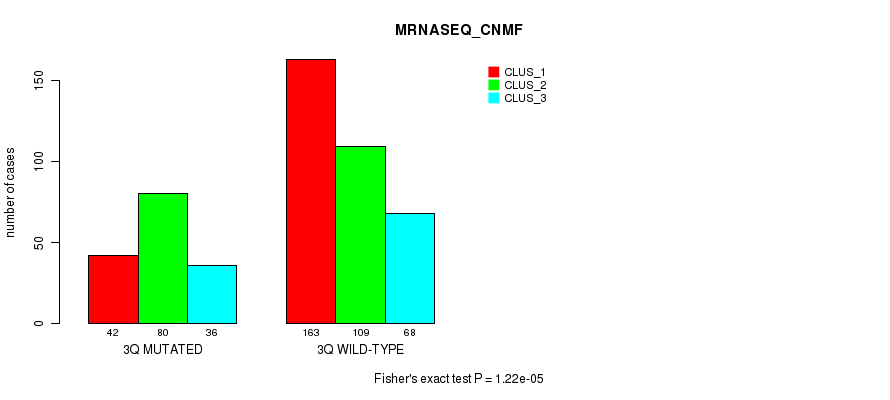
P value = 8.44e-06 (Fisher's exact test), Q value = 0.0032
Table S17. Gene #6: '3q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 3Q MUTATED | 46 | 89 | 23 |
| 3Q WILD-TYPE | 169 | 117 | 54 |
Figure S17. Get High-res Image Gene #6: '3q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
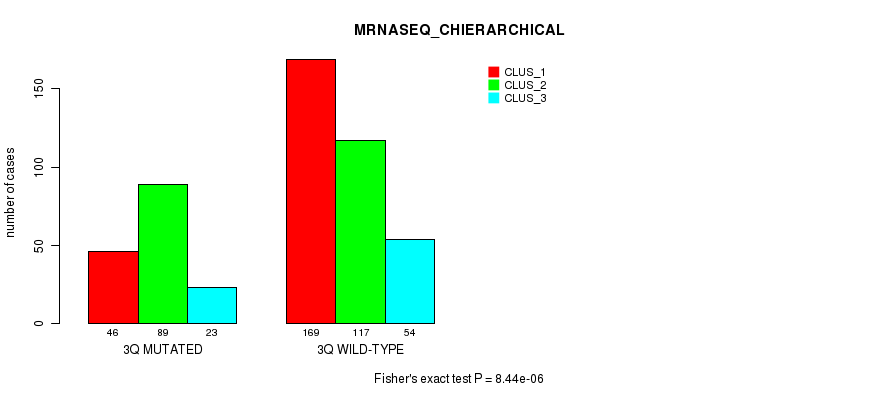
P value = 1.59e-05 (Fisher's exact test), Q value = 0.0058
Table S18. Gene #6: '3q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 3Q MUTATED | 34 | 46 | 75 |
| 3Q WILD-TYPE | 84 | 157 | 90 |
Figure S18. Get High-res Image Gene #6: '3q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
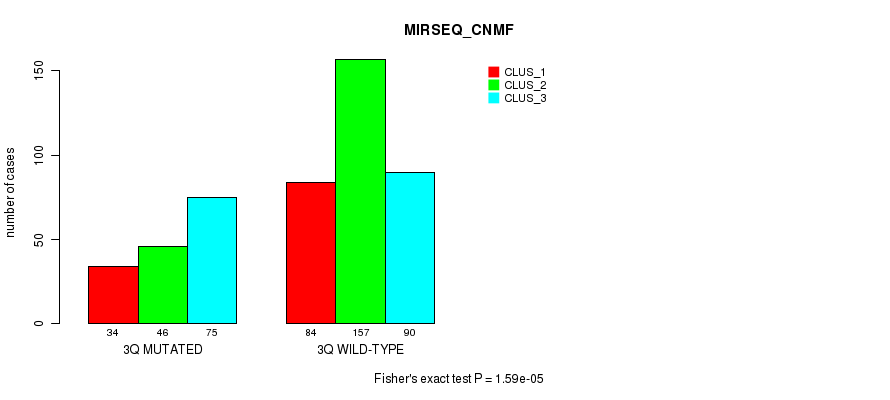
P value = 0.000598 (Fisher's exact test), Q value = 0.18
Table S19. Gene #6: '3q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 3Q MUTATED | 16 | 18 | 36 |
| 3Q WILD-TYPE | 37 | 77 | 42 |
Figure S19. Get High-res Image Gene #6: '3q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
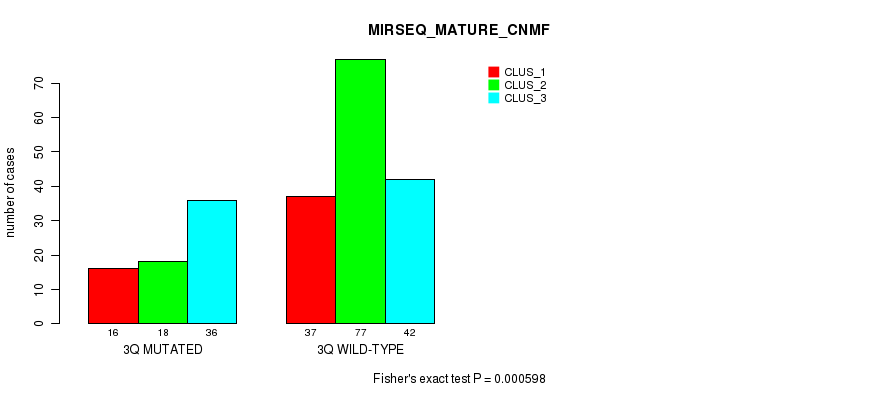
P value = 2.41e-10 (Fisher's exact test), Q value = 1e-07
Table S20. Gene #7: '4p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 4P MUTATED | 51 | 11 | 22 |
| 4P WILD-TYPE | 120 | 201 | 99 |
Figure S20. Get High-res Image Gene #7: '4p' versus Molecular Subtype #3: 'CN_CNMF'
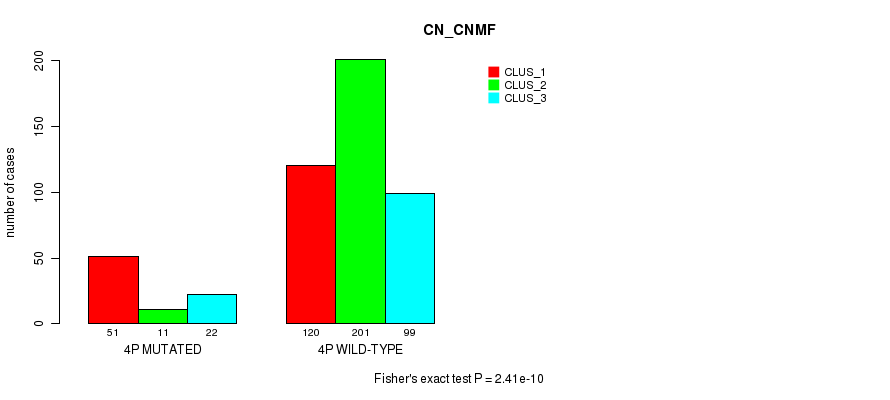
P value = 0.000658 (Fisher's exact test), Q value = 0.2
Table S21. Gene #7: '4p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 4P MUTATED | 31 | 13 | 10 |
| 4P WILD-TYPE | 69 | 101 | 66 |
Figure S21. Get High-res Image Gene #7: '4p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
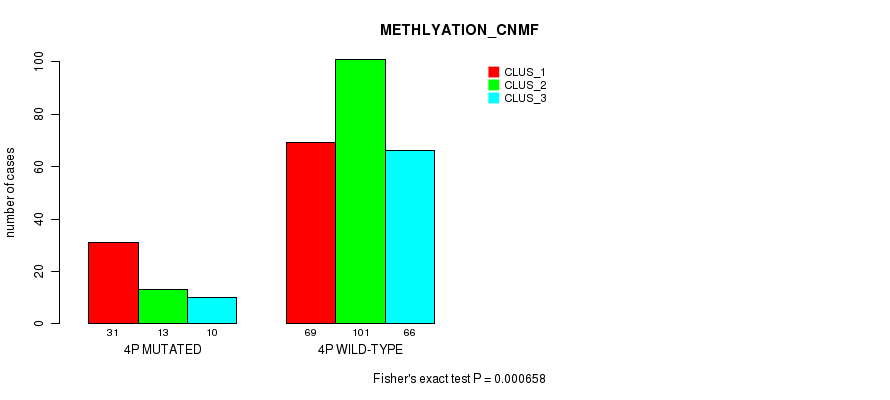
P value = 8.35e-13 (Fisher's exact test), Q value = 3.7e-10
Table S22. Gene #8: '4q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 4Q MUTATED | 51 | 7 | 20 |
| 4Q WILD-TYPE | 120 | 205 | 101 |
Figure S22. Get High-res Image Gene #8: '4q' versus Molecular Subtype #3: 'CN_CNMF'
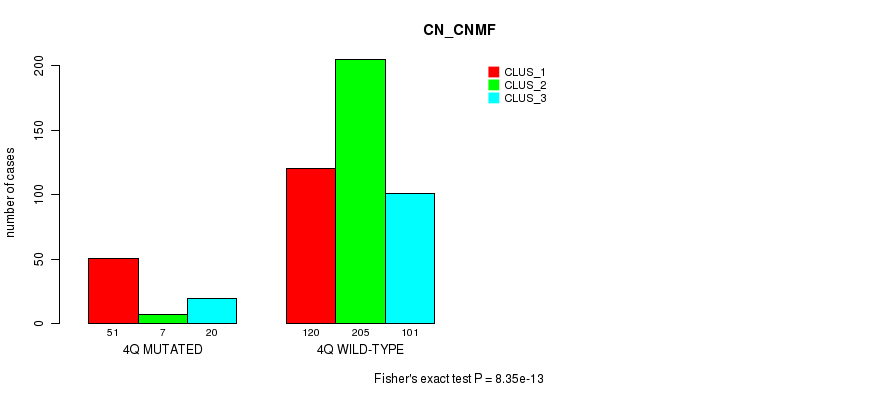
P value = 3.75e-05 (Fisher's exact test), Q value = 0.013
Table S23. Gene #9: '5p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 5P MUTATED | 86 | 59 | 47 |
| 5P WILD-TYPE | 85 | 153 | 74 |
Figure S23. Get High-res Image Gene #9: '5p' versus Molecular Subtype #3: 'CN_CNMF'
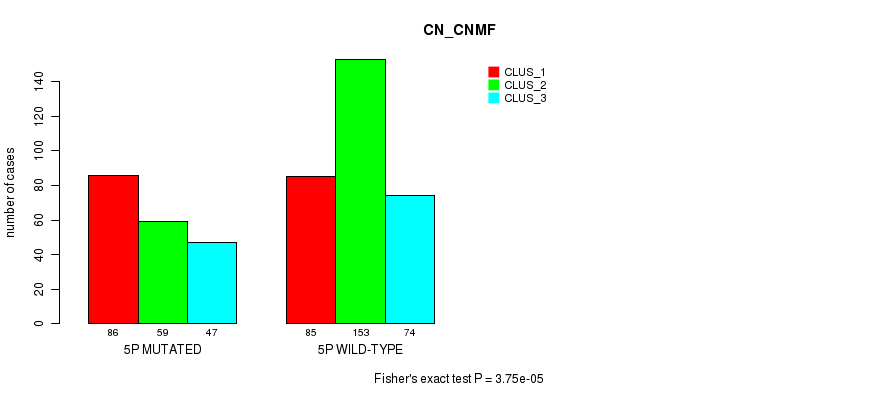
P value = 5.28e-18 (Fisher's exact test), Q value = 2.5e-15
Table S24. Gene #11: '6p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 6P MUTATED | 69 | 10 | 31 |
| 6P WILD-TYPE | 102 | 202 | 90 |
Figure S24. Get High-res Image Gene #11: '6p' versus Molecular Subtype #3: 'CN_CNMF'
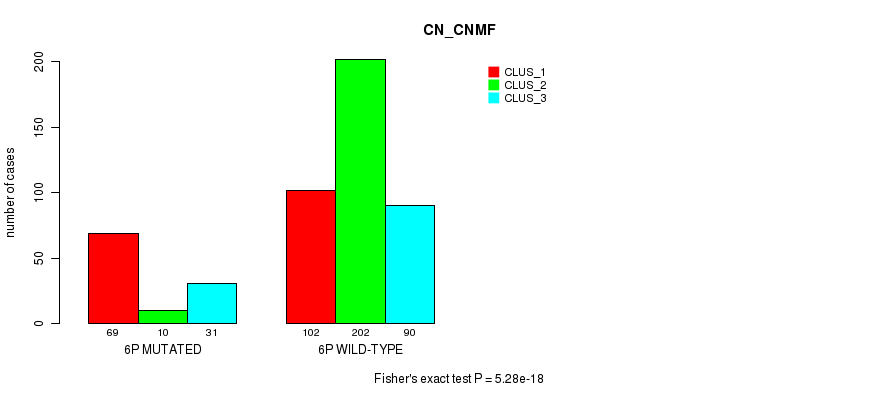
P value = 1.07e-06 (Chi-square test), Q value = 0.00042
Table S25. Gene #11: '6p' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 6P MUTATED | 18 | 8 | 17 | 23 | 6 | 27 |
| 6P WILD-TYPE | 82 | 80 | 67 | 52 | 37 | 30 |
Figure S25. Get High-res Image Gene #11: '6p' versus Molecular Subtype #5: 'RPPA_CNMF'
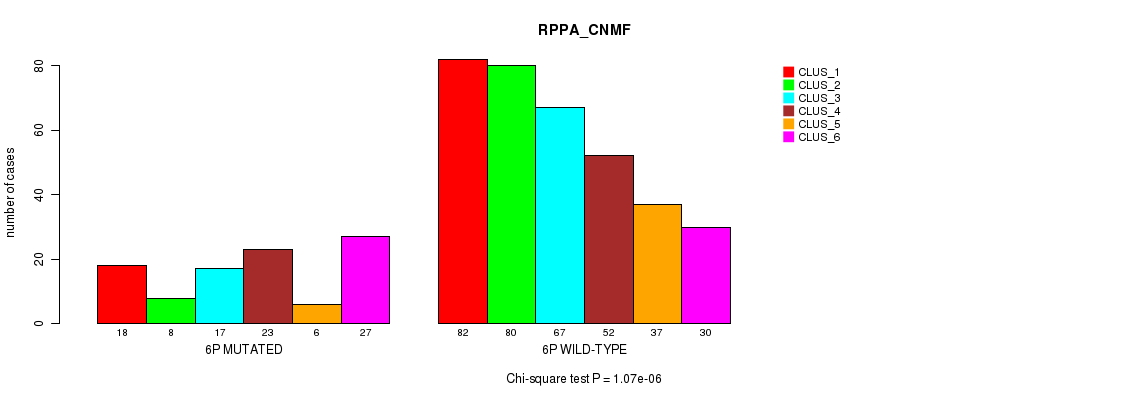
P value = 1.78e-06 (Fisher's exact test), Q value = 0.00069
Table S26. Gene #11: '6p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 6P MUTATED | 25 | 63 | 19 |
| 6P WILD-TYPE | 180 | 126 | 85 |
Figure S26. Get High-res Image Gene #11: '6p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
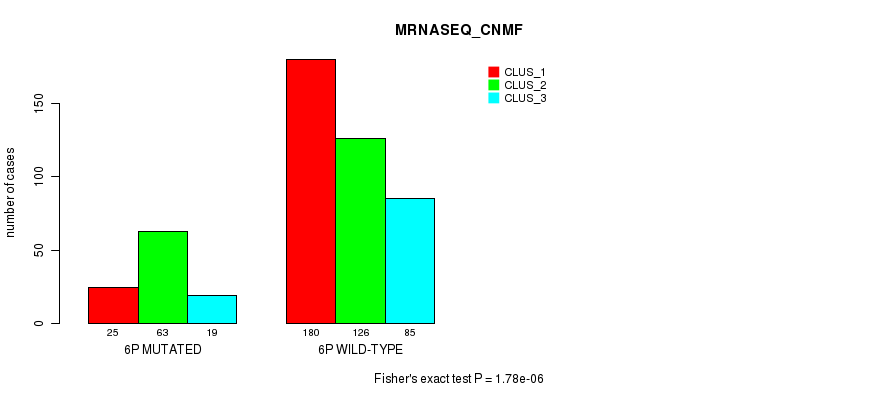
P value = 3.47e-06 (Fisher's exact test), Q value = 0.0013
Table S27. Gene #11: '6p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 6P MUTATED | 26 | 66 | 15 |
| 6P WILD-TYPE | 189 | 140 | 62 |
Figure S27. Get High-res Image Gene #11: '6p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
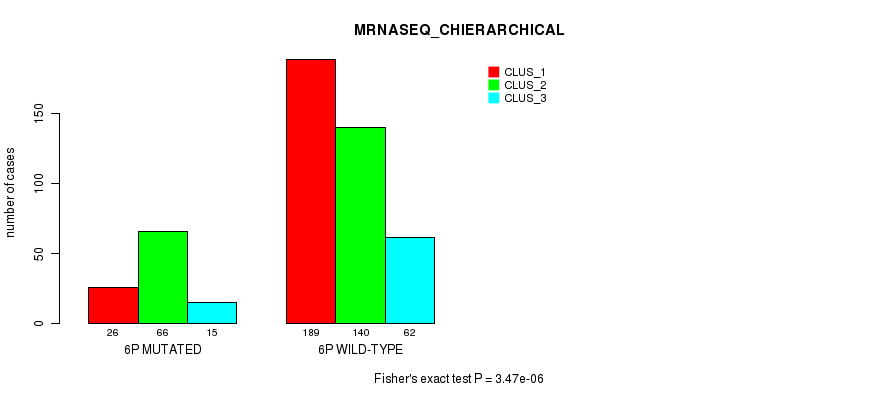
P value = 9.15e-10 (Fisher's exact test), Q value = 3.8e-07
Table S28. Gene #11: '6p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 6P MUTATED | 19 | 23 | 64 |
| 6P WILD-TYPE | 99 | 180 | 101 |
Figure S28. Get High-res Image Gene #11: '6p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
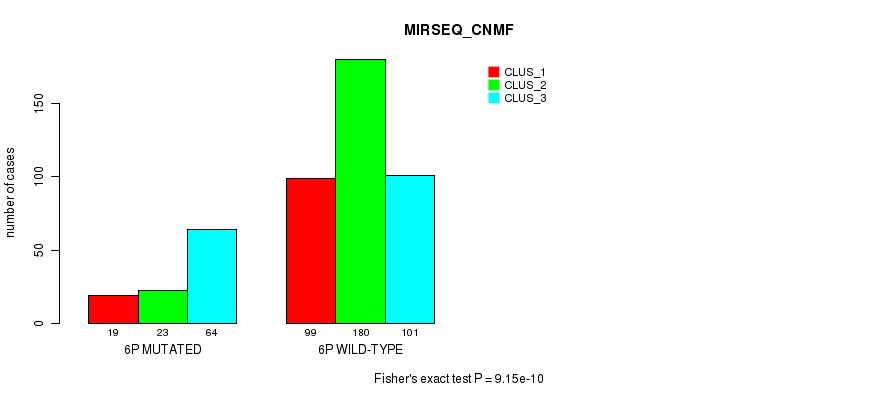
P value = 2.38e-05 (Fisher's exact test), Q value = 0.0085
Table S29. Gene #11: '6p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 6P MUTATED | 41 | 19 | 46 |
| 6P WILD-TYPE | 223 | 21 | 136 |
Figure S29. Get High-res Image Gene #11: '6p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
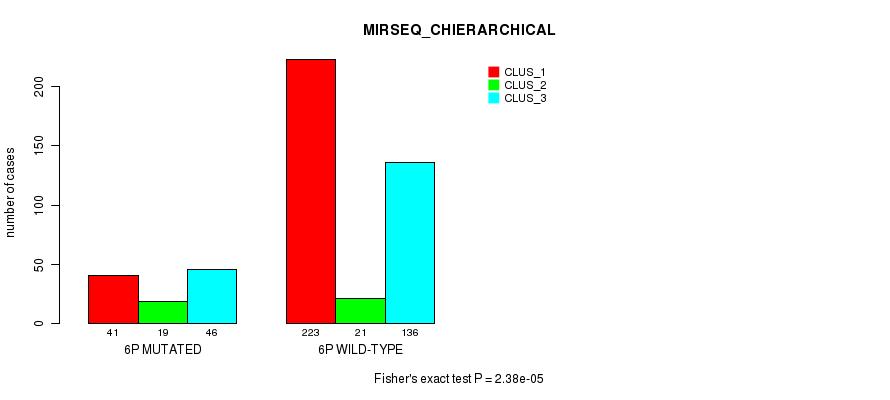
P value = 7.8e-07 (Fisher's exact test), Q value = 0.00031
Table S30. Gene #11: '6p' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 6P MUTATED | 10 | 6 | 30 |
| 6P WILD-TYPE | 43 | 89 | 48 |
Figure S30. Get High-res Image Gene #11: '6p' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
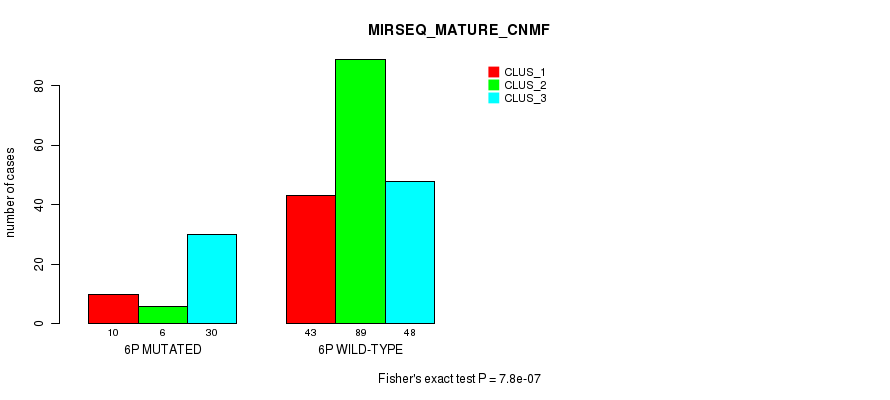
P value = 1.01e-17 (Fisher's exact test), Q value = 4.7e-15
Table S31. Gene #12: '6q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 6Q MUTATED | 84 | 20 | 31 |
| 6Q WILD-TYPE | 87 | 192 | 90 |
Figure S31. Get High-res Image Gene #12: '6q' versus Molecular Subtype #3: 'CN_CNMF'
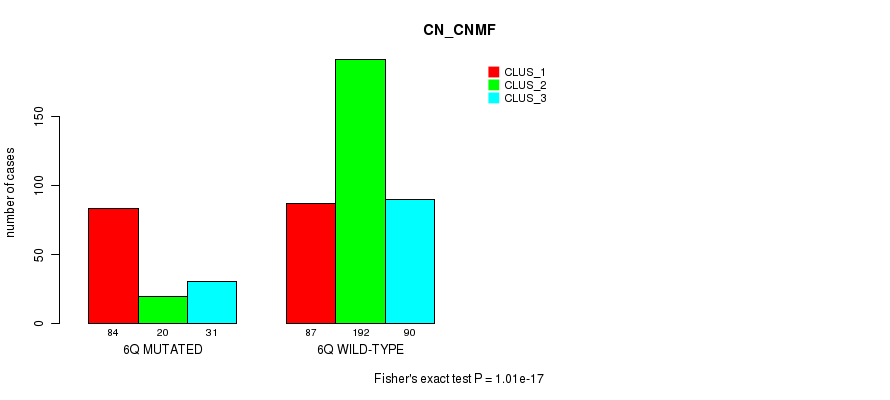
P value = 2.1e-05 (Chi-square test), Q value = 0.0076
Table S32. Gene #12: '6q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 6Q MUTATED | 29 | 8 | 24 | 24 | 8 | 27 |
| 6Q WILD-TYPE | 71 | 80 | 60 | 51 | 35 | 30 |
Figure S32. Get High-res Image Gene #12: '6q' versus Molecular Subtype #5: 'RPPA_CNMF'
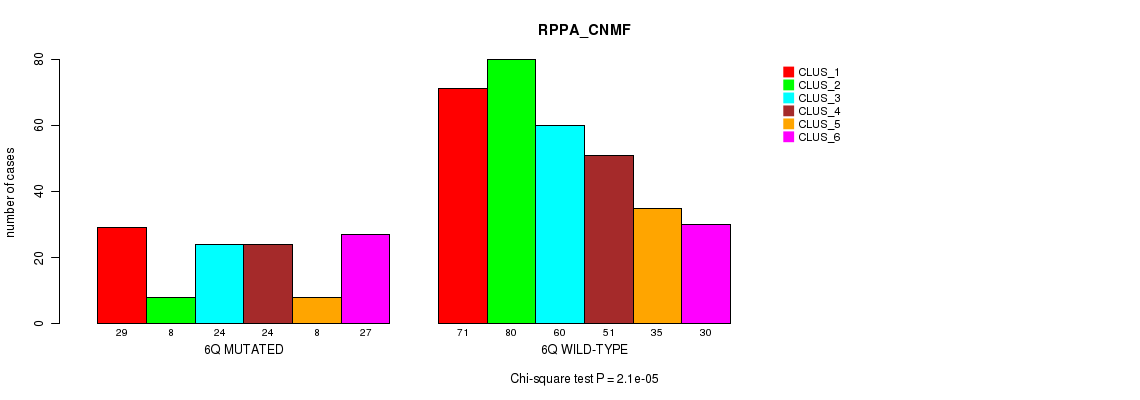
P value = 4.41e-07 (Fisher's exact test), Q value = 0.00018
Table S33. Gene #12: '6q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 6Q MUTATED | 23 | 38 | 70 |
| 6Q WILD-TYPE | 95 | 165 | 95 |
Figure S33. Get High-res Image Gene #12: '6q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
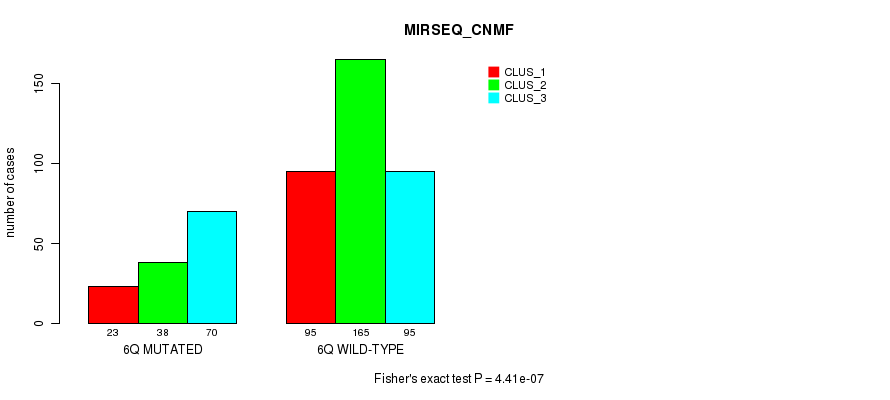
P value = 0.000418 (Fisher's exact test), Q value = 0.13
Table S34. Gene #12: '6q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 6Q MUTATED | 58 | 21 | 52 |
| 6Q WILD-TYPE | 206 | 19 | 130 |
Figure S34. Get High-res Image Gene #12: '6q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
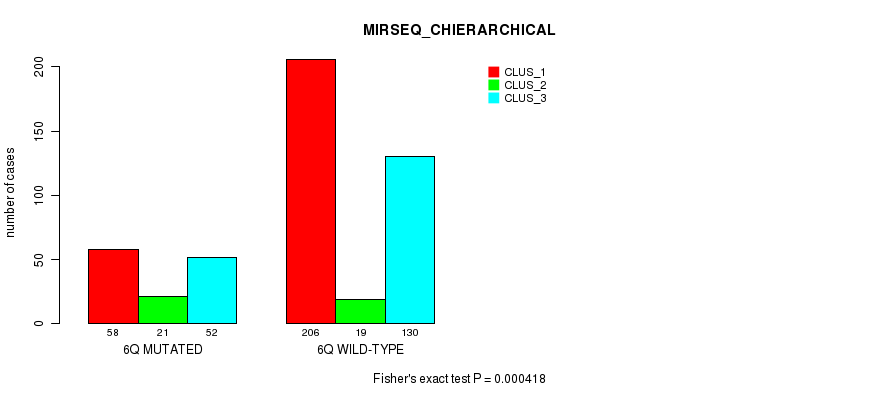
P value = 5.88e-05 (Fisher's exact test), Q value = 0.02
Table S35. Gene #12: '6q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 6Q MUTATED | 13 | 14 | 35 |
| 6Q WILD-TYPE | 40 | 81 | 43 |
Figure S35. Get High-res Image Gene #12: '6q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
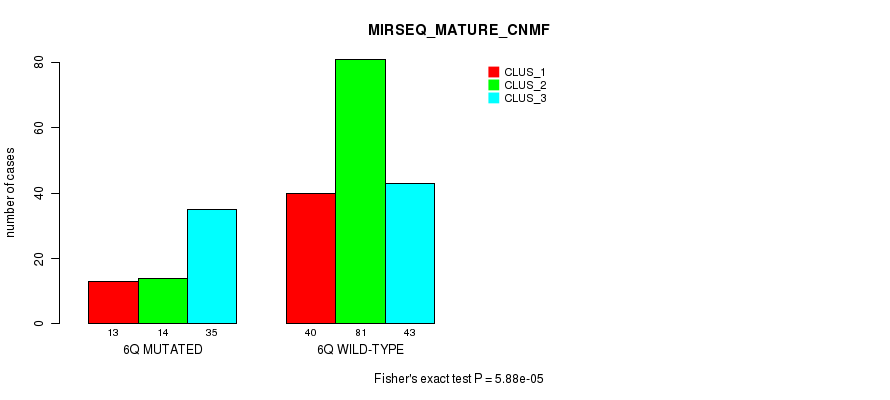
P value = 7.13e-14 (Fisher's exact test), Q value = 3.2e-11
Table S36. Gene #13: '7p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 7P MUTATED | 88 | 33 | 49 |
| 7P WILD-TYPE | 83 | 179 | 72 |
Figure S36. Get High-res Image Gene #13: '7p' versus Molecular Subtype #3: 'CN_CNMF'
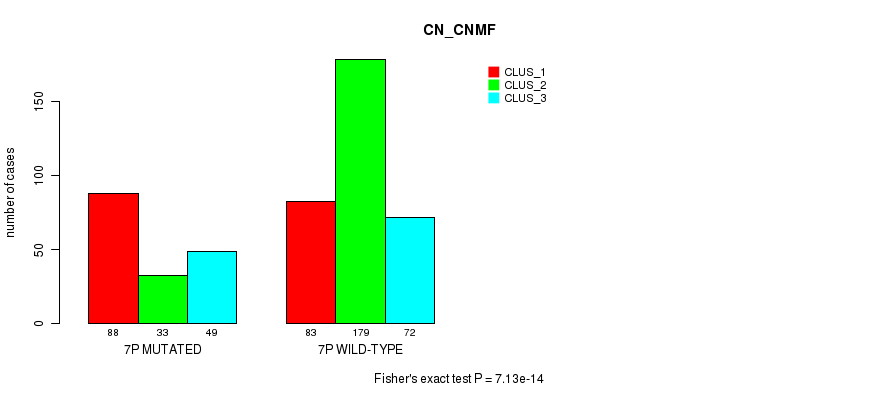
P value = 3.22e-06 (Chi-square test), Q value = 0.0012
Table S37. Gene #13: '7p' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 7P MUTATED | 21 | 19 | 34 | 34 | 11 | 32 |
| 7P WILD-TYPE | 79 | 69 | 50 | 41 | 32 | 25 |
Figure S37. Get High-res Image Gene #13: '7p' versus Molecular Subtype #5: 'RPPA_CNMF'
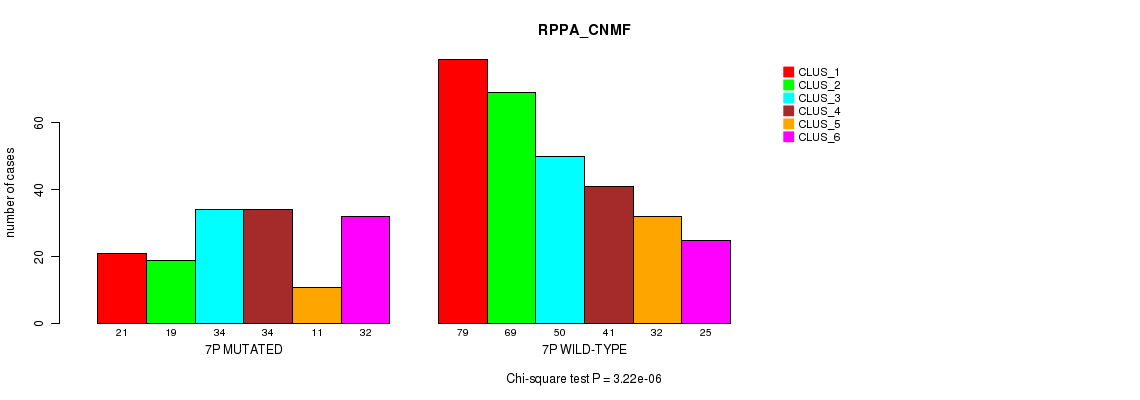
P value = 0.000147 (Fisher's exact test), Q value = 0.048
Table S38. Gene #13: '7p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 7P MUTATED | 52 | 85 | 31 |
| 7P WILD-TYPE | 153 | 104 | 73 |
Figure S38. Get High-res Image Gene #13: '7p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
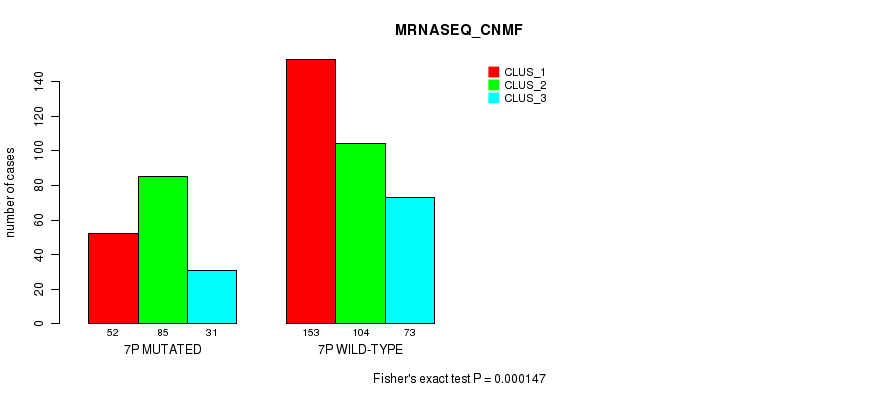
P value = 2.11e-13 (Fisher's exact test), Q value = 9.4e-11
Table S39. Gene #14: '7q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 7Q MUTATED | 87 | 34 | 51 |
| 7Q WILD-TYPE | 84 | 178 | 70 |
Figure S39. Get High-res Image Gene #14: '7q' versus Molecular Subtype #3: 'CN_CNMF'
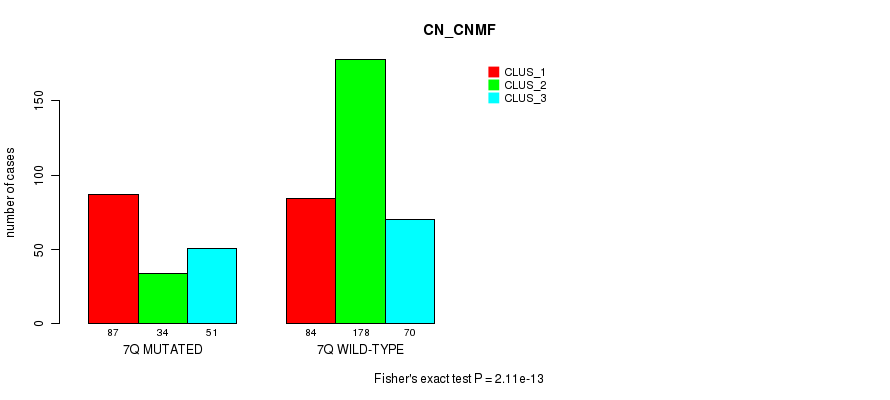
P value = 4.52e-06 (Chi-square test), Q value = 0.0017
Table S40. Gene #14: '7q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 7Q MUTATED | 21 | 21 | 35 | 34 | 10 | 32 |
| 7Q WILD-TYPE | 79 | 67 | 49 | 41 | 33 | 25 |
Figure S40. Get High-res Image Gene #14: '7q' versus Molecular Subtype #5: 'RPPA_CNMF'
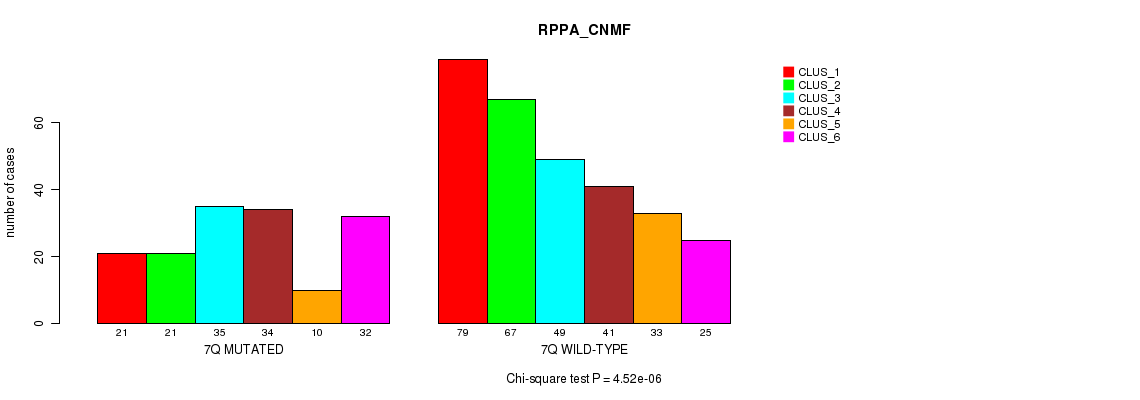
P value = 0.000615 (Fisher's exact test), Q value = 0.19
Table S41. Gene #14: '7q' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 186 | 152 | 109 |
| 7Q MUTATED | 45 | 61 | 47 |
| 7Q WILD-TYPE | 141 | 91 | 62 |
Figure S41. Get High-res Image Gene #14: '7q' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
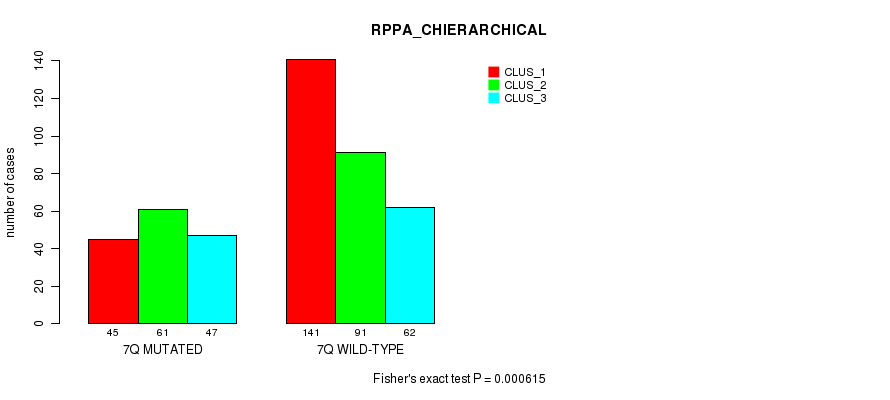
P value = 0.000108 (Fisher's exact test), Q value = 0.036
Table S42. Gene #14: '7q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 7Q MUTATED | 52 | 86 | 32 |
| 7Q WILD-TYPE | 153 | 103 | 72 |
Figure S42. Get High-res Image Gene #14: '7q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
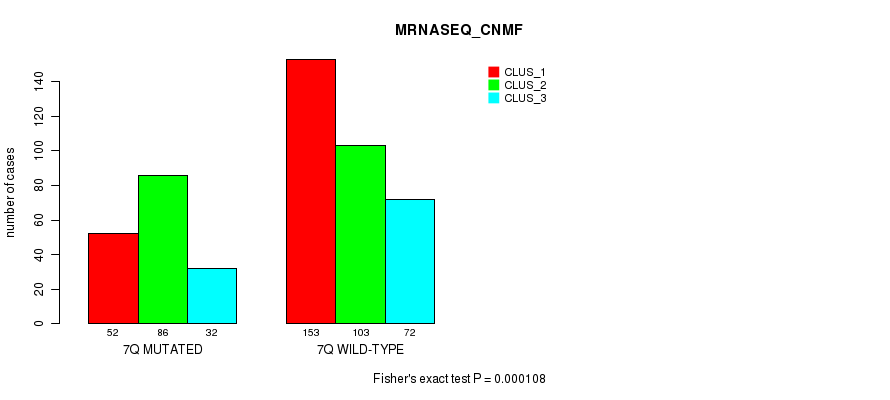
P value = 0.000597 (Fisher's exact test), Q value = 0.18
Table S43. Gene #14: '7q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 7Q MUTATED | 56 | 90 | 24 |
| 7Q WILD-TYPE | 159 | 116 | 53 |
Figure S43. Get High-res Image Gene #14: '7q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
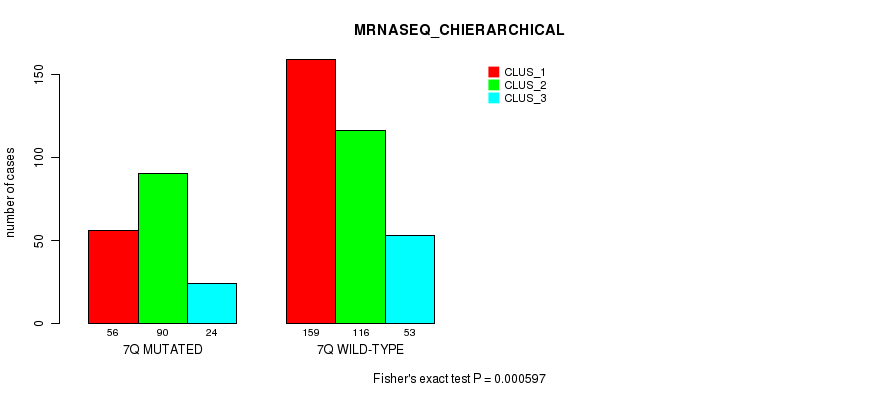
P value = 8.91e-12 (Fisher's exact test), Q value = 3.8e-09
Table S44. Gene #15: '8p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 8P MUTATED | 90 | 39 | 36 |
| 8P WILD-TYPE | 81 | 173 | 85 |
Figure S44. Get High-res Image Gene #15: '8p' versus Molecular Subtype #3: 'CN_CNMF'
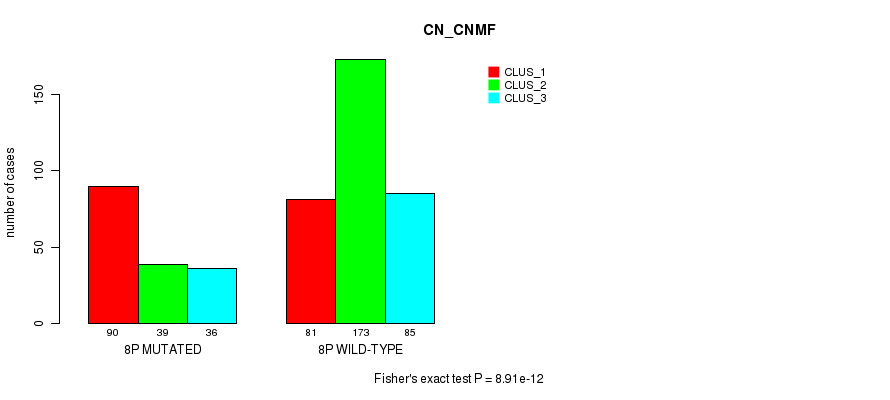
P value = 0.000424 (Fisher's exact test), Q value = 0.13
Table S45. Gene #15: '8p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 8P MUTATED | 55 | 82 | 26 |
| 8P WILD-TYPE | 150 | 107 | 78 |
Figure S45. Get High-res Image Gene #15: '8p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
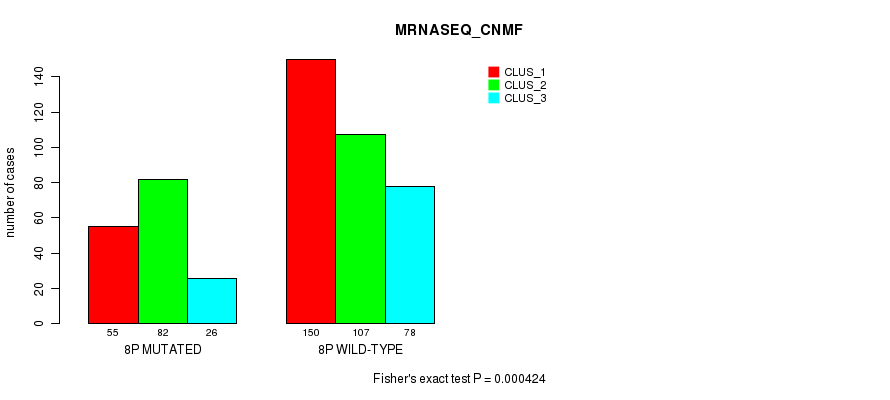
P value = 0.000826 (Fisher's exact test), Q value = 0.24
Table S46. Gene #15: '8p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 8P MUTATED | 56 | 87 | 20 |
| 8P WILD-TYPE | 159 | 119 | 57 |
Figure S46. Get High-res Image Gene #15: '8p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
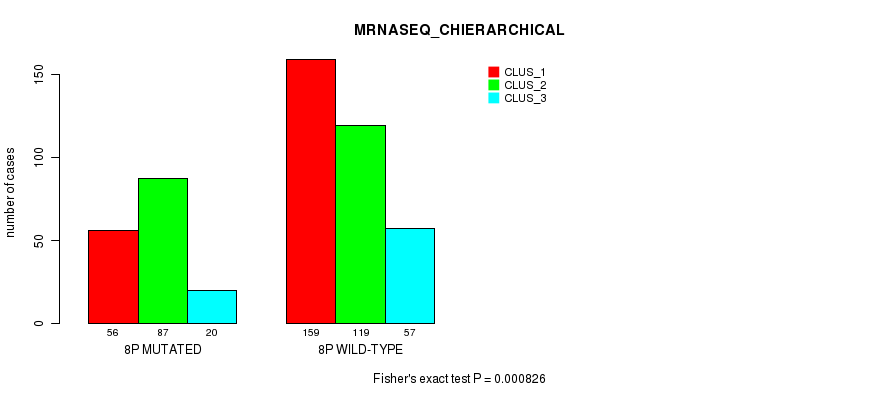
P value = 1.36e-05 (Fisher's exact test), Q value = 0.005
Table S47. Gene #15: '8p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 8P MUTATED | 35 | 49 | 78 |
| 8P WILD-TYPE | 83 | 154 | 87 |
Figure S47. Get High-res Image Gene #15: '8p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
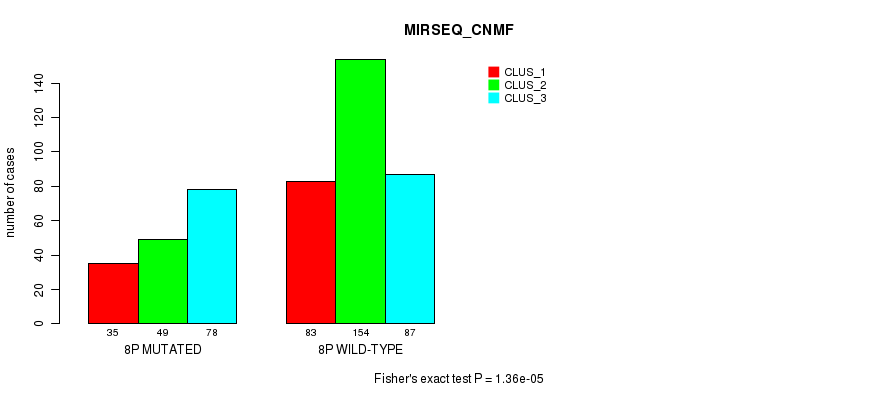
P value = 4.83e-15 (Fisher's exact test), Q value = 2.2e-12
Table S48. Gene #16: '8q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 8Q MUTATED | 76 | 19 | 29 |
| 8Q WILD-TYPE | 95 | 193 | 92 |
Figure S48. Get High-res Image Gene #16: '8q' versus Molecular Subtype #3: 'CN_CNMF'
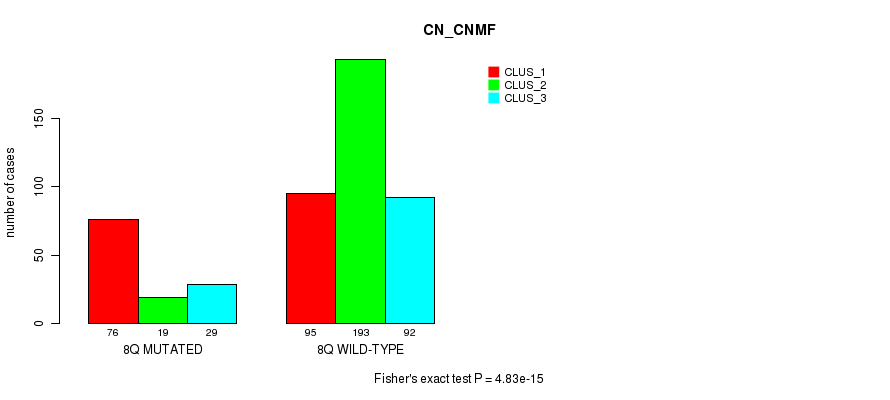
P value = 9.92e-05 (Fisher's exact test), Q value = 0.033
Table S49. Gene #16: '8q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 8Q MUTATED | 41 | 19 | 14 |
| 8Q WILD-TYPE | 59 | 95 | 62 |
Figure S49. Get High-res Image Gene #16: '8q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
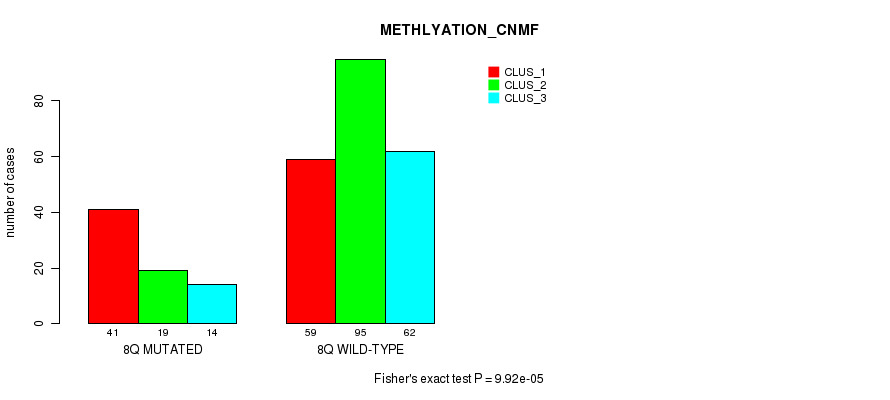
P value = 1.19e-05 (Chi-square test), Q value = 0.0044
Table S50. Gene #16: '8q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 8Q MUTATED | 11 | 22 | 30 | 19 | 5 | 25 |
| 8Q WILD-TYPE | 89 | 66 | 54 | 56 | 38 | 32 |
Figure S50. Get High-res Image Gene #16: '8q' versus Molecular Subtype #5: 'RPPA_CNMF'
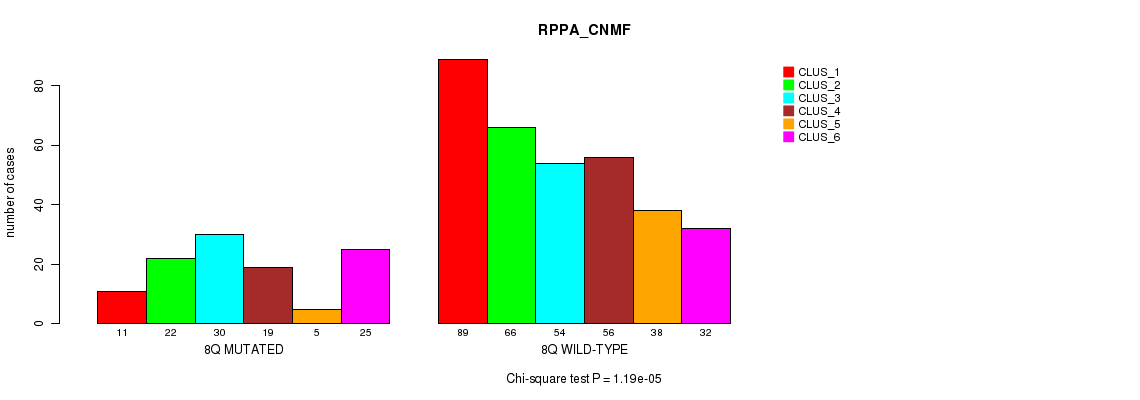
P value = 1.03e-05 (Fisher's exact test), Q value = 0.0039
Table S51. Gene #16: '8q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 8Q MUTATED | 35 | 69 | 18 |
| 8Q WILD-TYPE | 170 | 120 | 86 |
Figure S51. Get High-res Image Gene #16: '8q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
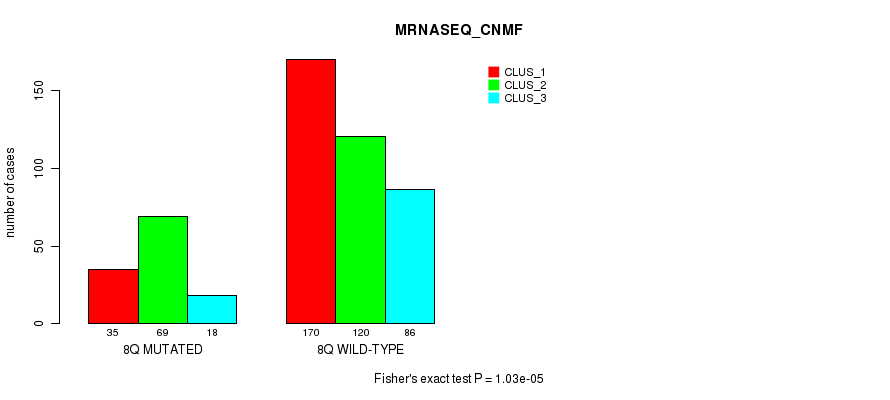
P value = 8.99e-05 (Fisher's exact test), Q value = 0.03
Table S52. Gene #16: '8q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 8Q MUTATED | 39 | 71 | 12 |
| 8Q WILD-TYPE | 176 | 135 | 65 |
Figure S52. Get High-res Image Gene #16: '8q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
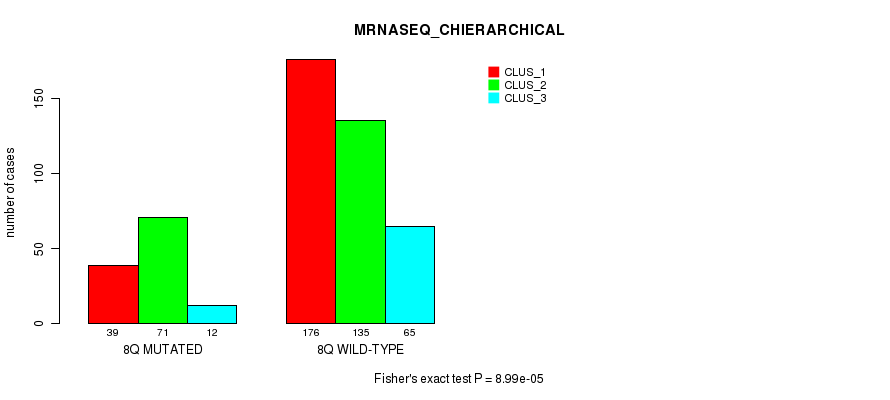
P value = 5.15e-06 (Fisher's exact test), Q value = 0.002
Table S53. Gene #16: '8q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 8Q MUTATED | 27 | 32 | 63 |
| 8Q WILD-TYPE | 91 | 171 | 102 |
Figure S53. Get High-res Image Gene #16: '8q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
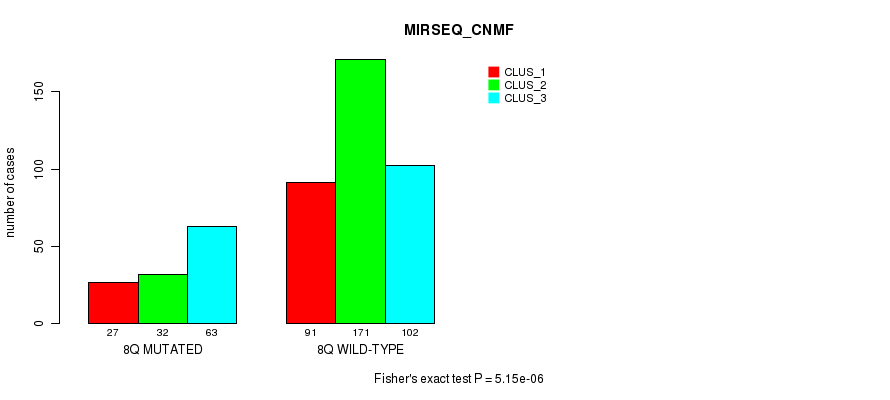
P value = 1.58e-05 (Fisher's exact test), Q value = 0.0058
Table S54. Gene #16: '8q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 8Q MUTATED | 13 | 10 | 32 |
| 8Q WILD-TYPE | 40 | 85 | 46 |
Figure S54. Get High-res Image Gene #16: '8q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
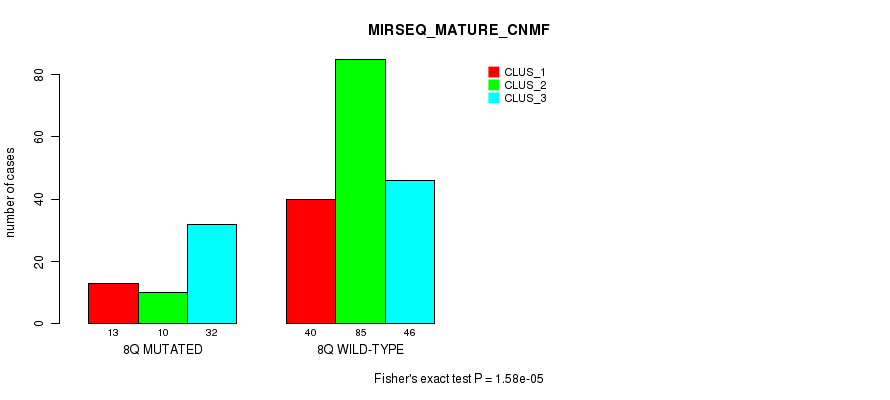
P value = 6.96e-05 (Fisher's exact test), Q value = 0.024
Table S55. Gene #16: '8q' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 86 | 69 | 71 |
| 8Q MUTATED | 8 | 25 | 22 |
| 8Q WILD-TYPE | 78 | 44 | 49 |
Figure S55. Get High-res Image Gene #16: '8q' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
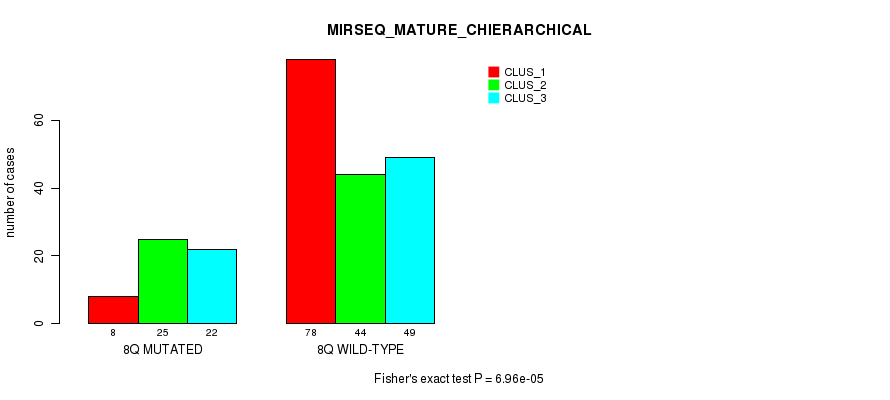
P value = 0.00044 (Fisher's exact test), Q value = 0.14
Table S56. Gene #17: '9p' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 14 | 32 | 24 |
| 9P MUTATED | 2 | 4 | 14 |
| 9P WILD-TYPE | 12 | 28 | 10 |
Figure S56. Get High-res Image Gene #17: '9p' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
P value = 6.08e-41 (Fisher's exact test), Q value = 2.9e-38
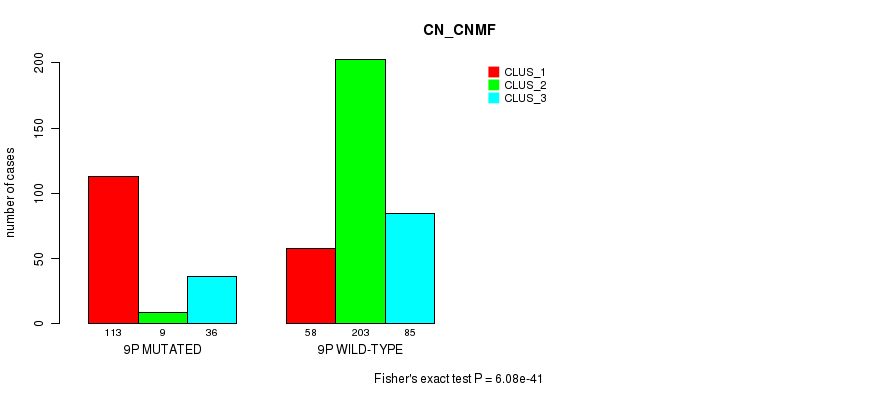
Table S57. Gene #17: '9p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 9P MUTATED | 113 | 9 | 36 |
| 9P WILD-TYPE | 58 | 203 | 85 |
Figure S57. Get High-res Image Gene #17: '9p' versus Molecular Subtype #3: 'CN_CNMF'
P value = 3.82e-17 (Fisher's exact test), Q value = 1.8e-14
Table S58. Gene #17: '9p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 9P MUTATED | 62 | 9 | 26 |
| 9P WILD-TYPE | 38 | 105 | 50 |
Figure S58. Get High-res Image Gene #17: '9p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
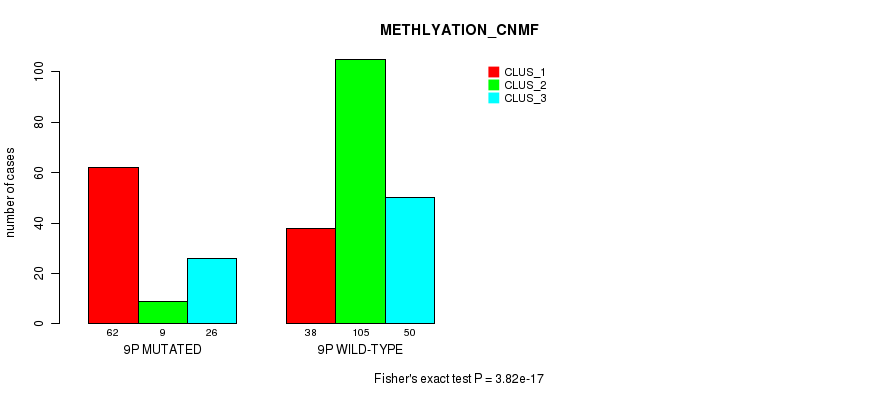
P value = 2.54e-05 (Chi-square test), Q value = 0.0091
Table S59. Gene #17: '9p' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 9P MUTATED | 21 | 25 | 32 | 21 | 11 | 34 |
| 9P WILD-TYPE | 79 | 63 | 52 | 54 | 32 | 23 |
Figure S59. Get High-res Image Gene #17: '9p' versus Molecular Subtype #5: 'RPPA_CNMF'
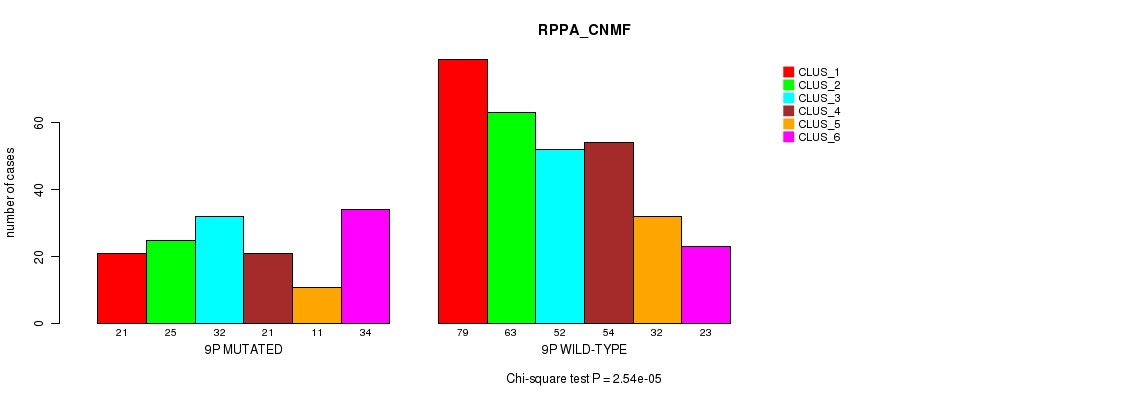
P value = 6.25e-20 (Fisher's exact test), Q value = 3e-17
Table S60. Gene #17: '9p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 9P MUTATED | 30 | 106 | 19 |
| 9P WILD-TYPE | 175 | 83 | 85 |
Figure S60. Get High-res Image Gene #17: '9p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
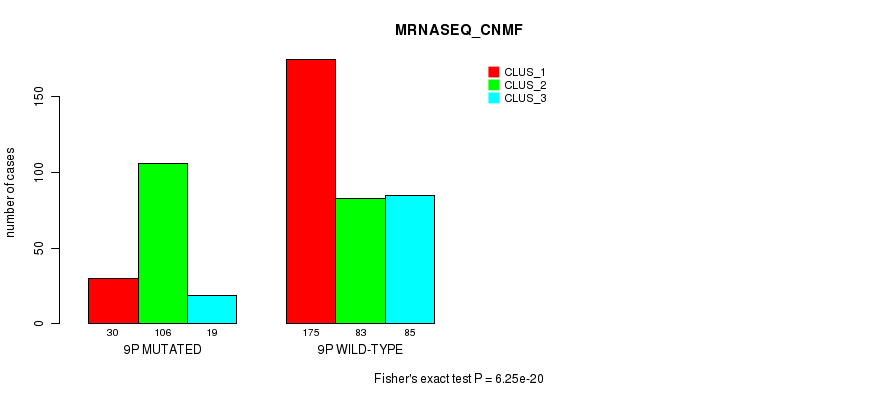
P value = 5.43e-21 (Fisher's exact test), Q value = 2.6e-18
Table S61. Gene #17: '9p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 9P MUTATED | 30 | 113 | 12 |
| 9P WILD-TYPE | 185 | 93 | 65 |
Figure S61. Get High-res Image Gene #17: '9p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
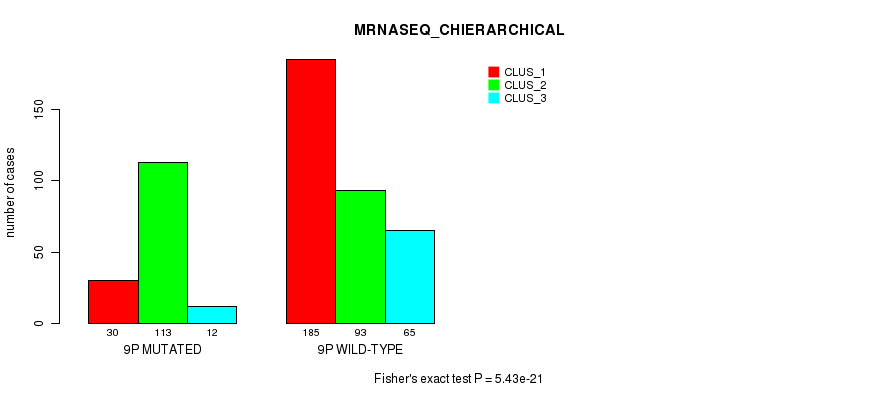
P value = 8.53e-13 (Fisher's exact test), Q value = 3.8e-10
Table S62. Gene #17: '9p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 9P MUTATED | 32 | 34 | 87 |
| 9P WILD-TYPE | 86 | 169 | 78 |
Figure S62. Get High-res Image Gene #17: '9p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
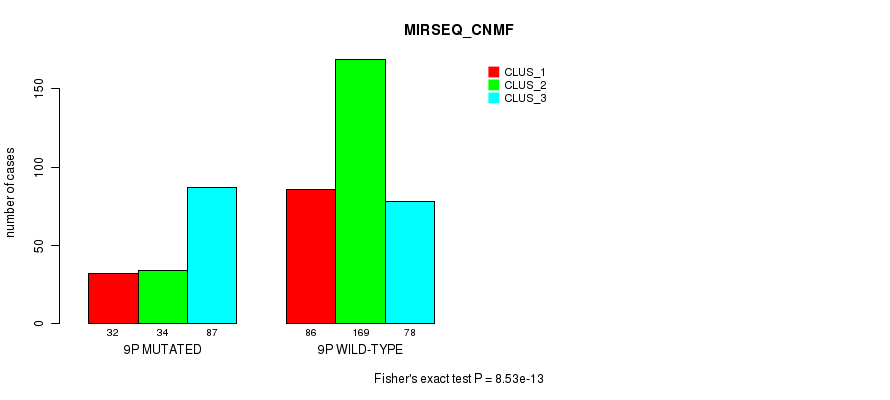
P value = 0.00011 (Fisher's exact test), Q value = 0.036
Table S63. Gene #17: '9p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 9P MUTATED | 62 | 19 | 72 |
| 9P WILD-TYPE | 202 | 21 | 110 |
Figure S63. Get High-res Image Gene #17: '9p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
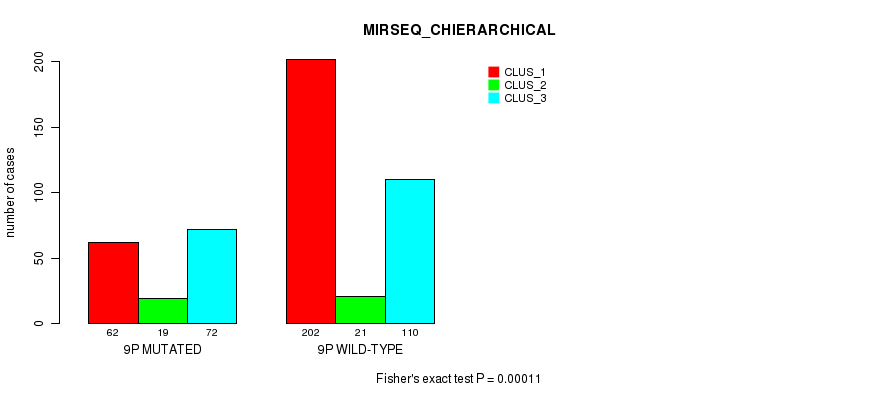
P value = 3.3e-08 (Fisher's exact test), Q value = 1.3e-05
Table S64. Gene #17: '9p' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 9P MUTATED | 21 | 12 | 41 |
| 9P WILD-TYPE | 32 | 83 | 37 |
Figure S64. Get High-res Image Gene #17: '9p' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
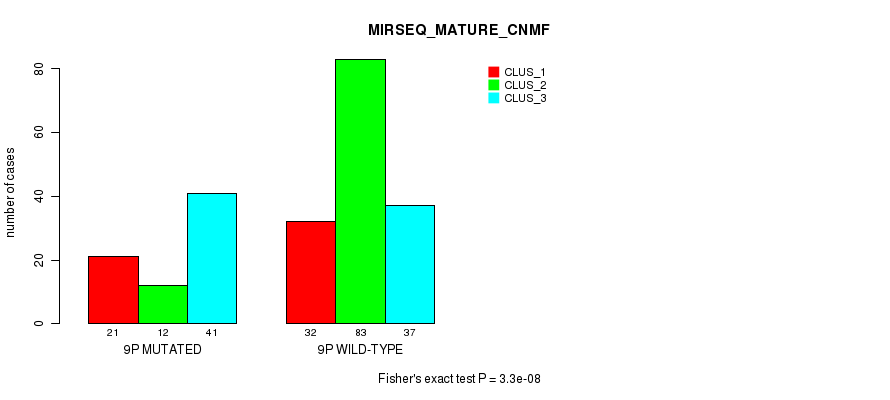
P value = 0.000175 (Fisher's exact test), Q value = 0.057
Table S65. Gene #17: '9p' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 86 | 69 | 71 |
| 9P MUTATED | 15 | 33 | 26 |
| 9P WILD-TYPE | 71 | 36 | 45 |
Figure S65. Get High-res Image Gene #17: '9p' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
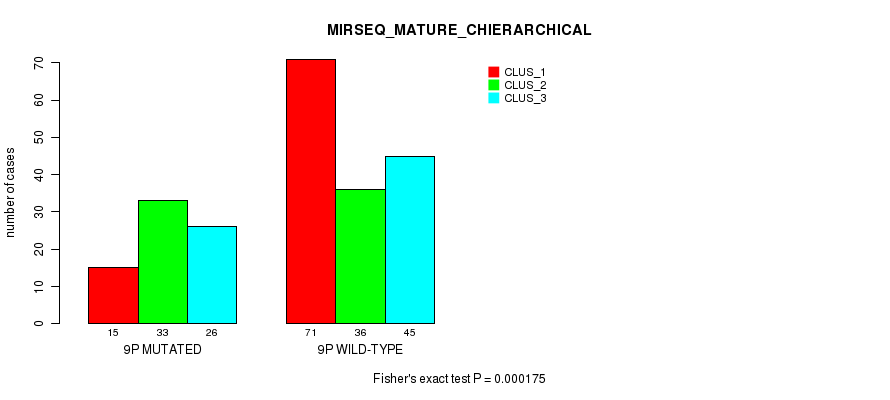
P value = 0.000826 (Fisher's exact test), Q value = 0.24
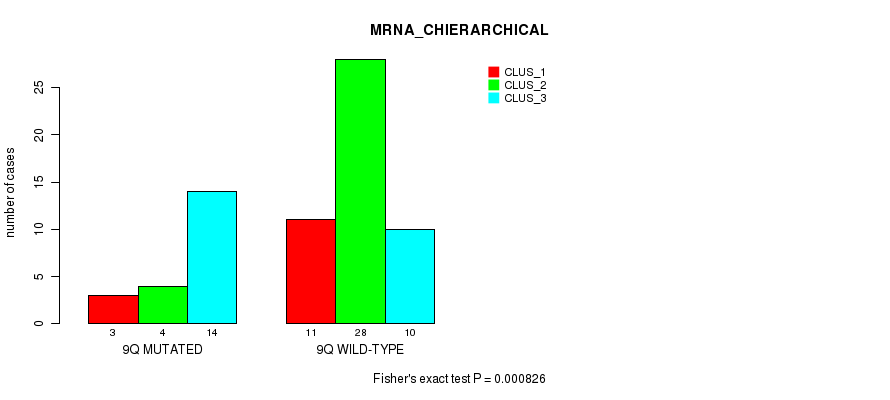
Table S66. Gene #18: '9q' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 14 | 32 | 24 |
| 9Q MUTATED | 3 | 4 | 14 |
| 9Q WILD-TYPE | 11 | 28 | 10 |
Figure S66. Get High-res Image Gene #18: '9q' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
P value = 1.48e-42 (Fisher's exact test), Q value = 7.1e-40
Table S67. Gene #18: '9q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 9Q MUTATED | 114 | 8 | 37 |
| 9Q WILD-TYPE | 57 | 204 | 84 |
Figure S67. Get High-res Image Gene #18: '9q' versus Molecular Subtype #3: 'CN_CNMF'
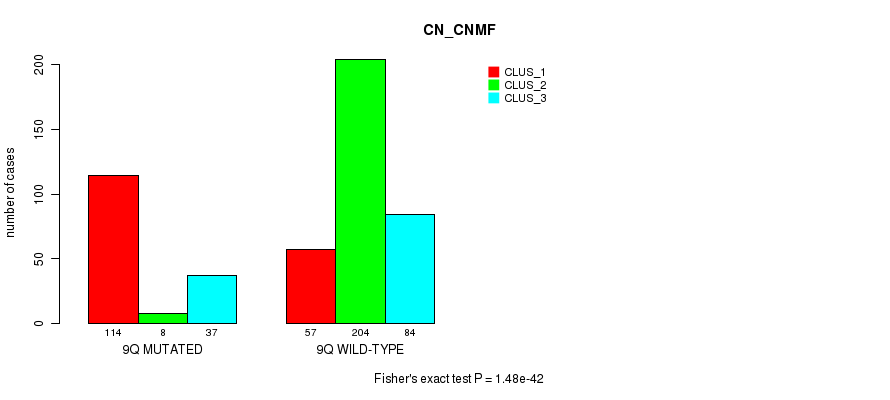
P value = 5.8e-18 (Fisher's exact test), Q value = 2.7e-15
Table S68. Gene #18: '9q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 9Q MUTATED | 62 | 8 | 27 |
| 9Q WILD-TYPE | 38 | 106 | 49 |
Figure S68. Get High-res Image Gene #18: '9q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
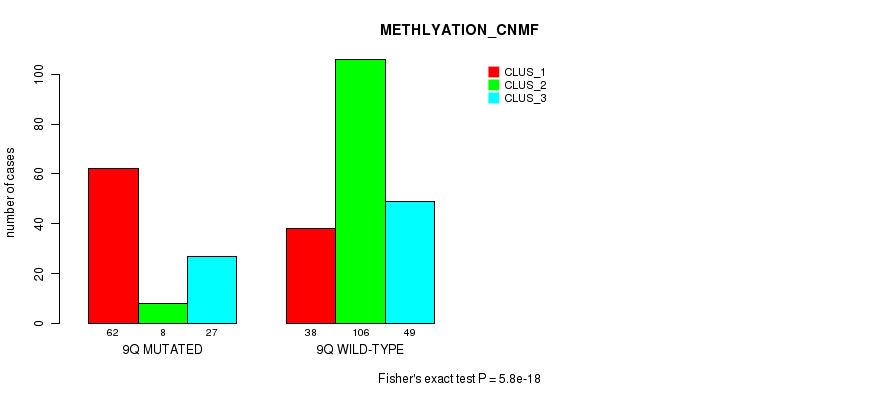
P value = 5.31e-07 (Chi-square test), Q value = 0.00021
Table S69. Gene #18: '9q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 9Q MUTATED | 18 | 26 | 35 | 21 | 10 | 35 |
| 9Q WILD-TYPE | 82 | 62 | 49 | 54 | 33 | 22 |
Figure S69. Get High-res Image Gene #18: '9q' versus Molecular Subtype #5: 'RPPA_CNMF'
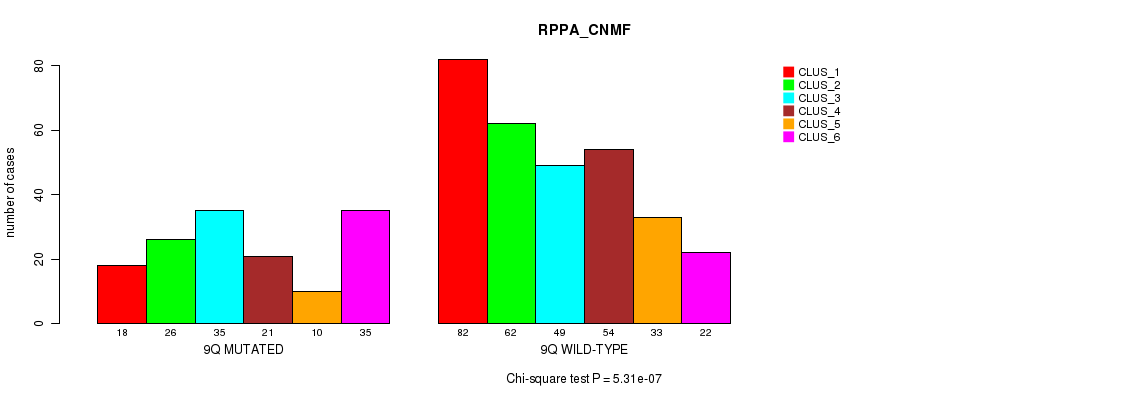
P value = 0.000429 (Fisher's exact test), Q value = 0.13
Table S70. Gene #18: '9q' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 186 | 152 | 109 |
| 9Q MUTATED | 42 | 56 | 47 |
| 9Q WILD-TYPE | 144 | 96 | 62 |
Figure S70. Get High-res Image Gene #18: '9q' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
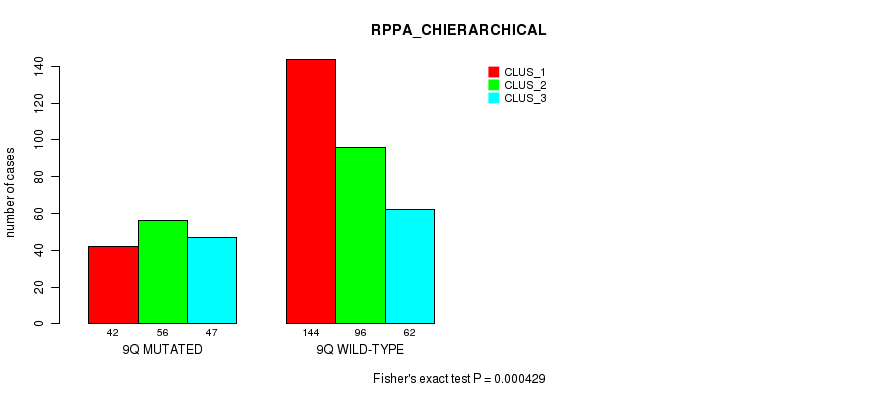
P value = 6.51e-22 (Fisher's exact test), Q value = 3.1e-19
Table S71. Gene #18: '9q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 9Q MUTATED | 28 | 109 | 20 |
| 9Q WILD-TYPE | 177 | 80 | 84 |
Figure S71. Get High-res Image Gene #18: '9q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
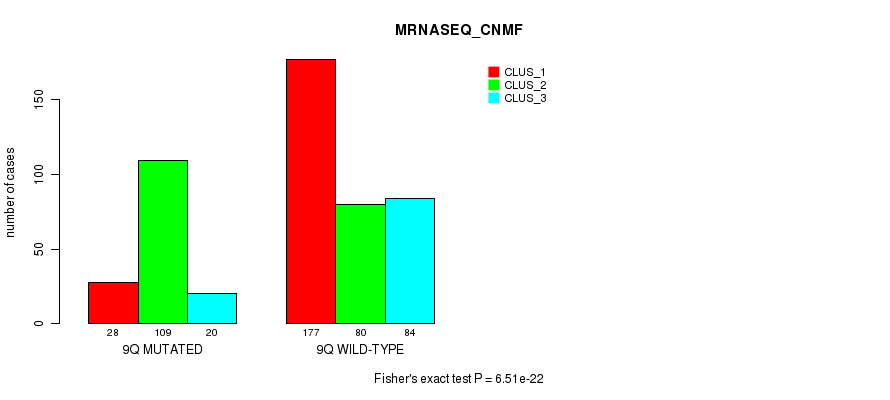
P value = 7.16e-23 (Fisher's exact test), Q value = 3.4e-20
Table S72. Gene #18: '9q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 9Q MUTATED | 28 | 116 | 13 |
| 9Q WILD-TYPE | 187 | 90 | 64 |
Figure S72. Get High-res Image Gene #18: '9q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
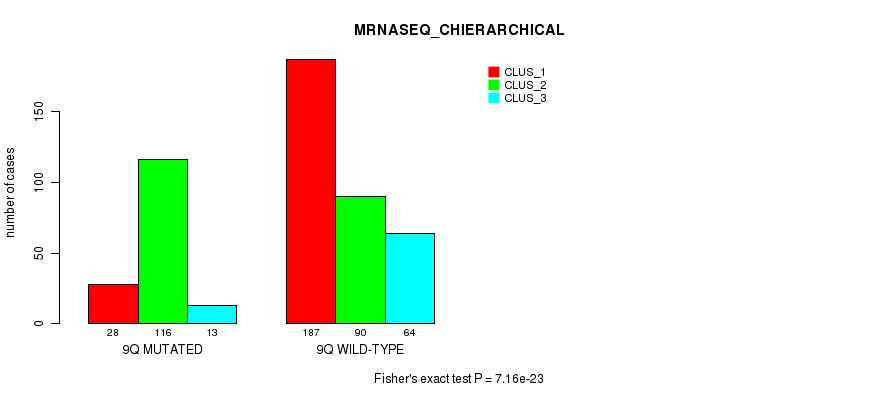
P value = 1.15e-15 (Fisher's exact test), Q value = 5.3e-13
Table S73. Gene #18: '9q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 9Q MUTATED | 31 | 31 | 91 |
| 9Q WILD-TYPE | 87 | 172 | 74 |
Figure S73. Get High-res Image Gene #18: '9q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
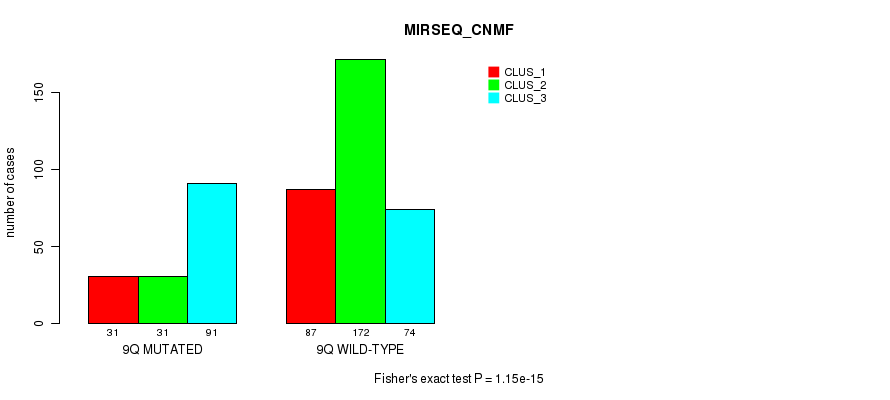
P value = 3.6e-05 (Fisher's exact test), Q value = 0.013
Table S74. Gene #18: '9q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 9Q MUTATED | 61 | 20 | 72 |
| 9Q WILD-TYPE | 203 | 20 | 110 |
Figure S74. Get High-res Image Gene #18: '9q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
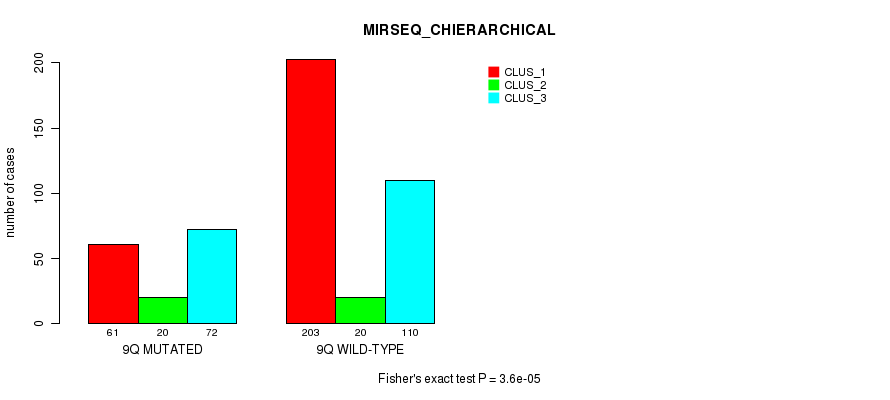
P value = 1.47e-08 (Fisher's exact test), Q value = 6e-06
Table S75. Gene #18: '9q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 9Q MUTATED | 21 | 12 | 42 |
| 9Q WILD-TYPE | 32 | 83 | 36 |
Figure S75. Get High-res Image Gene #18: '9q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
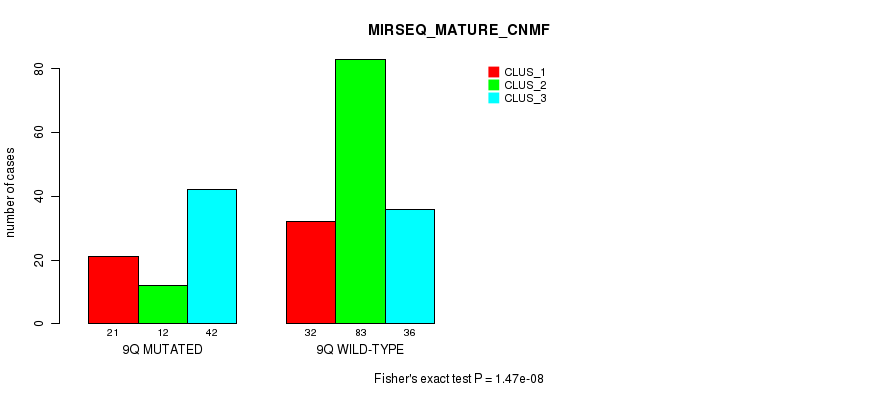
P value = 3.44e-06 (Fisher's exact test), Q value = 0.0013
Table S76. Gene #18: '9q' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 86 | 69 | 71 |
| 9Q MUTATED | 13 | 36 | 26 |
| 9Q WILD-TYPE | 73 | 33 | 45 |
Figure S76. Get High-res Image Gene #18: '9q' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
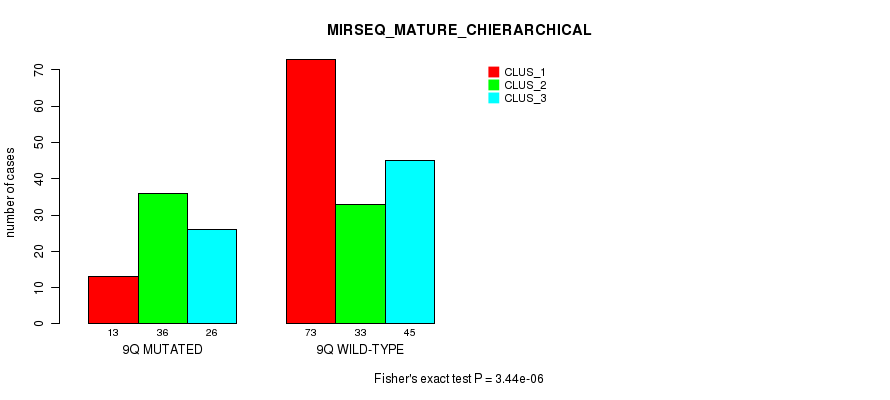
P value = 2.02e-12 (Fisher's exact test), Q value = 8.8e-10
Table S77. Gene #19: '10p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 10P MUTATED | 47 | 7 | 28 |
| 10P WILD-TYPE | 124 | 205 | 93 |
Figure S77. Get High-res Image Gene #19: '10p' versus Molecular Subtype #3: 'CN_CNMF'
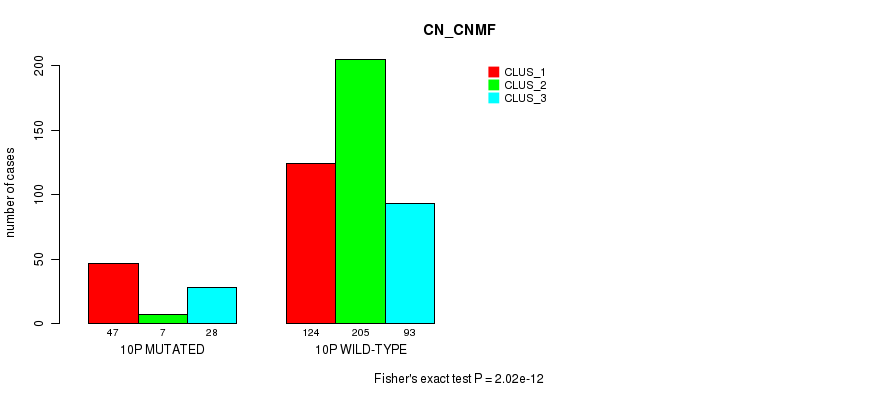
P value = 0.000829 (Fisher's exact test), Q value = 0.24
Table S78. Gene #19: '10p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 10P MUTATED | 33 | 15 | 32 |
| 10P WILD-TYPE | 231 | 25 | 150 |
Figure S78. Get High-res Image Gene #19: '10p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
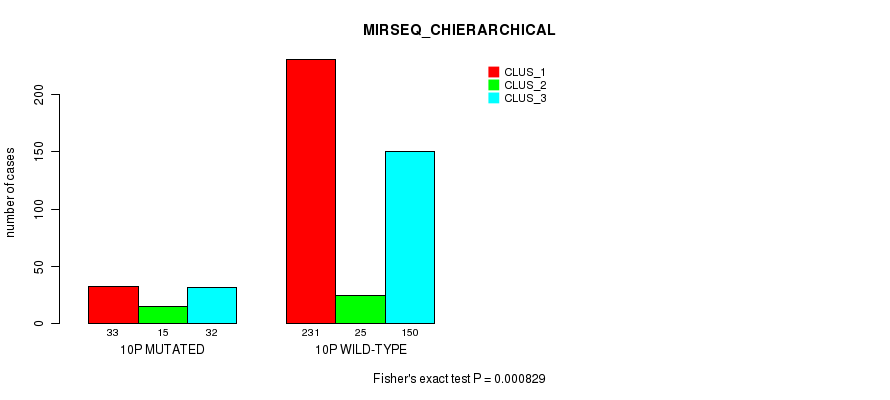
P value = 4.45e-12 (Fisher's exact test), Q value = 1.9e-09
Table S79. Gene #20: '10q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 10Q MUTATED | 56 | 12 | 30 |
| 10Q WILD-TYPE | 115 | 200 | 91 |
Figure S79. Get High-res Image Gene #20: '10q' versus Molecular Subtype #3: 'CN_CNMF'
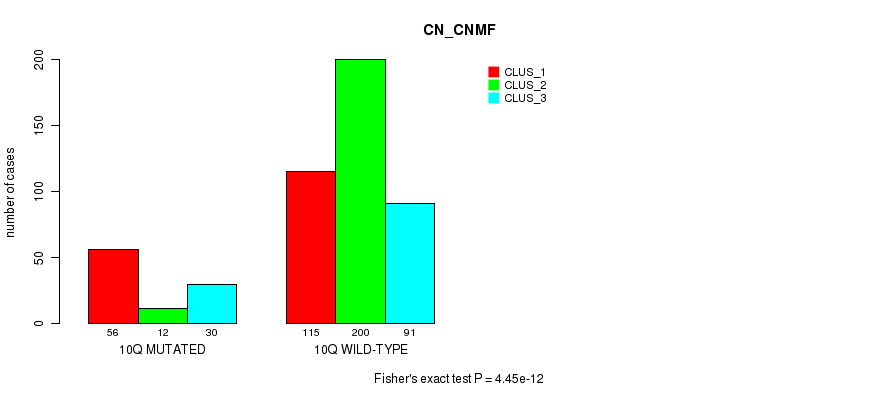
P value = 9.37e-11 (Fisher's exact test), Q value = 4e-08
Table S80. Gene #21: '11p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 11P MUTATED | 30 | 1 | 17 |
| 11P WILD-TYPE | 141 | 211 | 104 |
Figure S80. Get High-res Image Gene #21: '11p' versus Molecular Subtype #3: 'CN_CNMF'
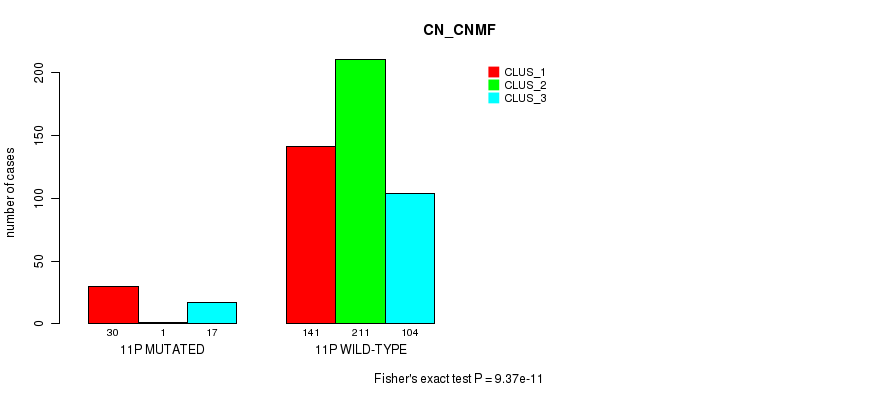
P value = 2.9e-10 (Fisher's exact test), Q value = 1.2e-07
Table S81. Gene #22: '11q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 11Q MUTATED | 32 | 2 | 17 |
| 11Q WILD-TYPE | 139 | 210 | 104 |
Figure S81. Get High-res Image Gene #22: '11q' versus Molecular Subtype #3: 'CN_CNMF'
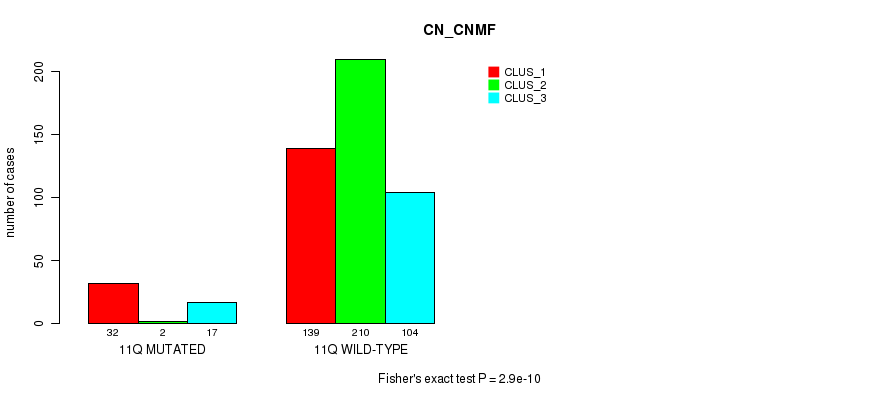
P value = 1.66e-13 (Fisher's exact test), Q value = 7.4e-11
Table S82. Gene #23: '12p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 12P MUTATED | 70 | 18 | 33 |
| 12P WILD-TYPE | 101 | 194 | 88 |
Figure S82. Get High-res Image Gene #23: '12p' versus Molecular Subtype #3: 'CN_CNMF'
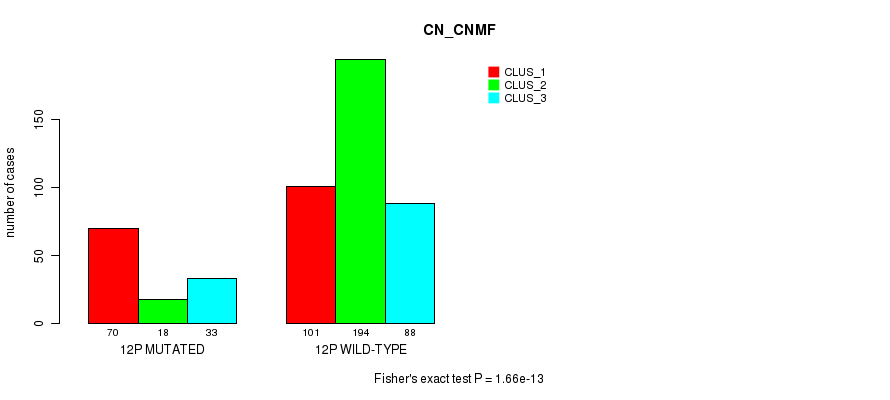
P value = 0.000101 (Fisher's exact test), Q value = 0.034
Table S83. Gene #23: '12p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 12P MUTATED | 38 | 15 | 22 |
| 12P WILD-TYPE | 62 | 99 | 54 |
Figure S83. Get High-res Image Gene #23: '12p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
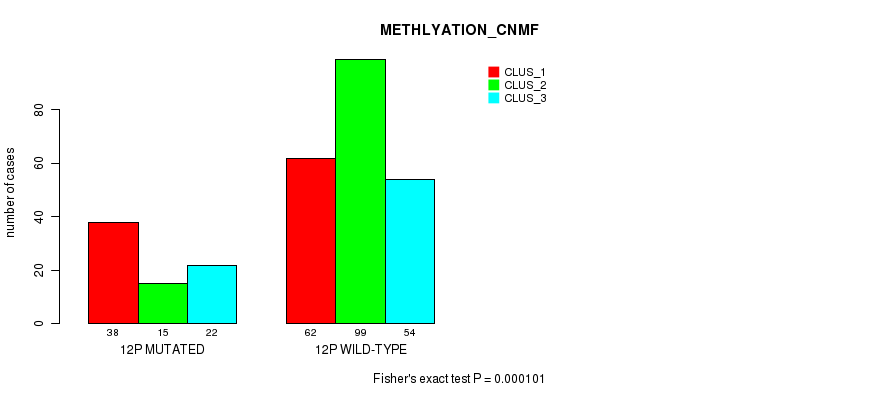
P value = 3.83e-12 (Chi-square test), Q value = 1.7e-09
Table S84. Gene #23: '12p' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 12P MUTATED | 4 | 17 | 31 | 15 | 7 | 31 |
| 12P WILD-TYPE | 96 | 71 | 53 | 60 | 36 | 26 |
Figure S84. Get High-res Image Gene #23: '12p' versus Molecular Subtype #5: 'RPPA_CNMF'
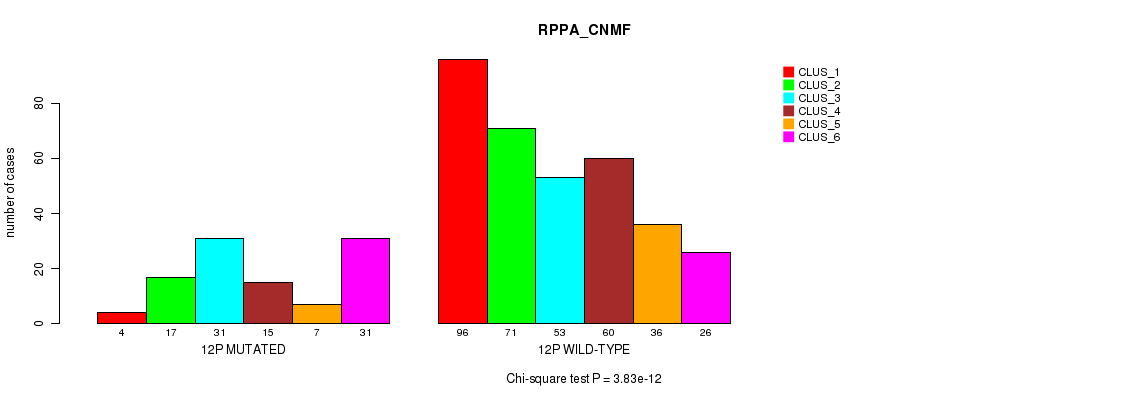
P value = 5.61e-11 (Fisher's exact test), Q value = 2.4e-08
Table S85. Gene #23: '12p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 12P MUTATED | 24 | 77 | 19 |
| 12P WILD-TYPE | 181 | 112 | 85 |
Figure S85. Get High-res Image Gene #23: '12p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
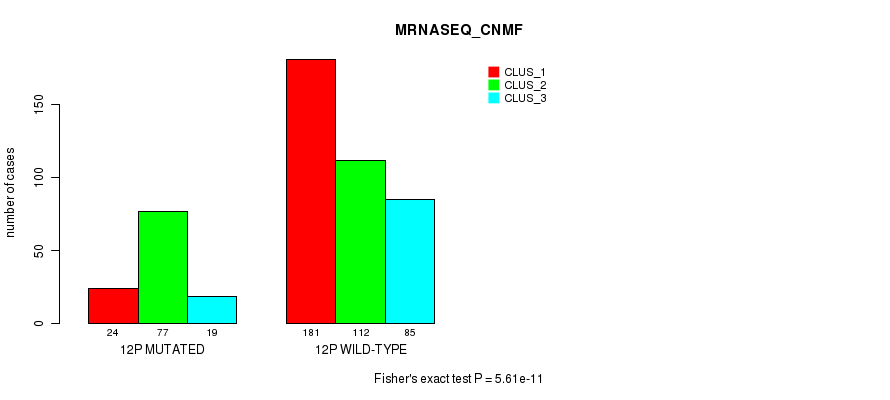
P value = 1.54e-10 (Fisher's exact test), Q value = 6.5e-08
Table S86. Gene #23: '12p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 12P MUTATED | 26 | 81 | 13 |
| 12P WILD-TYPE | 189 | 125 | 64 |
Figure S86. Get High-res Image Gene #23: '12p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
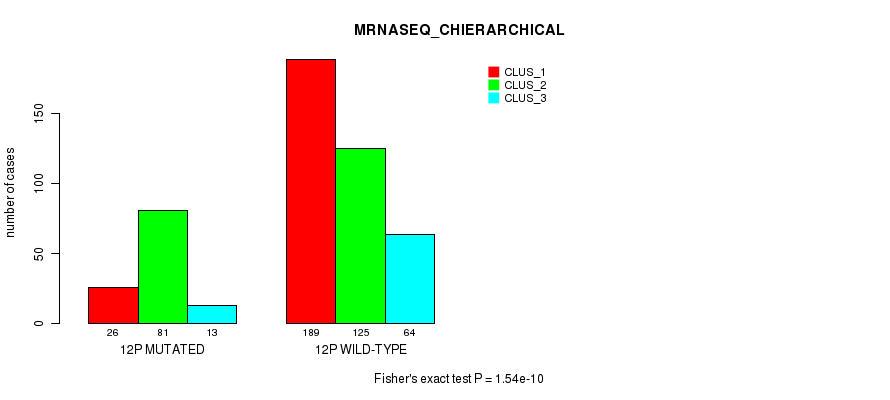
P value = 5.93e-09 (Fisher's exact test), Q value = 2.4e-06
Table S87. Gene #23: '12p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 12P MUTATED | 23 | 28 | 68 |
| 12P WILD-TYPE | 95 | 175 | 97 |
Figure S87. Get High-res Image Gene #23: '12p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
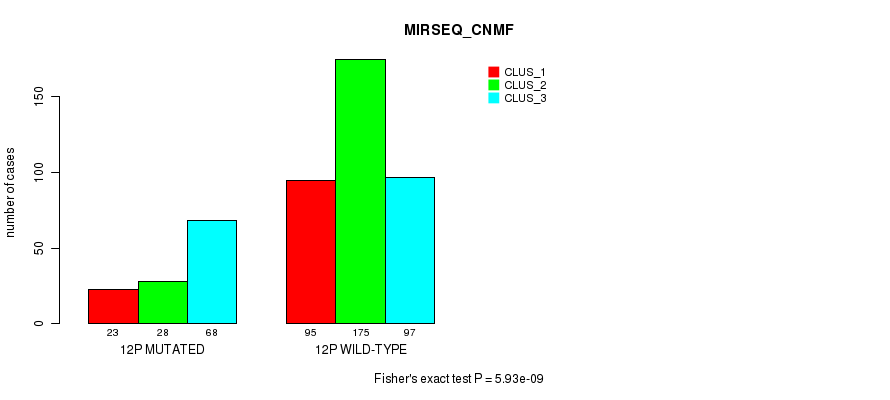
P value = 6.65e-07 (Fisher's exact test), Q value = 0.00026
Table S88. Gene #23: '12p' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 12P MUTATED | 10 | 8 | 33 |
| 12P WILD-TYPE | 43 | 87 | 45 |
Figure S88. Get High-res Image Gene #23: '12p' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
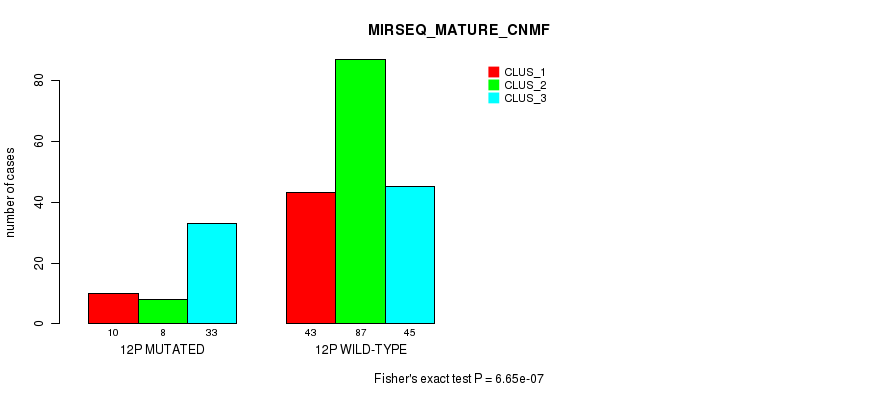
P value = 6.11e-14 (Fisher's exact test), Q value = 2.8e-11
Table S89. Gene #24: '12q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 12Q MUTATED | 71 | 18 | 34 |
| 12Q WILD-TYPE | 100 | 194 | 87 |
Figure S89. Get High-res Image Gene #24: '12q' versus Molecular Subtype #3: 'CN_CNMF'
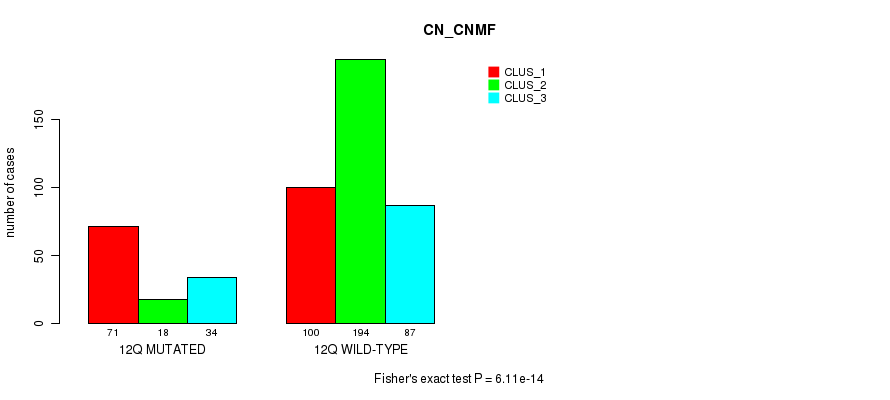
P value = 6.02e-05 (Fisher's exact test), Q value = 0.021
Table S90. Gene #24: '12q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 12Q MUTATED | 39 | 15 | 22 |
| 12Q WILD-TYPE | 61 | 99 | 54 |
Figure S90. Get High-res Image Gene #24: '12q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
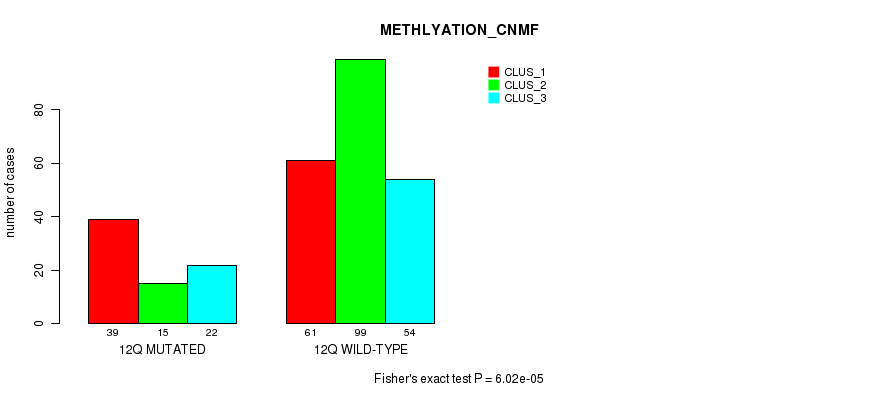
P value = 1.87e-13 (Chi-square test), Q value = 8.4e-11
Table S91. Gene #24: '12q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 12Q MUTATED | 4 | 17 | 31 | 15 | 7 | 33 |
| 12Q WILD-TYPE | 96 | 71 | 53 | 60 | 36 | 24 |
Figure S91. Get High-res Image Gene #24: '12q' versus Molecular Subtype #5: 'RPPA_CNMF'
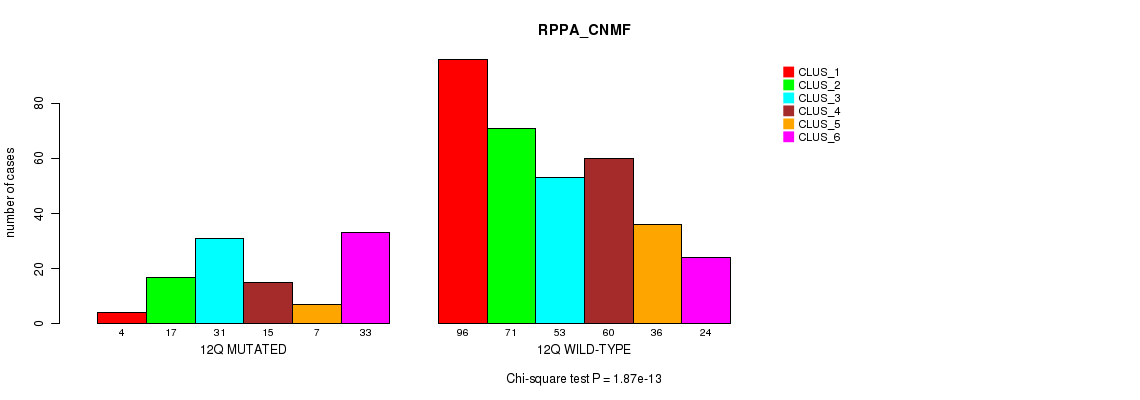
P value = 3.61e-11 (Fisher's exact test), Q value = 1.5e-08
Table S92. Gene #24: '12q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 12Q MUTATED | 24 | 78 | 20 |
| 12Q WILD-TYPE | 181 | 111 | 84 |
Figure S92. Get High-res Image Gene #24: '12q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
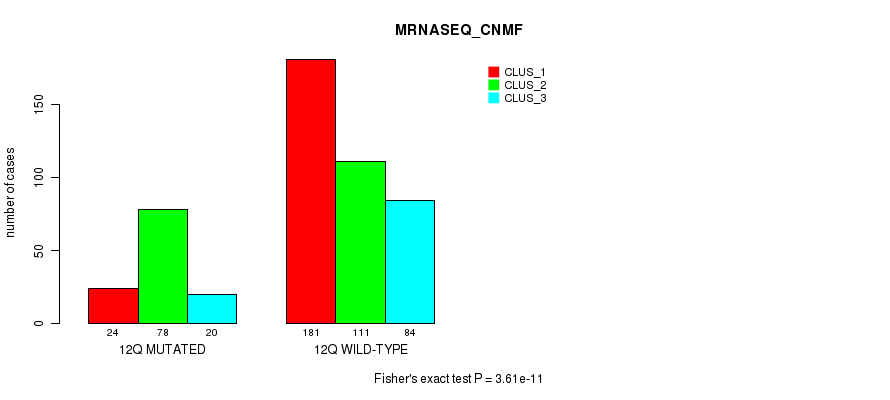
P value = 3.61e-11 (Fisher's exact test), Q value = 1.5e-08
Table S93. Gene #24: '12q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 12Q MUTATED | 26 | 83 | 13 |
| 12Q WILD-TYPE | 189 | 123 | 64 |
Figure S93. Get High-res Image Gene #24: '12q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
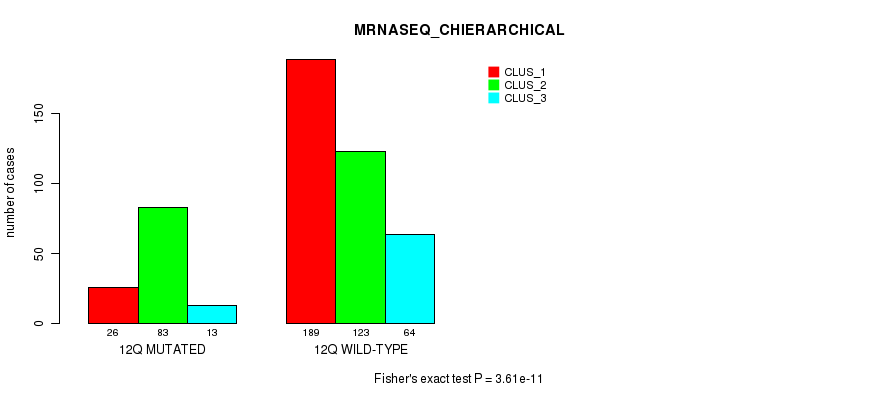
P value = 1.21e-09 (Fisher's exact test), Q value = 5e-07
Table S94. Gene #24: '12q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 12Q MUTATED | 23 | 28 | 70 |
| 12Q WILD-TYPE | 95 | 175 | 95 |
Figure S94. Get High-res Image Gene #24: '12q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
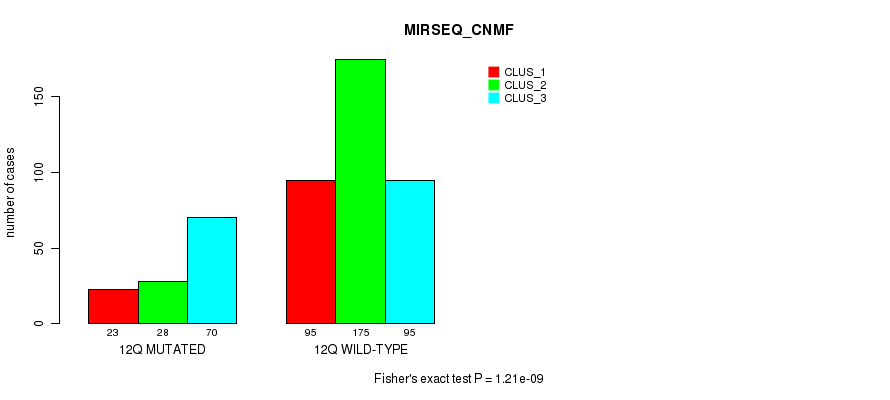
P value = 2.52e-07 (Fisher's exact test), Q value = 1e-04
Table S95. Gene #24: '12q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 12Q MUTATED | 10 | 8 | 34 |
| 12Q WILD-TYPE | 43 | 87 | 44 |
Figure S95. Get High-res Image Gene #24: '12q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
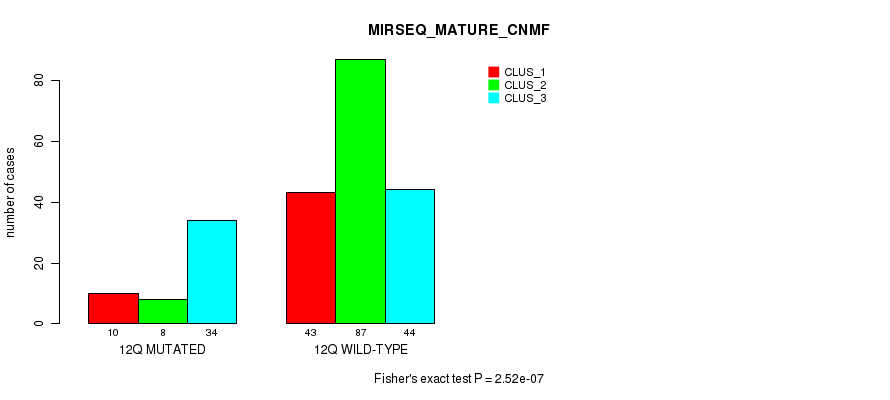
P value = 0.000654 (Fisher's exact test), Q value = 0.2
Table S96. Gene #24: '12q' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 86 | 69 | 71 |
| 12Q MUTATED | 10 | 26 | 16 |
| 12Q WILD-TYPE | 76 | 43 | 55 |
Figure S96. Get High-res Image Gene #24: '12q' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
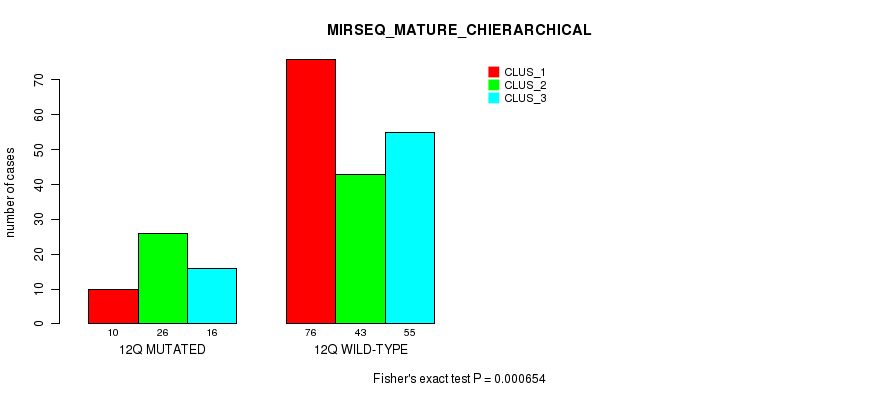
P value = 4.88e-10 (Fisher's exact test), Q value = 2e-07
Table S97. Gene #25: '13q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 13Q MUTATED | 48 | 12 | 31 |
| 13Q WILD-TYPE | 123 | 200 | 90 |
Figure S97. Get High-res Image Gene #25: '13q' versus Molecular Subtype #3: 'CN_CNMF'
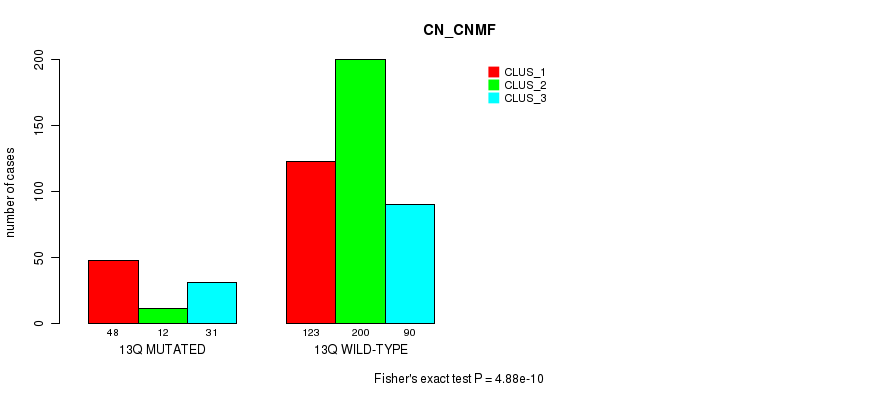
P value = 0.000741 (Fisher's exact test), Q value = 0.22
Table S98. Gene #25: '13q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 13Q MUTATED | 30 | 12 | 20 |
| 13Q WILD-TYPE | 70 | 102 | 56 |
Figure S98. Get High-res Image Gene #25: '13q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
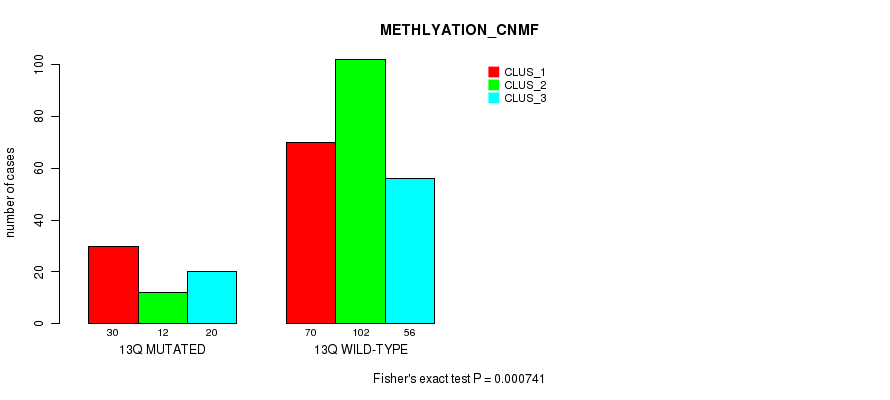
P value = 5.85e-05 (Chi-square test), Q value = 0.02
Table S99. Gene #25: '13q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 13Q MUTATED | 6 | 16 | 19 | 15 | 4 | 21 |
| 13Q WILD-TYPE | 94 | 72 | 65 | 60 | 39 | 36 |
Figure S99. Get High-res Image Gene #25: '13q' versus Molecular Subtype #5: 'RPPA_CNMF'
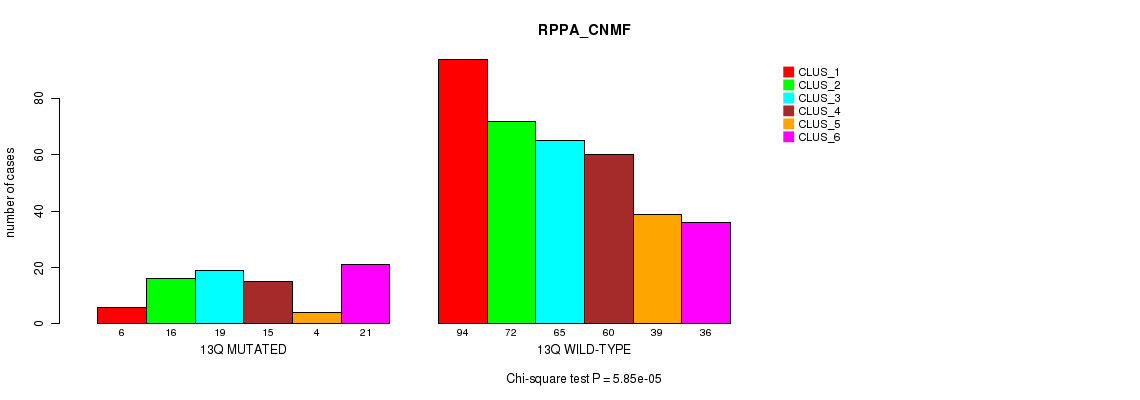
P value = 2.41e-05 (Fisher's exact test), Q value = 0.0086
Table S100. Gene #25: '13q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 13Q MUTATED | 19 | 50 | 22 |
| 13Q WILD-TYPE | 186 | 139 | 82 |
Figure S100. Get High-res Image Gene #25: '13q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
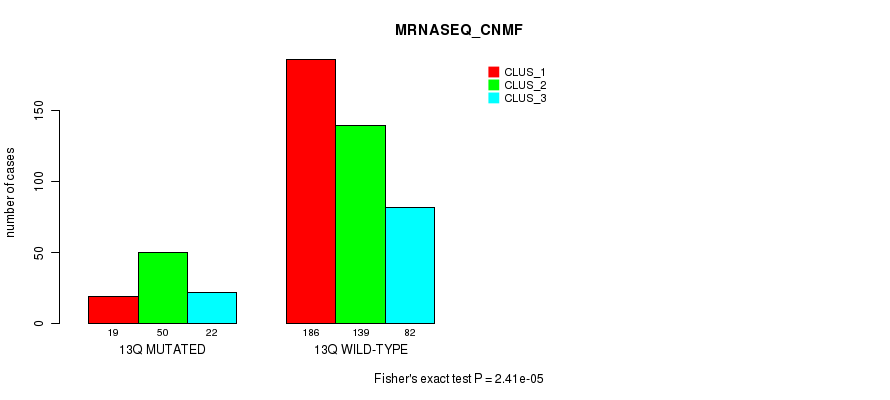
P value = 3.13e-06 (Fisher's exact test), Q value = 0.0012
Table S101. Gene #25: '13q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 13Q MUTATED | 19 | 56 | 16 |
| 13Q WILD-TYPE | 196 | 150 | 61 |
Figure S101. Get High-res Image Gene #25: '13q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
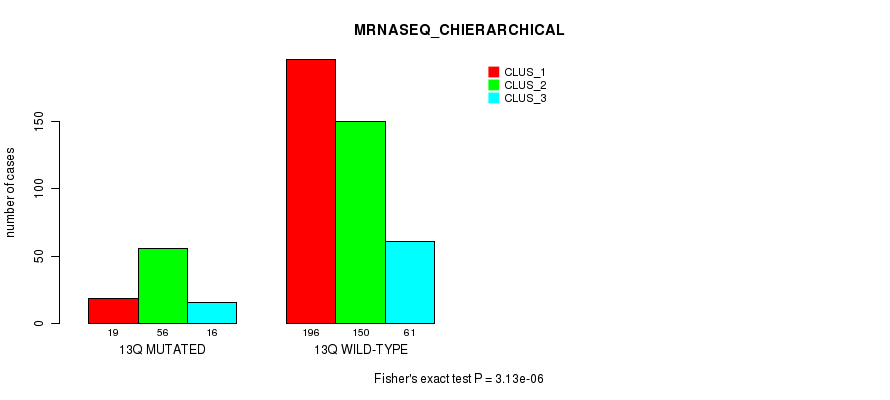
P value = 1.1e-05 (Fisher's exact test), Q value = 0.0041
Table S102. Gene #25: '13q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 13Q MUTATED | 22 | 19 | 47 |
| 13Q WILD-TYPE | 96 | 184 | 118 |
Figure S102. Get High-res Image Gene #25: '13q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
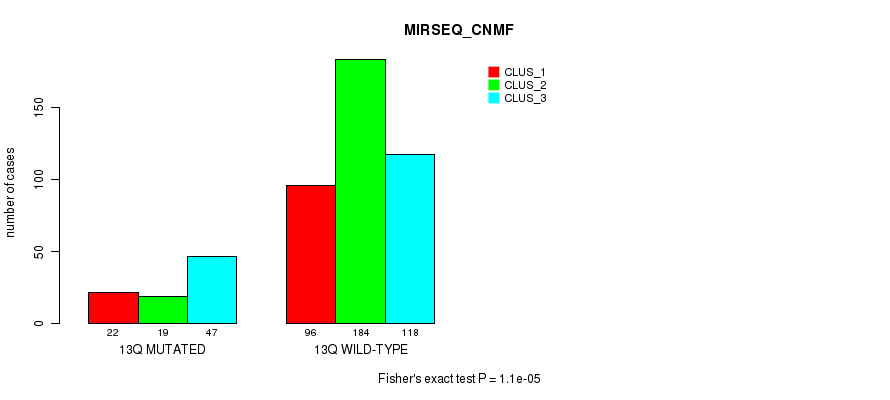
P value = 0.000303 (Fisher's exact test), Q value = 0.096
Table S103. Gene #25: '13q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 13Q MUTATED | 35 | 16 | 37 |
| 13Q WILD-TYPE | 229 | 24 | 145 |
Figure S103. Get High-res Image Gene #25: '13q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
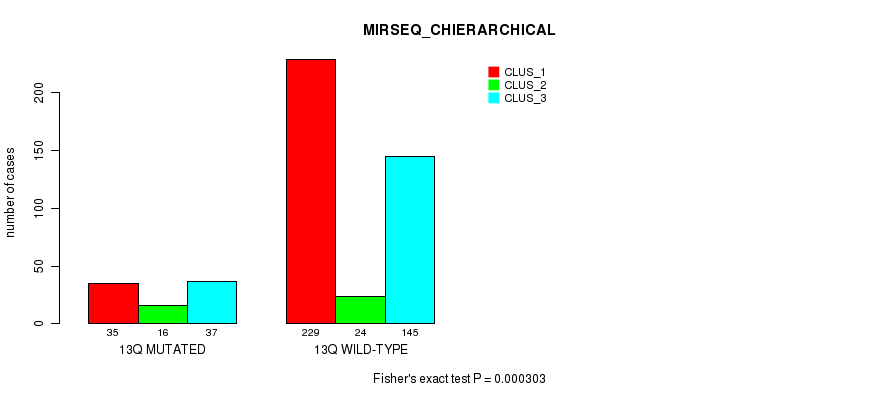
P value = 4.78e-05 (Fisher's exact test), Q value = 0.017
Table S104. Gene #26: '14q' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 24 | 13 |
| 14Q MUTATED | 10 | 20 | 3 |
| 14Q WILD-TYPE | 23 | 4 | 10 |
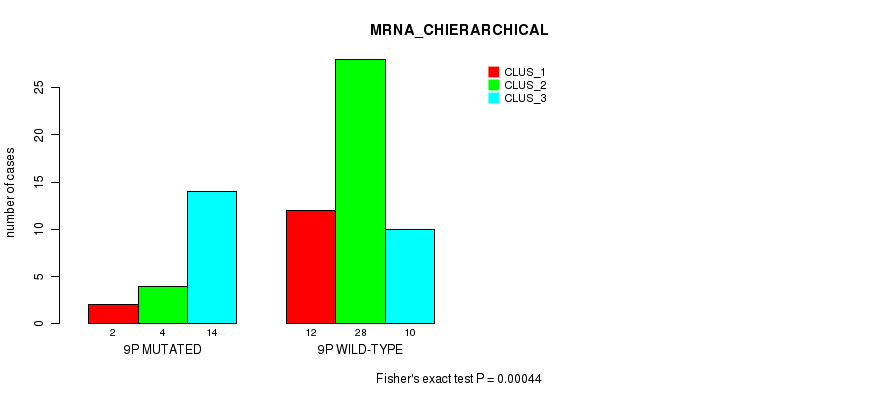
Figure S104. Get High-res Image Gene #26: '14q' versus Molecular Subtype #1: 'MRNA_CNMF'
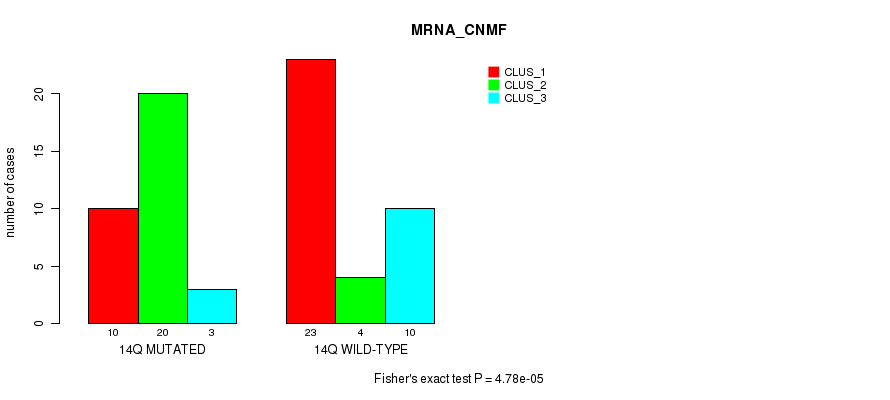
P value = 4.72e-05 (Fisher's exact test), Q value = 0.016
Table S105. Gene #26: '14q' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 14 | 32 | 24 |
| 14Q MUTATED | 4 | 9 | 20 |
| 14Q WILD-TYPE | 10 | 23 | 4 |
Figure S105. Get High-res Image Gene #26: '14q' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
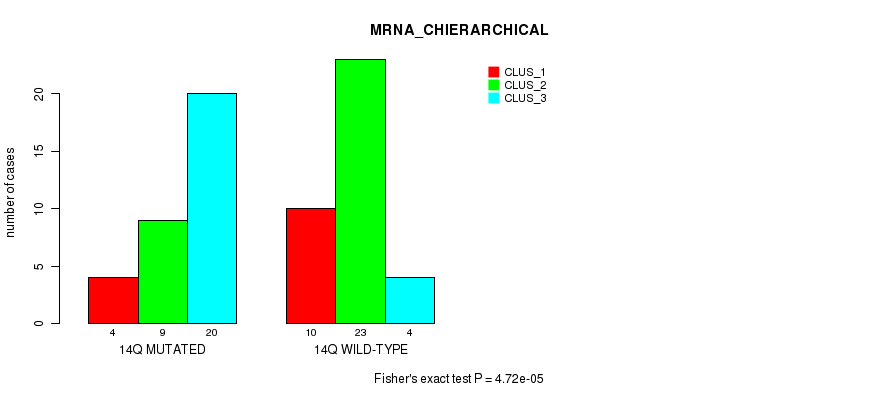
P value = 1.14e-19 (Fisher's exact test), Q value = 5.4e-17
Table S106. Gene #26: '14q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 14Q MUTATED | 123 | 53 | 51 |
| 14Q WILD-TYPE | 48 | 159 | 70 |
Figure S106. Get High-res Image Gene #26: '14q' versus Molecular Subtype #3: 'CN_CNMF'
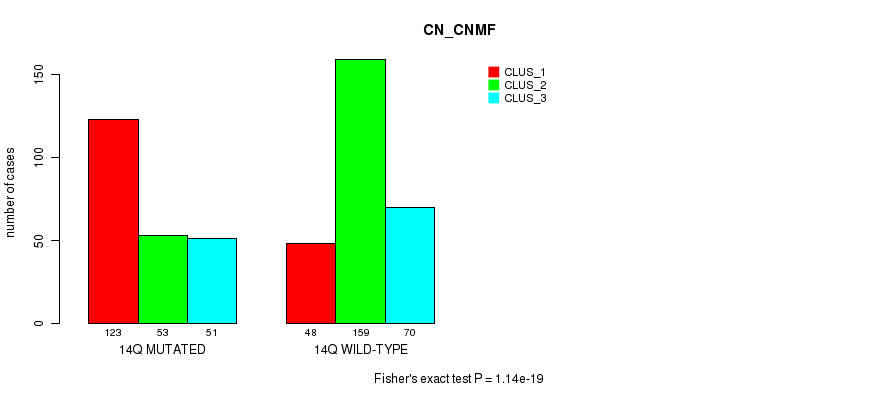
P value = 7.43e-09 (Fisher's exact test), Q value = 3e-06
Table S107. Gene #26: '14q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 14Q MUTATED | 62 | 26 | 40 |
| 14Q WILD-TYPE | 38 | 88 | 36 |
Figure S107. Get High-res Image Gene #26: '14q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
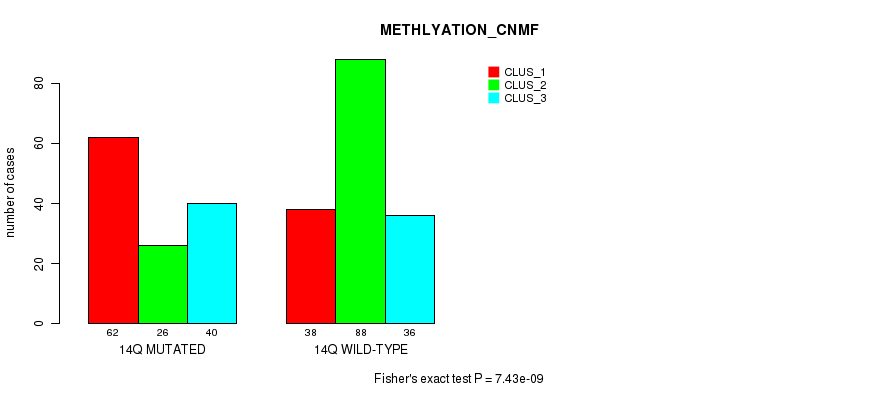
P value = 3.64e-13 (Chi-square test), Q value = 1.6e-10
Table S108. Gene #26: '14q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 14Q MUTATED | 28 | 30 | 63 | 30 | 13 | 41 |
| 14Q WILD-TYPE | 72 | 58 | 21 | 45 | 30 | 16 |
Figure S108. Get High-res Image Gene #26: '14q' versus Molecular Subtype #5: 'RPPA_CNMF'
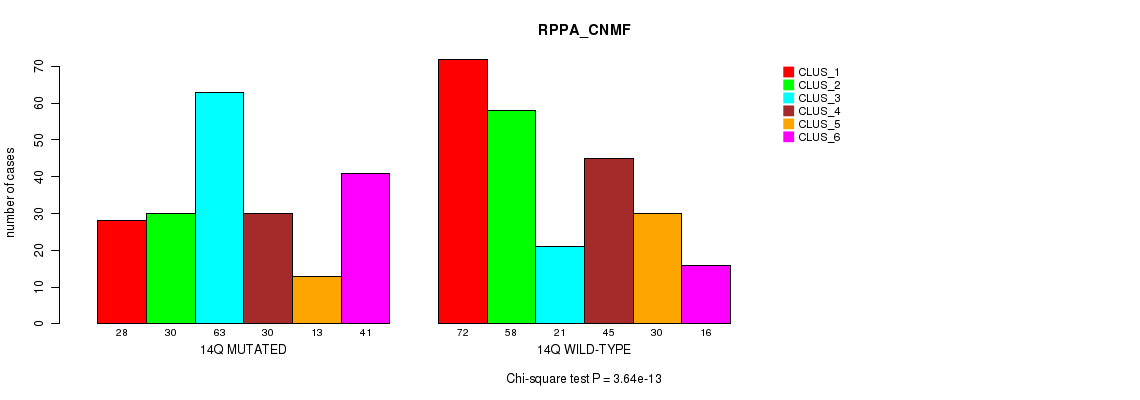
P value = 1.79e-07 (Fisher's exact test), Q value = 7.1e-05
Table S109. Gene #26: '14q' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 186 | 152 | 109 |
| 14Q MUTATED | 57 | 90 | 58 |
| 14Q WILD-TYPE | 129 | 62 | 51 |
Figure S109. Get High-res Image Gene #26: '14q' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
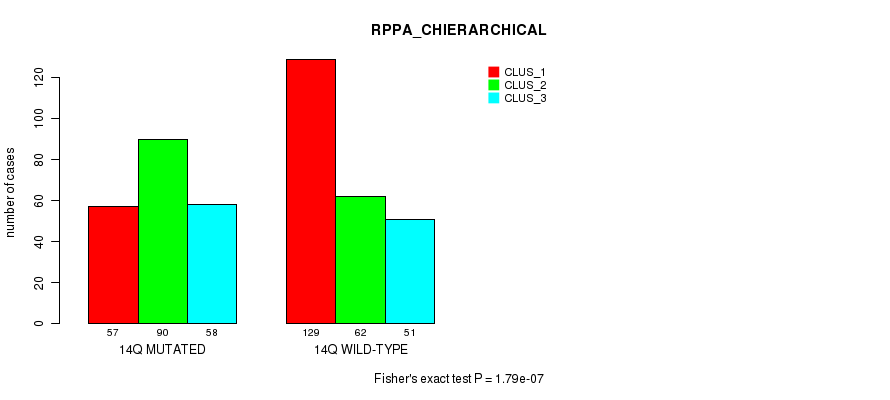
P value = 5.27e-18 (Fisher's exact test), Q value = 2.5e-15
Table S110. Gene #26: '14q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 14Q MUTATED | 67 | 131 | 24 |
| 14Q WILD-TYPE | 138 | 58 | 80 |
Figure S110. Get High-res Image Gene #26: '14q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
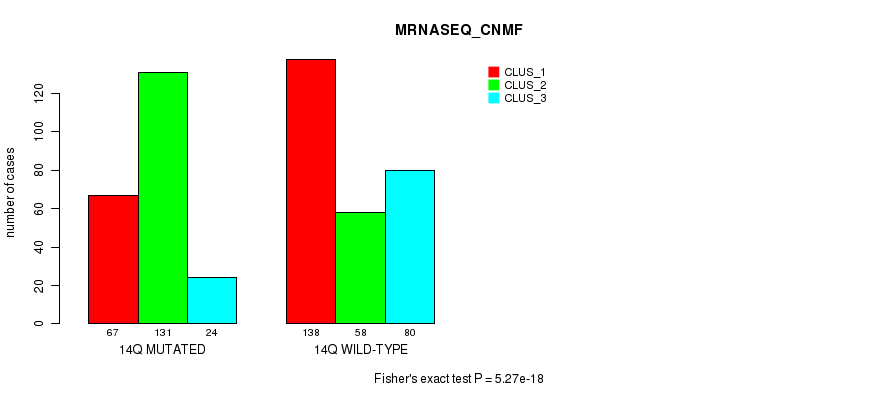
P value = 1.69e-15 (Fisher's exact test), Q value = 7.8e-13
Table S111. Gene #26: '14q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 14Q MUTATED | 68 | 136 | 18 |
| 14Q WILD-TYPE | 147 | 70 | 59 |
Figure S111. Get High-res Image Gene #26: '14q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
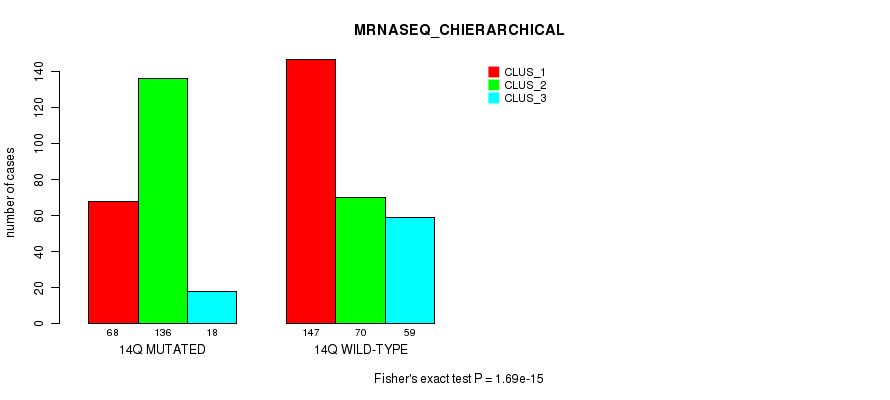
P value = 1.24e-14 (Fisher's exact test), Q value = 5.7e-12
Table S112. Gene #26: '14q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 14Q MUTATED | 39 | 65 | 116 |
| 14Q WILD-TYPE | 79 | 138 | 49 |
Figure S112. Get High-res Image Gene #26: '14q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
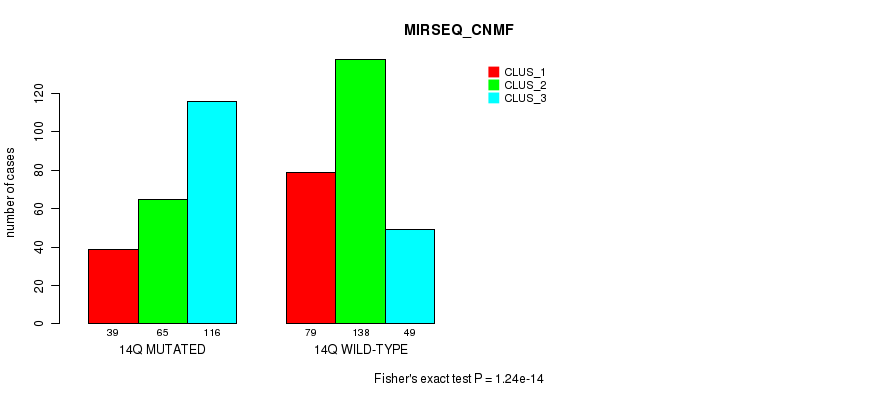
P value = 3e-04 (Fisher's exact test), Q value = 0.096
Table S113. Gene #26: '14q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 14Q MUTATED | 23 | 32 | 50 |
| 14Q WILD-TYPE | 30 | 63 | 28 |
Figure S113. Get High-res Image Gene #26: '14q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
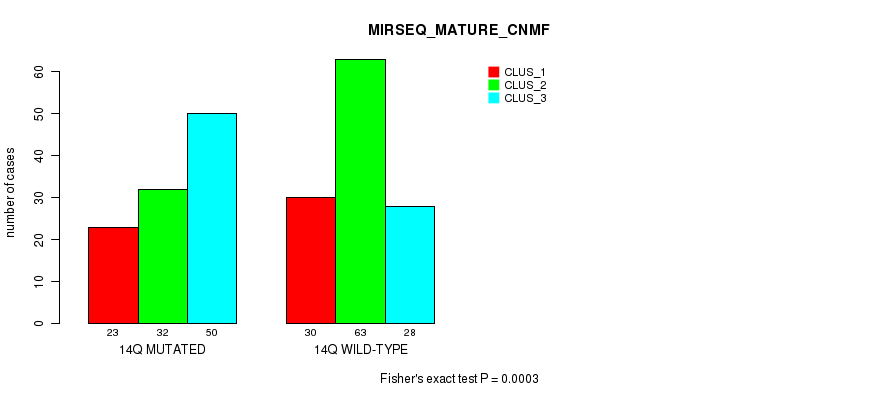
P value = 0.000568 (Fisher's exact test), Q value = 0.17
Table S114. Gene #26: '14q' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 86 | 69 | 71 |
| 14Q MUTATED | 30 | 45 | 30 |
| 14Q WILD-TYPE | 56 | 24 | 41 |
Figure S114. Get High-res Image Gene #26: '14q' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
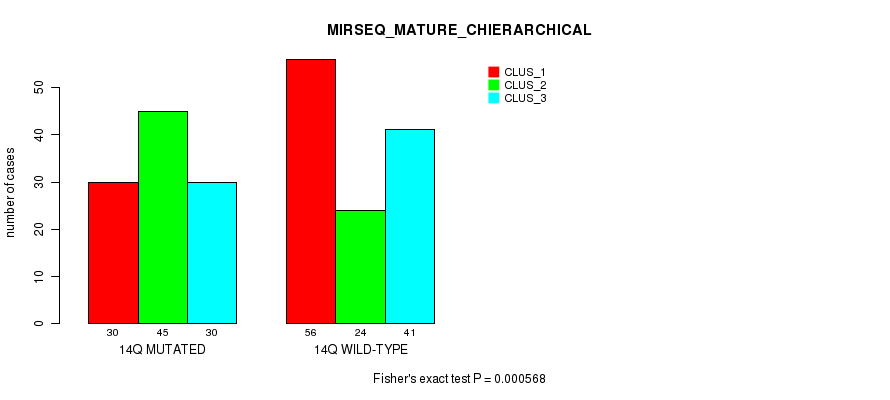
P value = 5.02e-12 (Fisher's exact test), Q value = 2.2e-09
Table S115. Gene #27: '15q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 15Q MUTATED | 40 | 3 | 18 |
| 15Q WILD-TYPE | 131 | 209 | 103 |
Figure S115. Get High-res Image Gene #27: '15q' versus Molecular Subtype #3: 'CN_CNMF'
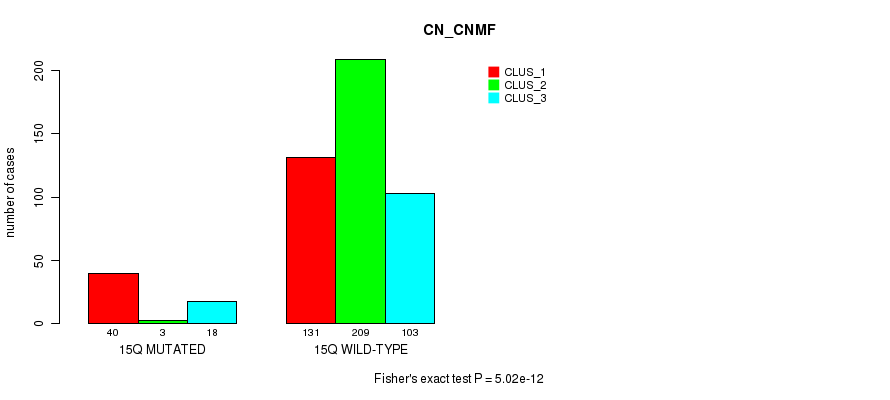
P value = 0.000472 (Fisher's exact test), Q value = 0.15
Table S116. Gene #27: '15q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 15Q MUTATED | 15 | 39 | 6 |
| 15Q WILD-TYPE | 200 | 167 | 71 |
Figure S116. Get High-res Image Gene #27: '15q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
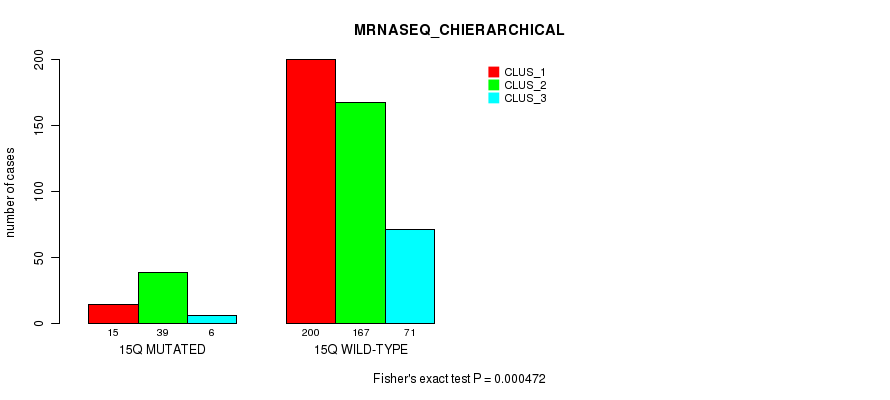
P value = 7.59e-05 (Fisher's exact test), Q value = 0.026
Table S117. Gene #28: '16p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 16P MUTATED | 54 | 29 | 32 |
| 16P WILD-TYPE | 117 | 183 | 89 |
Figure S117. Get High-res Image Gene #28: '16p' versus Molecular Subtype #3: 'CN_CNMF'
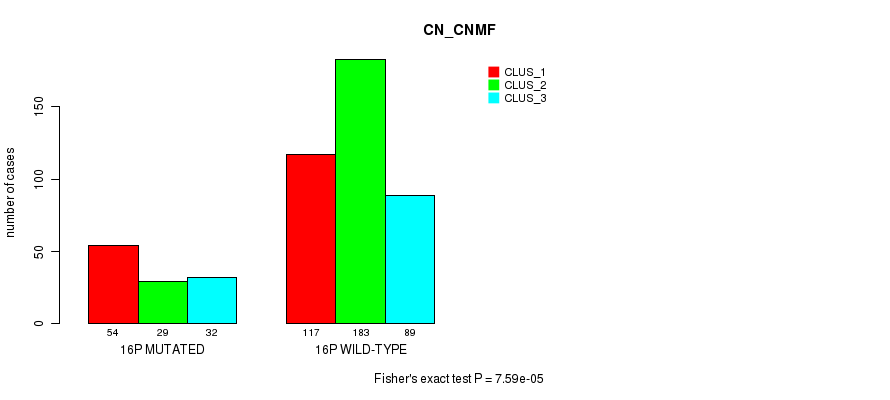
P value = 0.000225 (Fisher's exact test), Q value = 0.072
Table S118. Gene #28: '16p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 16P MUTATED | 31 | 60 | 24 |
| 16P WILD-TYPE | 184 | 146 | 53 |
Figure S118. Get High-res Image Gene #28: '16p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
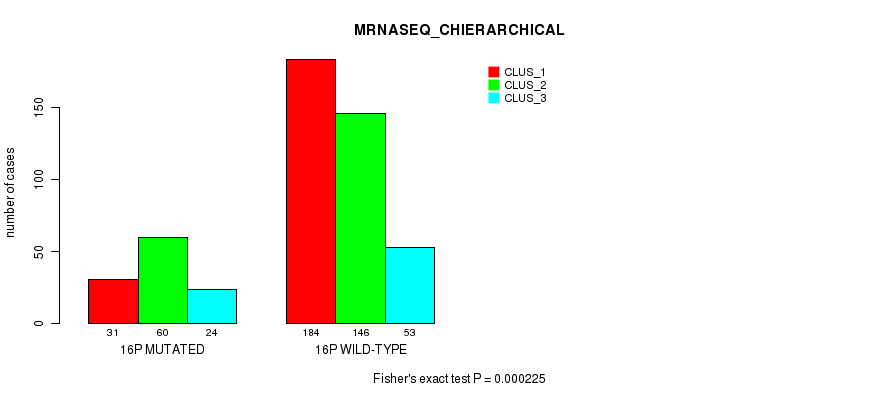
P value = 2.99e-06 (Fisher's exact test), Q value = 0.0012
Table S119. Gene #29: '16q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 16Q MUTATED | 55 | 25 | 32 |
| 16Q WILD-TYPE | 116 | 187 | 89 |
Figure S119. Get High-res Image Gene #29: '16q' versus Molecular Subtype #3: 'CN_CNMF'
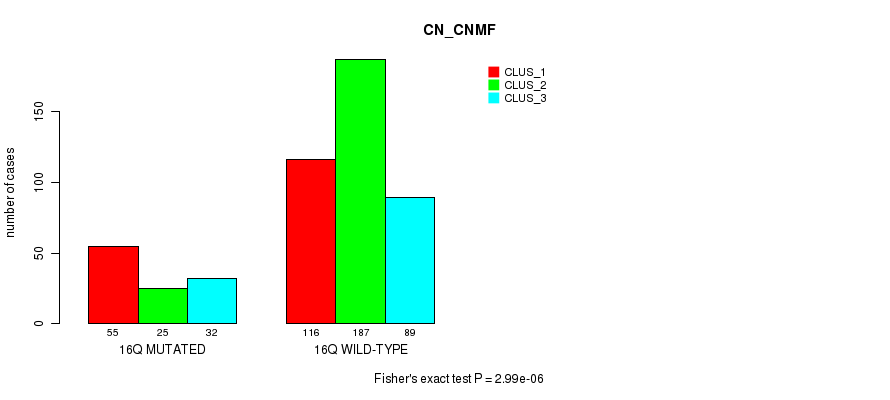
P value = 0.000244 (Fisher's exact test), Q value = 0.078
Table S120. Gene #29: '16q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 16Q MUTATED | 30 | 61 | 21 |
| 16Q WILD-TYPE | 185 | 145 | 56 |
Figure S120. Get High-res Image Gene #29: '16q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
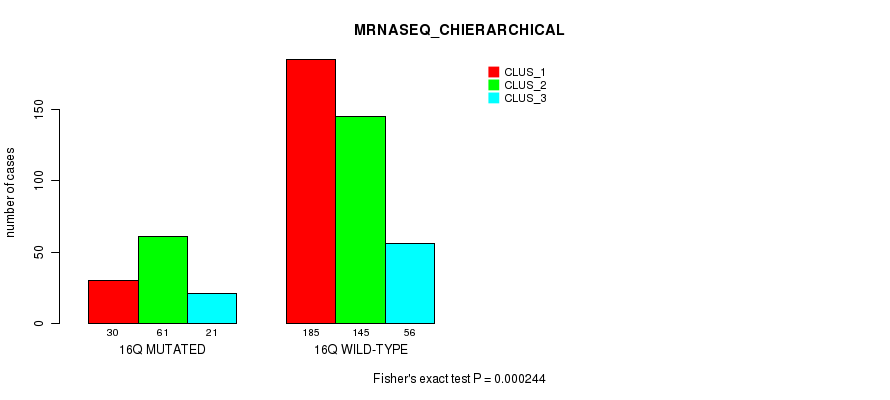
P value = 8.7e-06 (Fisher's exact test), Q value = 0.0033
Table S121. Gene #30: '17p' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 24 | 13 |
| 17P MUTATED | 1 | 5 | 9 |
| 17P WILD-TYPE | 32 | 19 | 4 |
Figure S121. Get High-res Image Gene #30: '17p' versus Molecular Subtype #1: 'MRNA_CNMF'
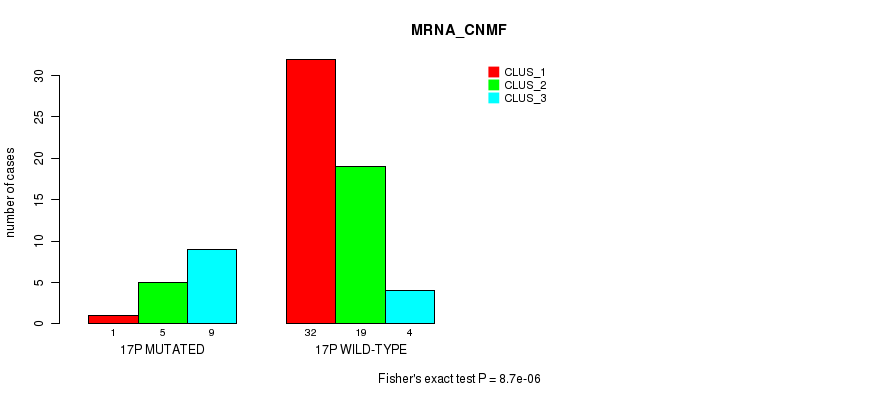
P value = 3.14e-05 (Fisher's exact test), Q value = 0.011
Table S122. Gene #30: '17p' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 14 | 32 | 24 |
| 17P MUTATED | 9 | 1 | 5 |
| 17P WILD-TYPE | 5 | 31 | 19 |
Figure S122. Get High-res Image Gene #30: '17p' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
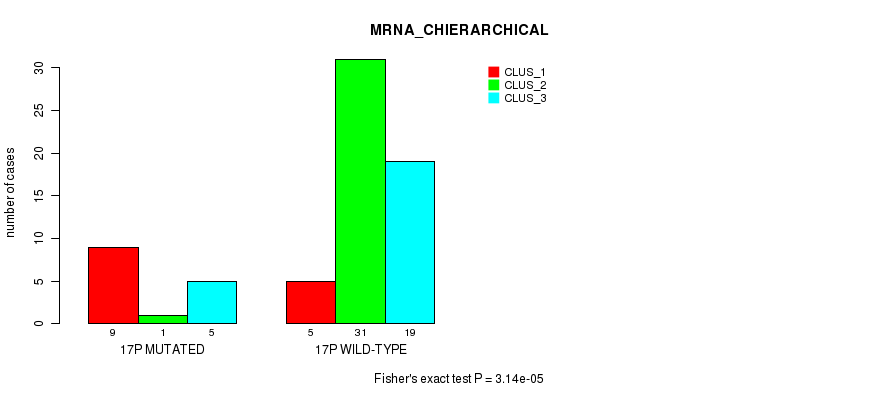
P value = 5.44e-15 (Fisher's exact test), Q value = 2.5e-12
Table S123. Gene #30: '17p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 17P MUTATED | 41 | 2 | 27 |
| 17P WILD-TYPE | 130 | 210 | 94 |
Figure S123. Get High-res Image Gene #30: '17p' versus Molecular Subtype #3: 'CN_CNMF'
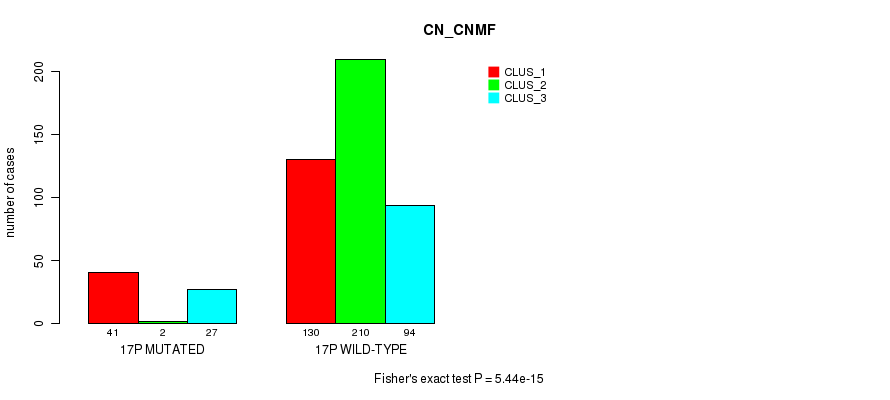
P value = 0.00033 (Fisher's exact test), Q value = 0.1
Table S124. Gene #30: '17p' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 186 | 152 | 109 |
| 17P MUTATED | 15 | 17 | 27 |
| 17P WILD-TYPE | 171 | 135 | 82 |
Figure S124. Get High-res Image Gene #30: '17p' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
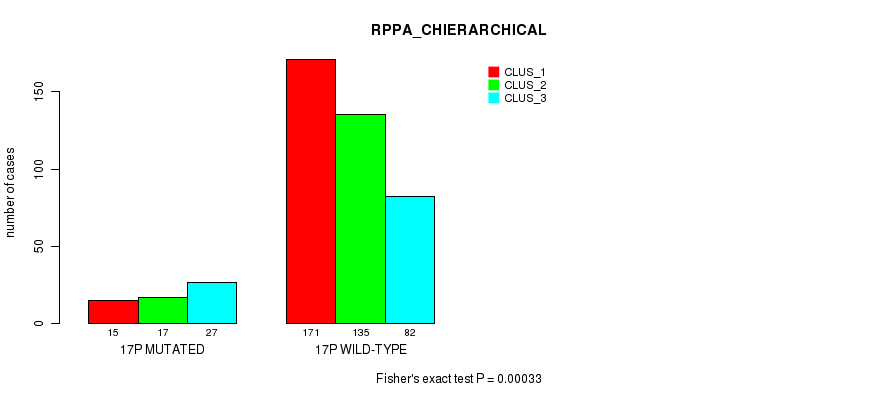
P value = 1.02e-06 (Fisher's exact test), Q value = 4e-04
Table S125. Gene #30: '17p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 17P MUTATED | 10 | 37 | 23 |
| 17P WILD-TYPE | 195 | 152 | 81 |
Figure S125. Get High-res Image Gene #30: '17p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
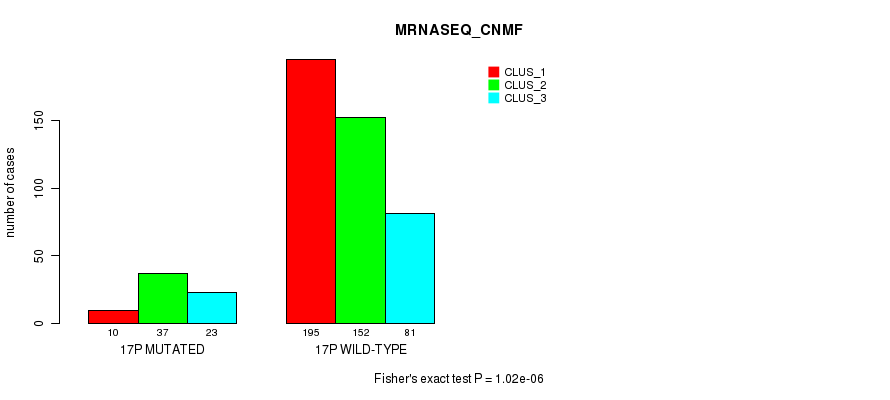
P value = 1.58e-08 (Fisher's exact test), Q value = 6.4e-06
Table S126. Gene #30: '17p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 17P MUTATED | 10 | 37 | 23 |
| 17P WILD-TYPE | 205 | 169 | 54 |
Figure S126. Get High-res Image Gene #30: '17p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
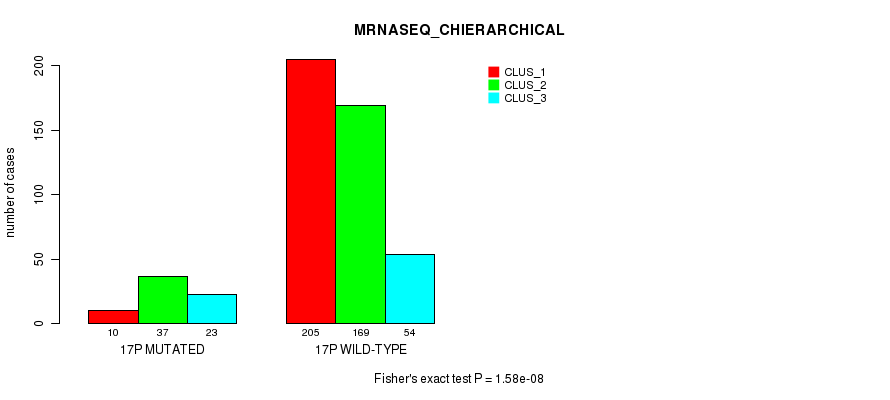
P value = 3.71e-08 (Fisher's exact test), Q value = 1.5e-05
Table S127. Gene #30: '17p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 17P MUTATED | 16 | 10 | 43 |
| 17P WILD-TYPE | 102 | 193 | 122 |
Figure S127. Get High-res Image Gene #30: '17p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
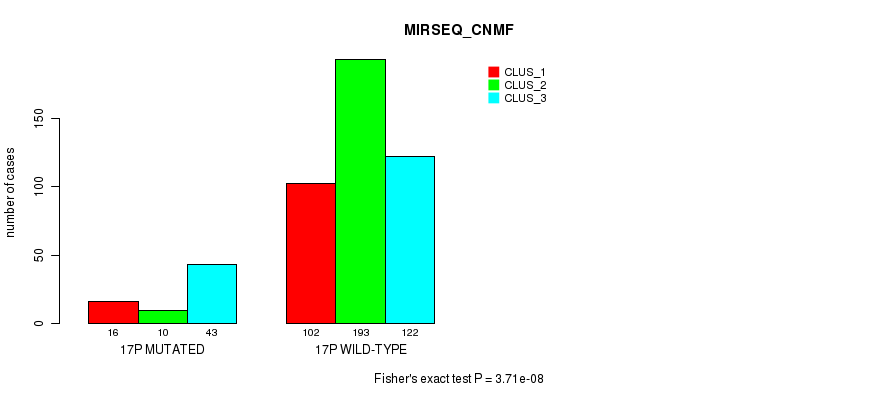
P value = 6.44e-11 (Fisher's exact test), Q value = 2.7e-08
Table S128. Gene #30: '17p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 17P MUTATED | 21 | 22 | 26 |
| 17P WILD-TYPE | 243 | 18 | 156 |
Figure S128. Get High-res Image Gene #30: '17p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
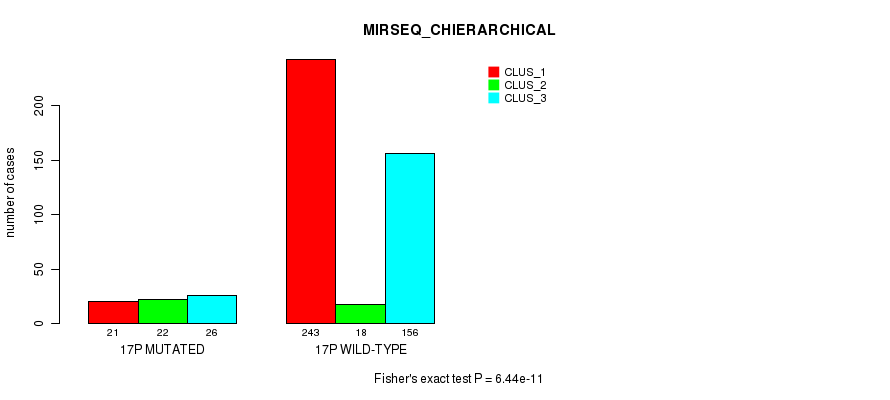
P value = 8.36e-05 (Fisher's exact test), Q value = 0.028
Table S129. Gene #31: '17q' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 24 | 13 |
| 17Q MUTATED | 1 | 5 | 8 |
| 17Q WILD-TYPE | 32 | 19 | 5 |
Figure S129. Get High-res Image Gene #31: '17q' versus Molecular Subtype #1: 'MRNA_CNMF'
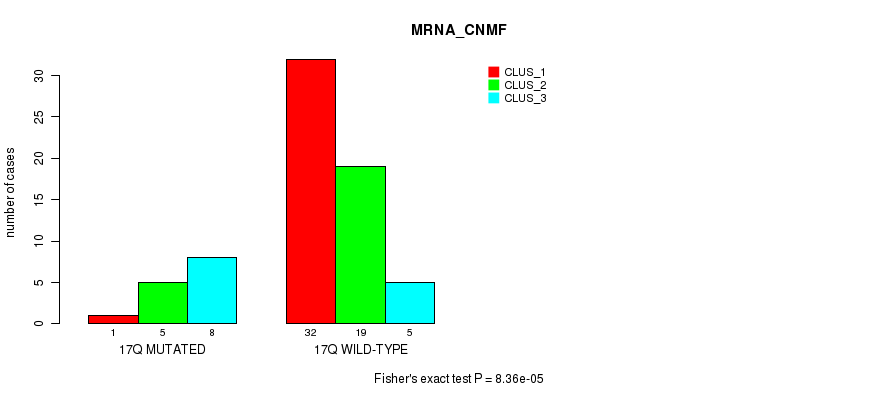
P value = 0.000175 (Fisher's exact test), Q value = 0.057
Table S130. Gene #31: '17q' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 14 | 32 | 24 |
| 17Q MUTATED | 8 | 1 | 5 |
| 17Q WILD-TYPE | 6 | 31 | 19 |
Figure S130. Get High-res Image Gene #31: '17q' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
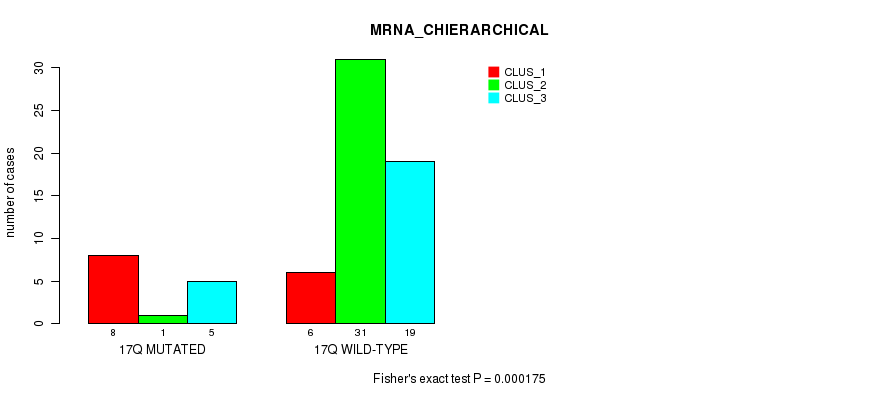
P value = 1.32e-10 (Fisher's exact test), Q value = 5.6e-08
Table S131. Gene #31: '17q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 17Q MUTATED | 31 | 3 | 24 |
| 17Q WILD-TYPE | 140 | 209 | 97 |
Figure S131. Get High-res Image Gene #31: '17q' versus Molecular Subtype #3: 'CN_CNMF'
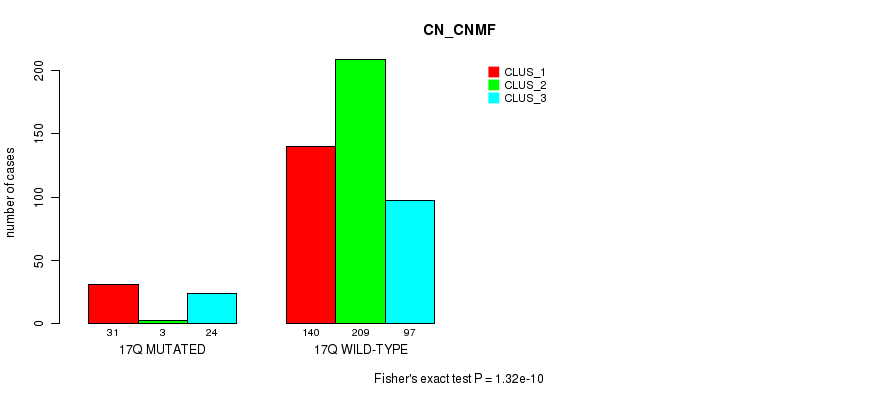
P value = 8.28e-05 (Fisher's exact test), Q value = 0.028
Table S132. Gene #31: '17q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 17Q MUTATED | 10 | 27 | 21 |
| 17Q WILD-TYPE | 195 | 162 | 83 |
Figure S132. Get High-res Image Gene #31: '17q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
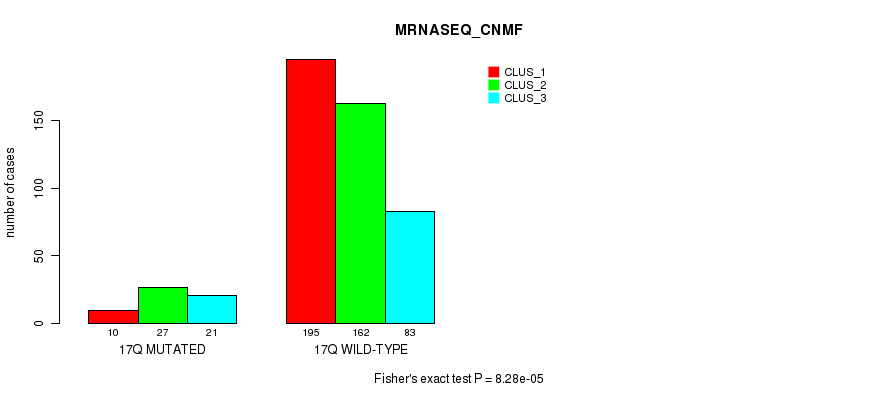
P value = 9.69e-07 (Fisher's exact test), Q value = 0.00038
Table S133. Gene #31: '17q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 17Q MUTATED | 10 | 27 | 21 |
| 17Q WILD-TYPE | 205 | 179 | 56 |
Figure S133. Get High-res Image Gene #31: '17q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
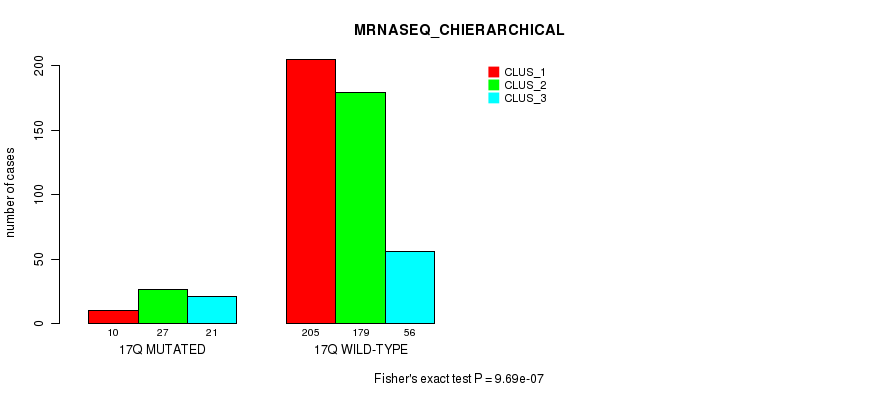
P value = 1.28e-05 (Fisher's exact test), Q value = 0.0047
Table S134. Gene #31: '17q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 17Q MUTATED | 15 | 9 | 33 |
| 17Q WILD-TYPE | 103 | 194 | 132 |
Figure S134. Get High-res Image Gene #31: '17q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
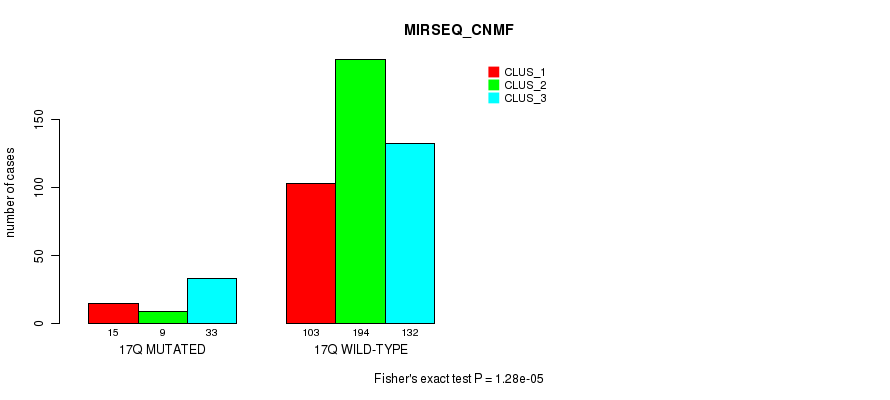
P value = 1.9e-10 (Fisher's exact test), Q value = 8e-08
Table S135. Gene #31: '17q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 17Q MUTATED | 17 | 20 | 20 |
| 17Q WILD-TYPE | 247 | 20 | 162 |
Figure S135. Get High-res Image Gene #31: '17q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
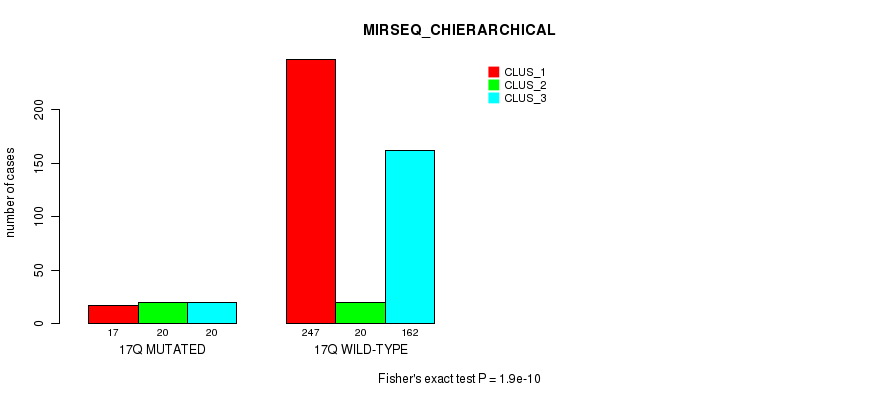
P value = 1.32e-13 (Fisher's exact test), Q value = 5.9e-11
Table S136. Gene #32: '18p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 18P MUTATED | 67 | 16 | 33 |
| 18P WILD-TYPE | 104 | 196 | 88 |
Figure S136. Get High-res Image Gene #32: '18p' versus Molecular Subtype #3: 'CN_CNMF'
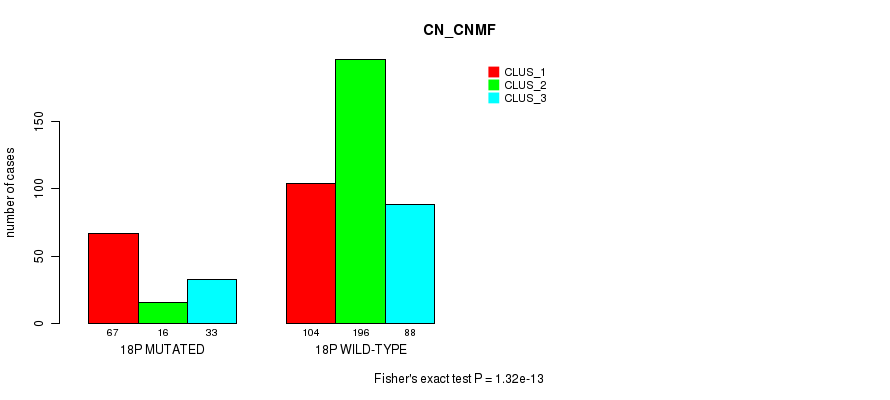
P value = 0.000198 (Fisher's exact test), Q value = 0.064
Table S137. Gene #32: '18p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 18P MUTATED | 32 | 13 | 24 |
| 18P WILD-TYPE | 68 | 101 | 52 |
Figure S137. Get High-res Image Gene #32: '18p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
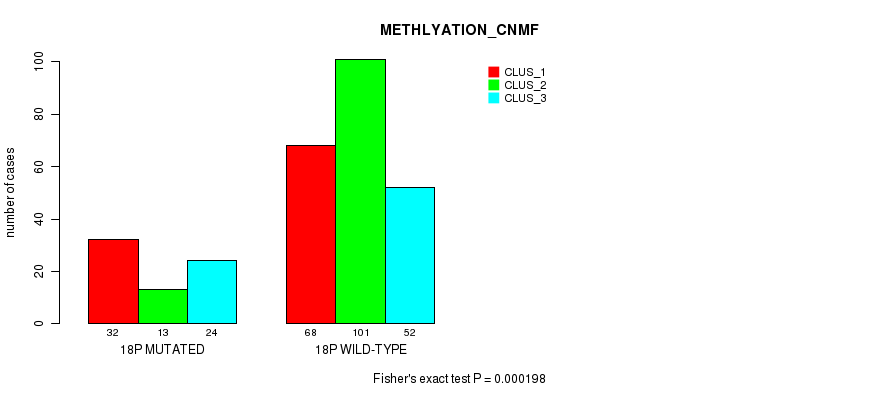
P value = 1.12e-05 (Chi-square test), Q value = 0.0042
Table S138. Gene #32: '18p' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 18P MUTATED | 12 | 17 | 27 | 18 | 5 | 26 |
| 18P WILD-TYPE | 88 | 71 | 57 | 57 | 38 | 31 |
Figure S138. Get High-res Image Gene #32: '18p' versus Molecular Subtype #5: 'RPPA_CNMF'
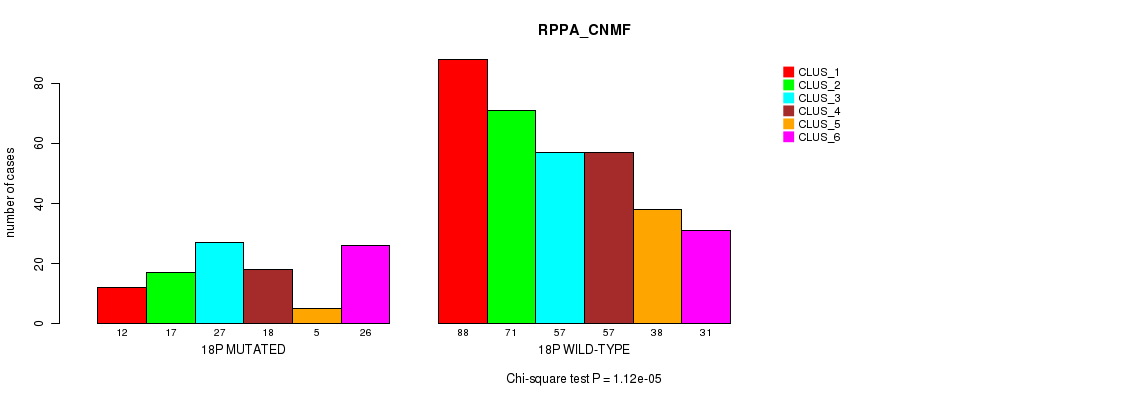
P value = 1.82e-14 (Fisher's exact test), Q value = 8.3e-12
Table S139. Gene #32: '18p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 18P MUTATED | 18 | 79 | 18 |
| 18P WILD-TYPE | 187 | 110 | 86 |
Figure S139. Get High-res Image Gene #32: '18p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
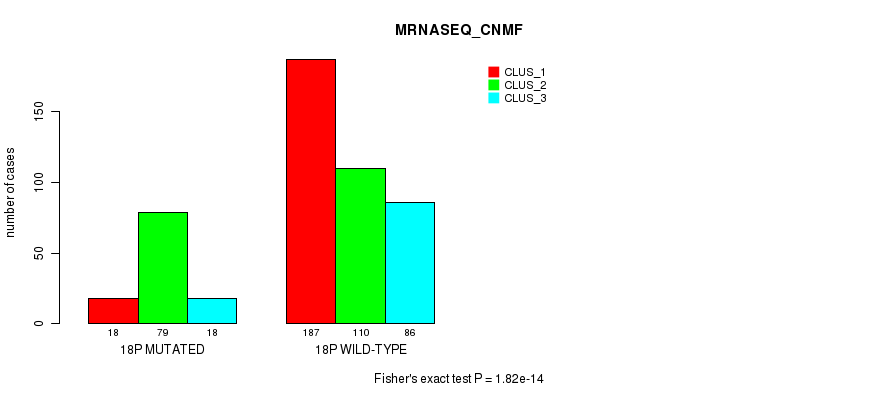
P value = 4.32e-12 (Fisher's exact test), Q value = 1.9e-09
Table S140. Gene #32: '18p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 18P MUTATED | 21 | 80 | 14 |
| 18P WILD-TYPE | 194 | 126 | 63 |
Figure S140. Get High-res Image Gene #32: '18p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
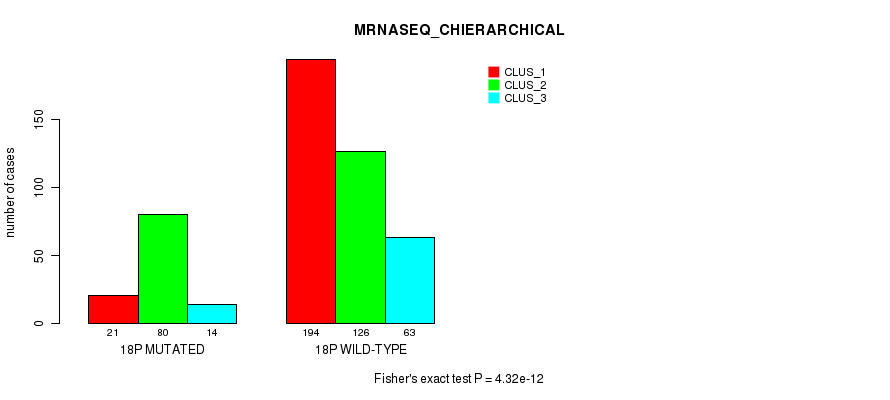
P value = 1.84e-09 (Fisher's exact test), Q value = 7.6e-07
Table S141. Gene #32: '18p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 18P MUTATED | 32 | 21 | 62 |
| 18P WILD-TYPE | 86 | 182 | 103 |
Figure S141. Get High-res Image Gene #32: '18p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
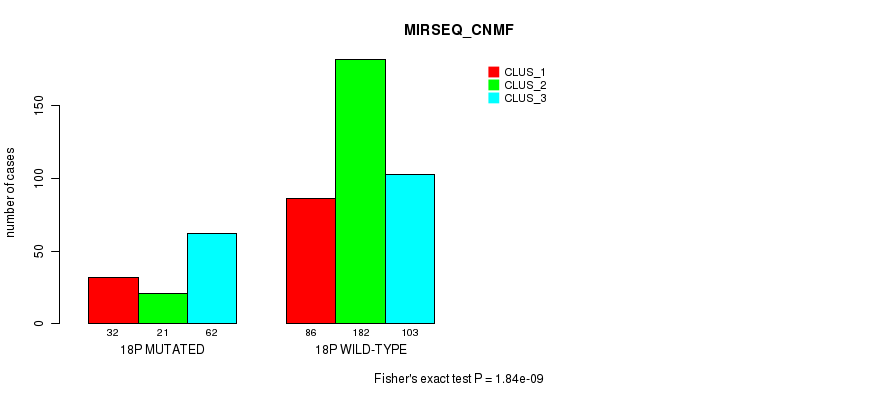
P value = 9.79e-07 (Fisher's exact test), Q value = 0.00038
Table S142. Gene #32: '18p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 18P MUTATED | 41 | 20 | 54 |
| 18P WILD-TYPE | 223 | 20 | 128 |
Figure S142. Get High-res Image Gene #32: '18p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
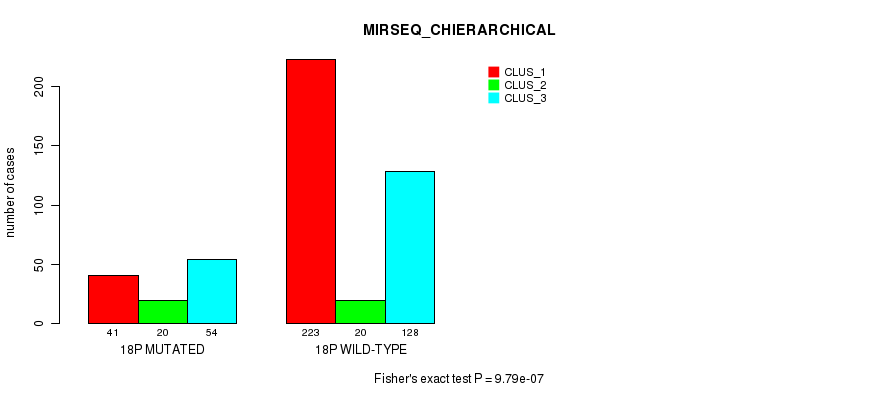
P value = 3.35e-05 (Fisher's exact test), Q value = 0.012
Table S143. Gene #32: '18p' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 18P MUTATED | 13 | 10 | 31 |
| 18P WILD-TYPE | 40 | 85 | 47 |
Figure S143. Get High-res Image Gene #32: '18p' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
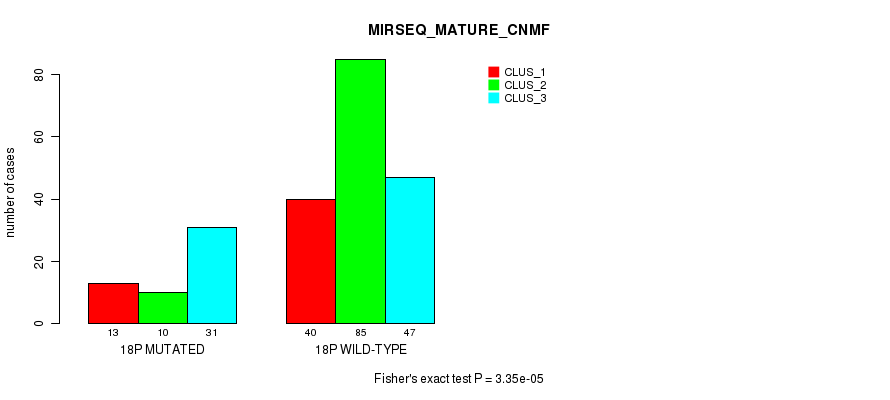
P value = 1.19e-13 (Fisher's exact test), Q value = 5.4e-11
Table S144. Gene #33: '18q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 18Q MUTATED | 69 | 17 | 32 |
| 18Q WILD-TYPE | 102 | 195 | 89 |
Figure S144. Get High-res Image Gene #33: '18q' versus Molecular Subtype #3: 'CN_CNMF'
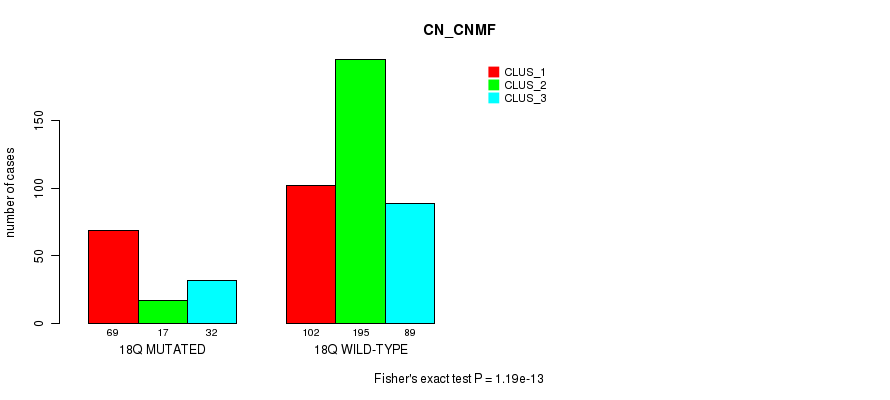
P value = 0.000451 (Fisher's exact test), Q value = 0.14
Table S145. Gene #33: '18q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 18Q MUTATED | 32 | 14 | 24 |
| 18Q WILD-TYPE | 68 | 100 | 52 |
Figure S145. Get High-res Image Gene #33: '18q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
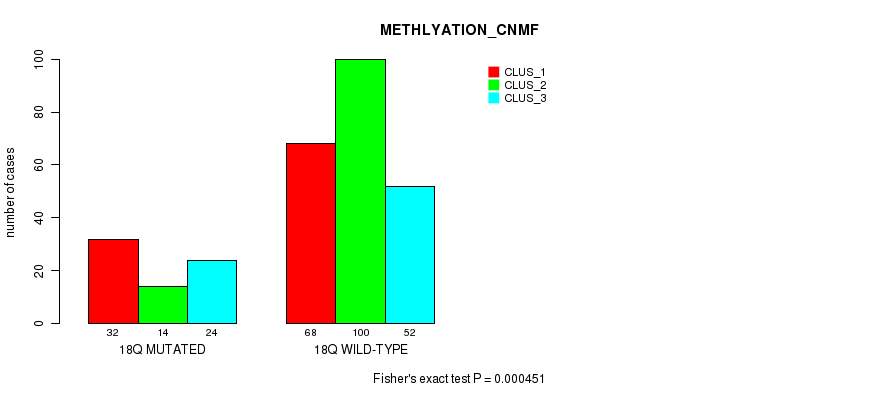
P value = 9.47e-06 (Chi-square test), Q value = 0.0035
Table S146. Gene #33: '18q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 18Q MUTATED | 12 | 16 | 27 | 19 | 7 | 27 |
| 18Q WILD-TYPE | 88 | 72 | 57 | 56 | 36 | 30 |
Figure S146. Get High-res Image Gene #33: '18q' versus Molecular Subtype #5: 'RPPA_CNMF'
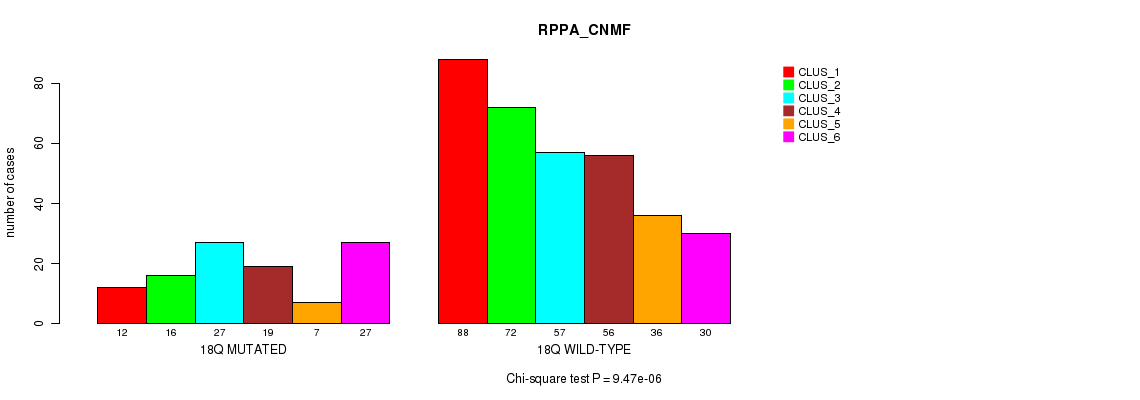
P value = 8.15e-14 (Fisher's exact test), Q value = 3.7e-11
Table S147. Gene #33: '18q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 18Q MUTATED | 19 | 79 | 19 |
| 18Q WILD-TYPE | 186 | 110 | 85 |
Figure S147. Get High-res Image Gene #33: '18q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
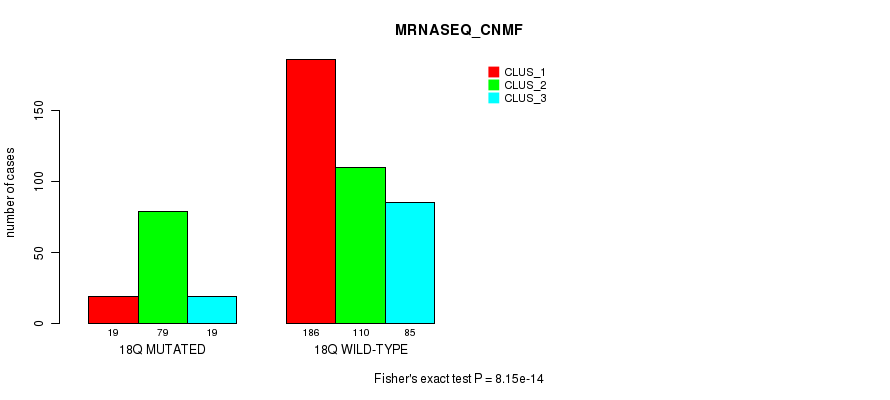
P value = 6.66e-12 (Fisher's exact test), Q value = 2.9e-09
Table S148. Gene #33: '18q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 18Q MUTATED | 21 | 80 | 16 |
| 18Q WILD-TYPE | 194 | 126 | 61 |
Figure S148. Get High-res Image Gene #33: '18q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
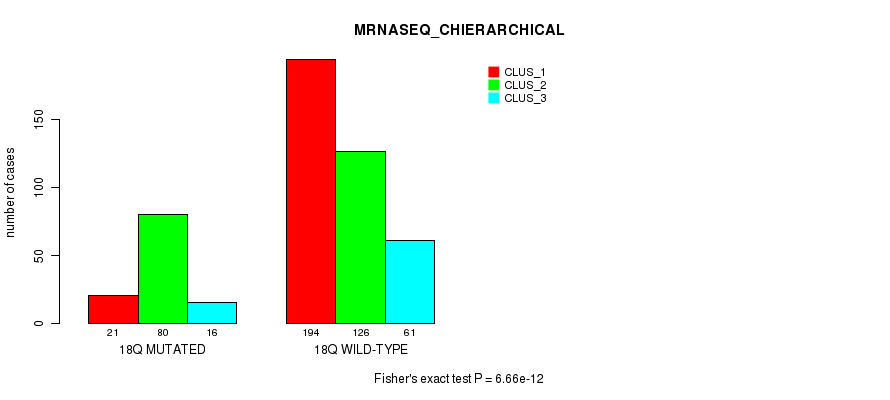
P value = 2.43e-09 (Fisher's exact test), Q value = 1e-06
Table S149. Gene #33: '18q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 18Q MUTATED | 32 | 22 | 63 |
| 18Q WILD-TYPE | 86 | 181 | 102 |
Figure S149. Get High-res Image Gene #33: '18q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
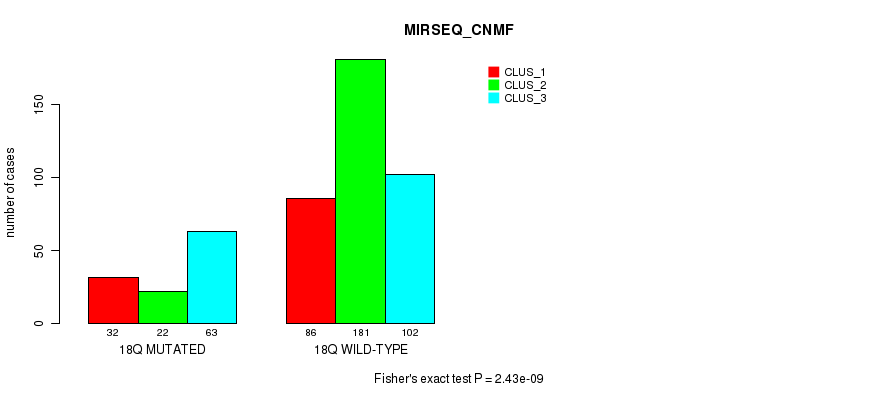
P value = 4.95e-07 (Fisher's exact test), Q value = 2e-04
Table S150. Gene #33: '18q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 18Q MUTATED | 42 | 21 | 54 |
| 18Q WILD-TYPE | 222 | 19 | 128 |
Figure S150. Get High-res Image Gene #33: '18q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
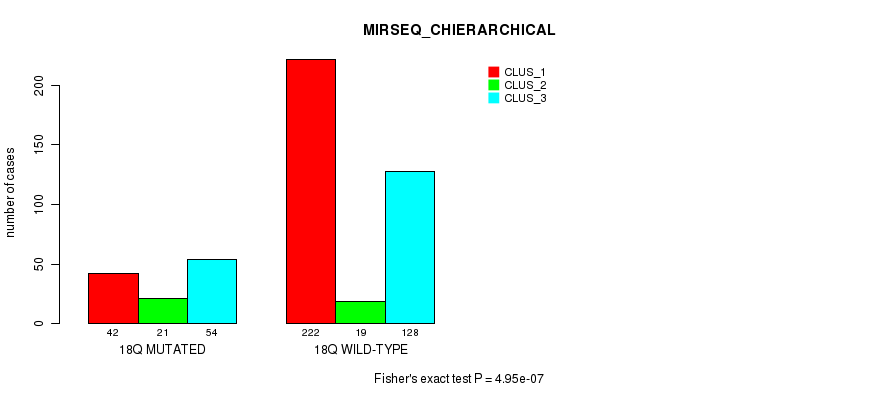
P value = 1.59e-05 (Fisher's exact test), Q value = 0.0058
Table S151. Gene #33: '18q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 18Q MUTATED | 12 | 10 | 32 |
| 18Q WILD-TYPE | 41 | 85 | 46 |
Figure S151. Get High-res Image Gene #33: '18q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
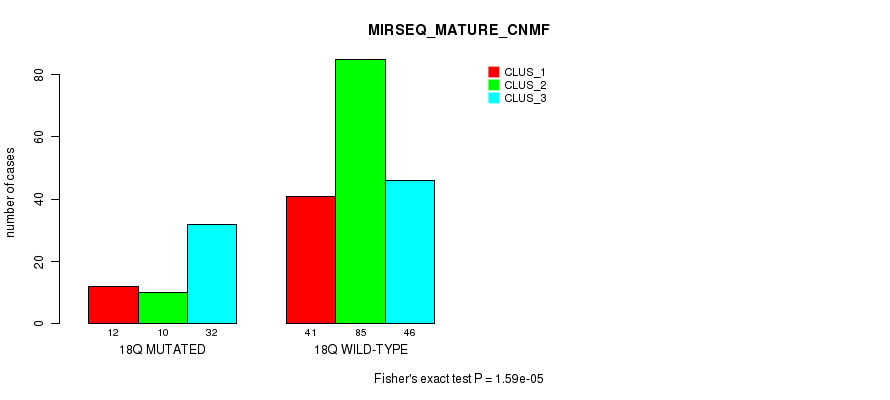
P value = 3.73e-13 (Fisher's exact test), Q value = 1.7e-10
Table S152. Gene #34: '19p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 19P MUTATED | 40 | 2 | 19 |
| 19P WILD-TYPE | 131 | 210 | 102 |
Figure S152. Get High-res Image Gene #34: '19p' versus Molecular Subtype #3: 'CN_CNMF'
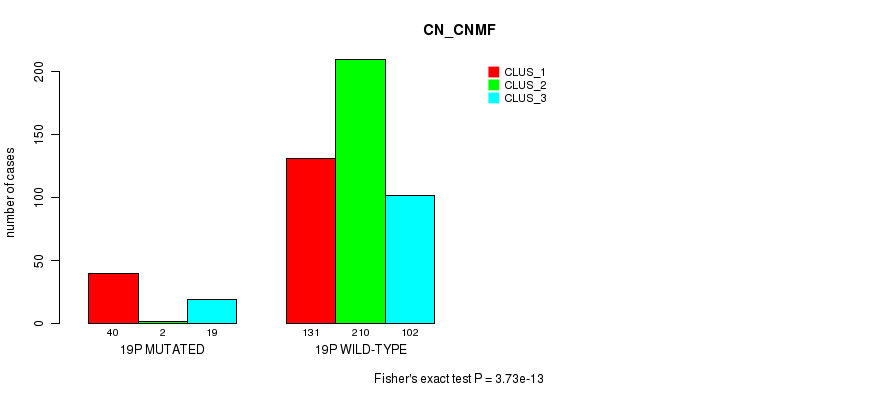
P value = 0.000771 (Fisher's exact test), Q value = 0.23
Table S153. Gene #34: '19p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 19P MUTATED | 24 | 7 | 9 |
| 19P WILD-TYPE | 76 | 107 | 67 |
Figure S153. Get High-res Image Gene #34: '19p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
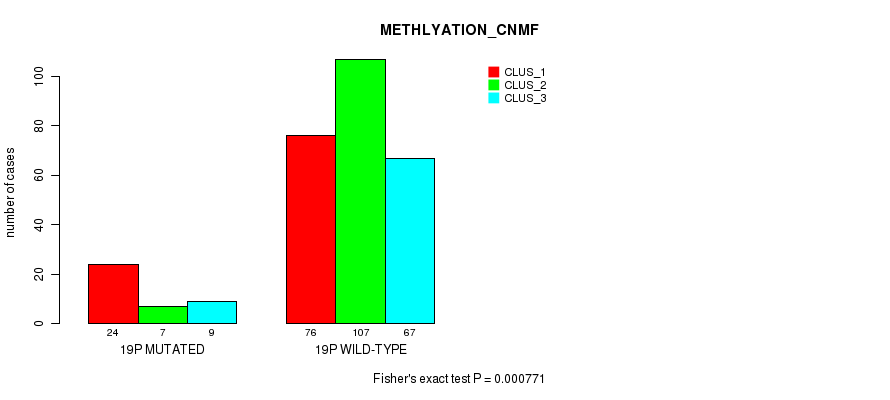
P value = 0.000682 (Fisher's exact test), Q value = 0.2
Table S154. Gene #34: '19p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 19P MUTATED | 12 | 33 | 16 |
| 19P WILD-TYPE | 193 | 156 | 88 |
Figure S154. Get High-res Image Gene #34: '19p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
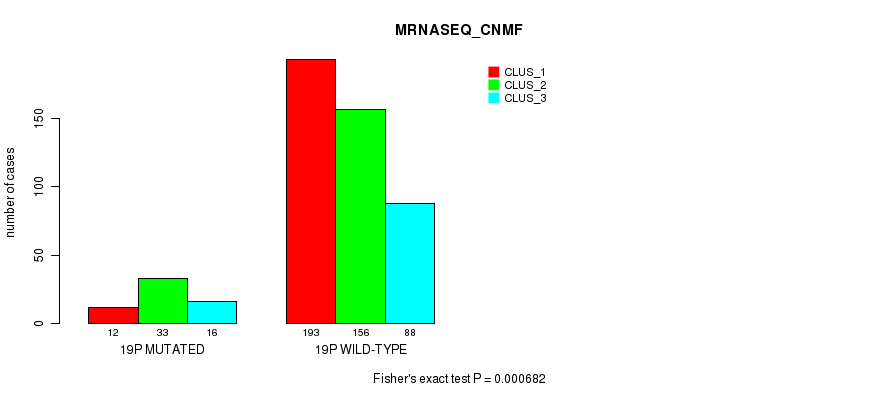
P value = 4.75e-05 (Fisher's exact test), Q value = 0.017
Table S155. Gene #34: '19p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 19P MUTATED | 15 | 11 | 34 |
| 19P WILD-TYPE | 103 | 192 | 131 |
Figure S155. Get High-res Image Gene #34: '19p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
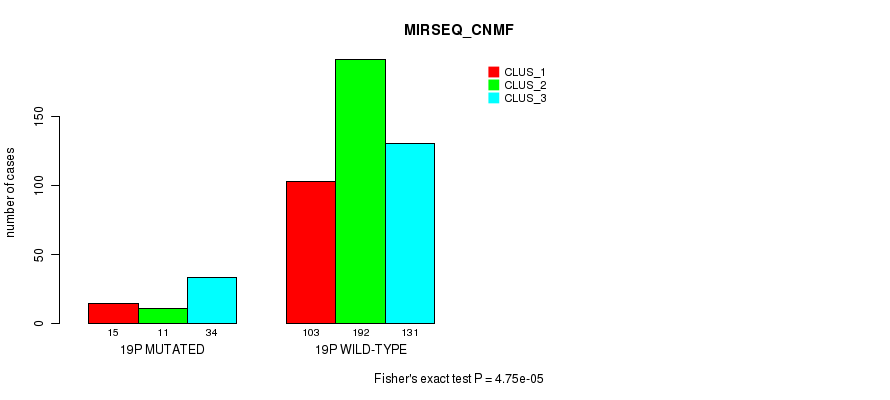
P value = 2.75e-13 (Fisher's exact test), Q value = 1.2e-10
Table S156. Gene #35: '19q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 19Q MUTATED | 41 | 2 | 17 |
| 19Q WILD-TYPE | 130 | 210 | 104 |
Figure S156. Get High-res Image Gene #35: '19q' versus Molecular Subtype #3: 'CN_CNMF'
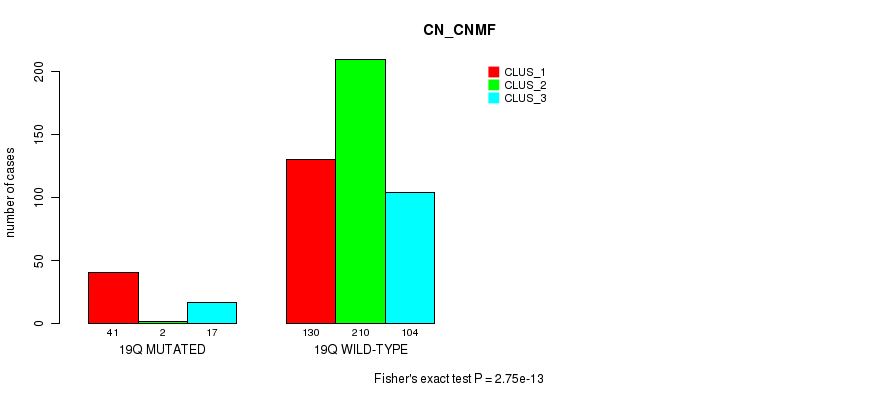
P value = 4.89e-05 (Fisher's exact test), Q value = 0.017
Table S157. Gene #35: '19q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 19Q MUTATED | 13 | 11 | 34 |
| 19Q WILD-TYPE | 105 | 192 | 131 |
Figure S157. Get High-res Image Gene #35: '19q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
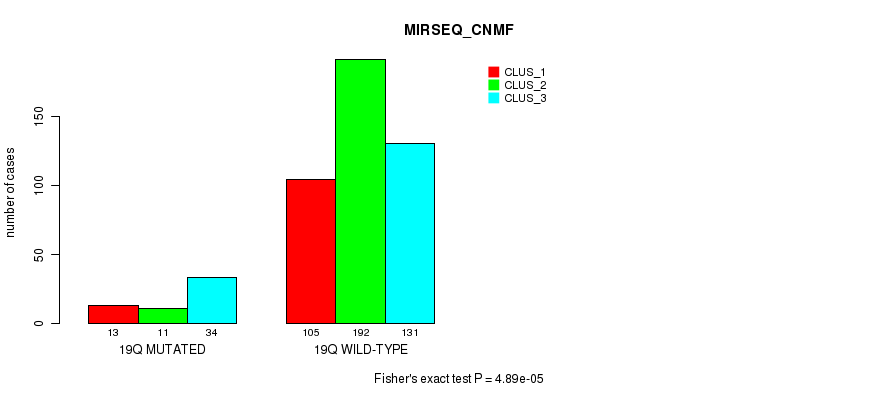
P value = 6.46e-22 (Fisher's exact test), Q value = 3.1e-19
Table S158. Gene #36: '20p' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 20P MUTATED | 73 | 8 | 37 |
| 20P WILD-TYPE | 98 | 204 | 84 |
Figure S158. Get High-res Image Gene #36: '20p' versus Molecular Subtype #3: 'CN_CNMF'
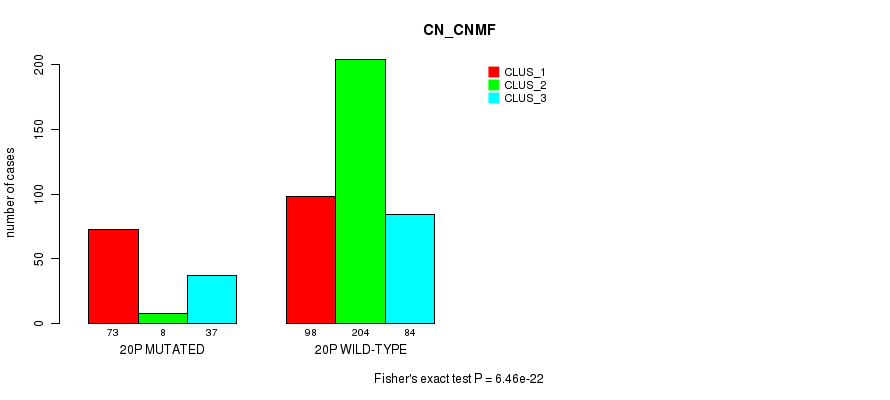
P value = 6.56e-05 (Fisher's exact test), Q value = 0.022
Table S159. Gene #36: '20p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 20P MUTATED | 35 | 12 | 19 |
| 20P WILD-TYPE | 65 | 102 | 57 |
Figure S159. Get High-res Image Gene #36: '20p' versus Molecular Subtype #4: 'METHLYATION_CNMF'
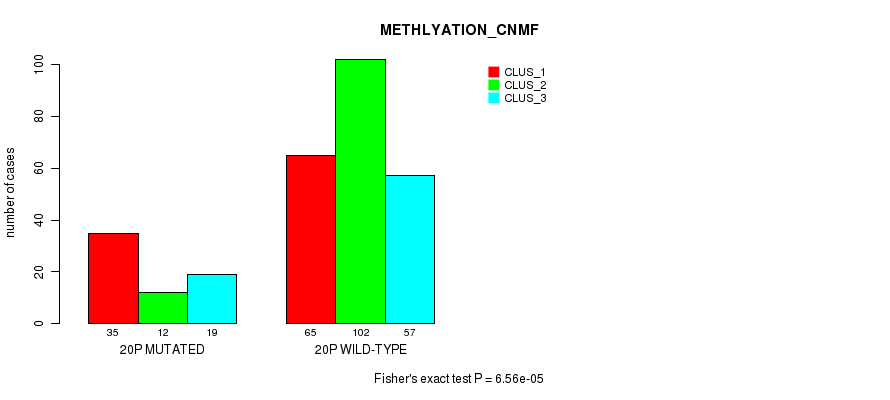
P value = 6.31e-09 (Chi-square test), Q value = 2.6e-06
Table S160. Gene #36: '20p' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 20P MUTATED | 7 | 22 | 20 | 18 | 7 | 31 |
| 20P WILD-TYPE | 93 | 66 | 64 | 57 | 36 | 26 |
Figure S160. Get High-res Image Gene #36: '20p' versus Molecular Subtype #5: 'RPPA_CNMF'
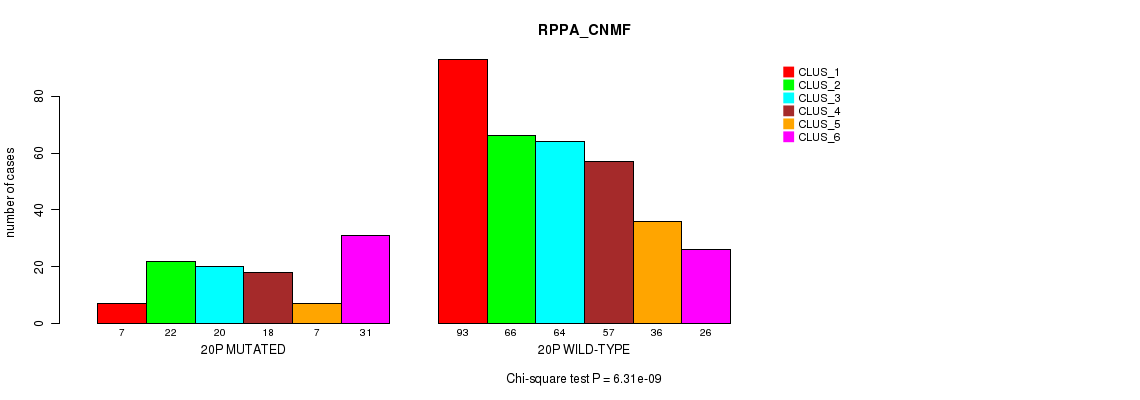
P value = 9.78e-15 (Fisher's exact test), Q value = 4.5e-12
Table S161. Gene #36: '20p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 20P MUTATED | 19 | 81 | 18 |
| 20P WILD-TYPE | 186 | 108 | 86 |
Figure S161. Get High-res Image Gene #36: '20p' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
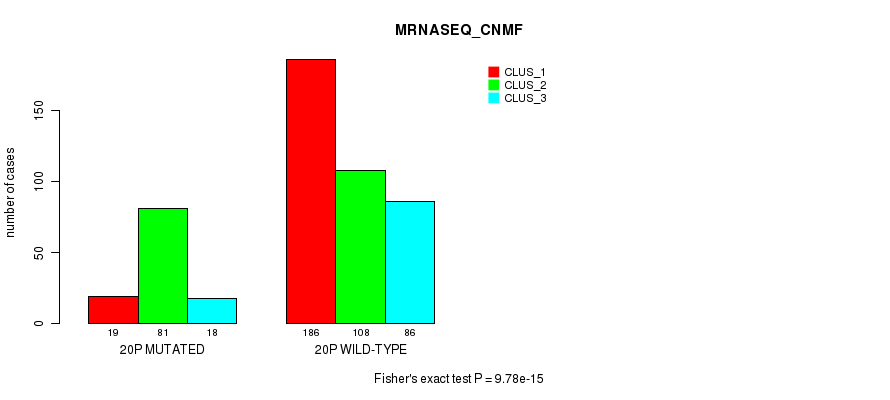
P value = 8.68e-13 (Fisher's exact test), Q value = 3.8e-10
Table S162. Gene #36: '20p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 20P MUTATED | 22 | 83 | 13 |
| 20P WILD-TYPE | 193 | 123 | 64 |
Figure S162. Get High-res Image Gene #36: '20p' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
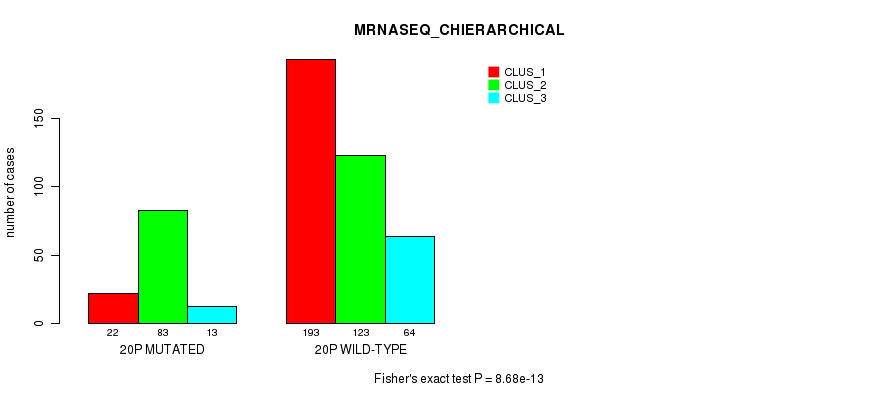
P value = 1.7e-12 (Fisher's exact test), Q value = 7.5e-10
Table S163. Gene #36: '20p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 20P MUTATED | 29 | 19 | 69 |
| 20P WILD-TYPE | 89 | 184 | 96 |
Figure S163. Get High-res Image Gene #36: '20p' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
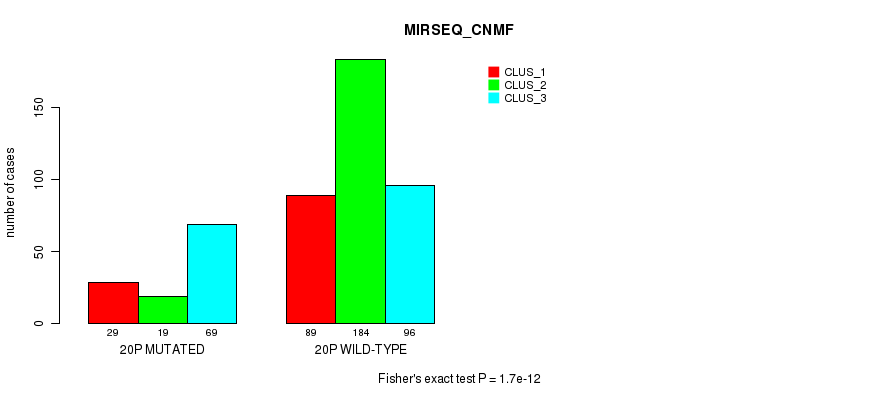
P value = 3.4e-05 (Fisher's exact test), Q value = 0.012
Table S164. Gene #36: '20p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 20P MUTATED | 43 | 16 | 58 |
| 20P WILD-TYPE | 221 | 24 | 124 |
Figure S164. Get High-res Image Gene #36: '20p' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
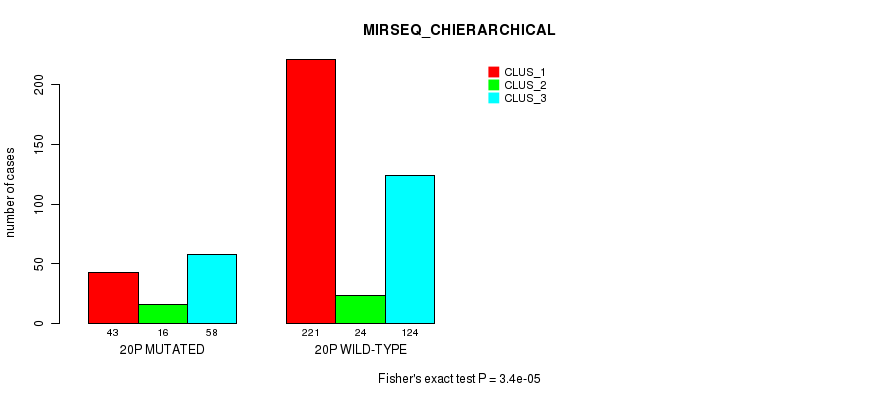
P value = 1.29e-07 (Fisher's exact test), Q value = 5.2e-05
Table S165. Gene #36: '20p' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 20P MUTATED | 10 | 6 | 32 |
| 20P WILD-TYPE | 43 | 89 | 46 |
Figure S165. Get High-res Image Gene #36: '20p' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
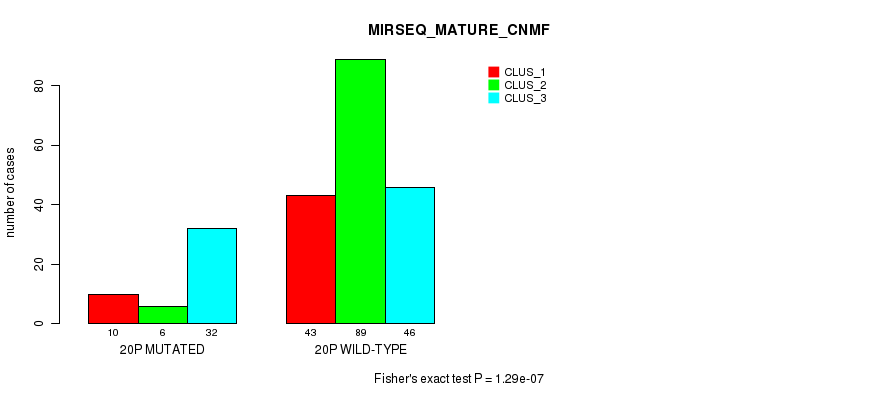
P value = 5.26e-24 (Fisher's exact test), Q value = 2.5e-21
Table S166. Gene #37: '20q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 20Q MUTATED | 74 | 6 | 36 |
| 20Q WILD-TYPE | 97 | 206 | 85 |
Figure S166. Get High-res Image Gene #37: '20q' versus Molecular Subtype #3: 'CN_CNMF'
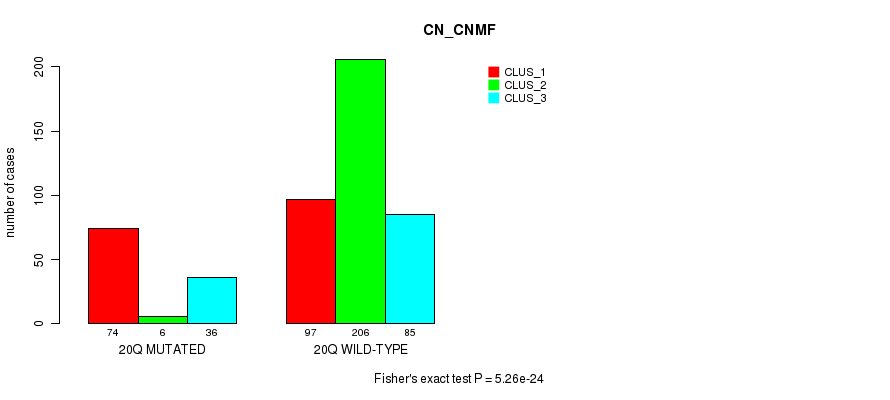
P value = 5.62e-05 (Fisher's exact test), Q value = 0.019
Table S167. Gene #37: '20q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 20Q MUTATED | 34 | 11 | 18 |
| 20Q WILD-TYPE | 66 | 103 | 58 |
Figure S167. Get High-res Image Gene #37: '20q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
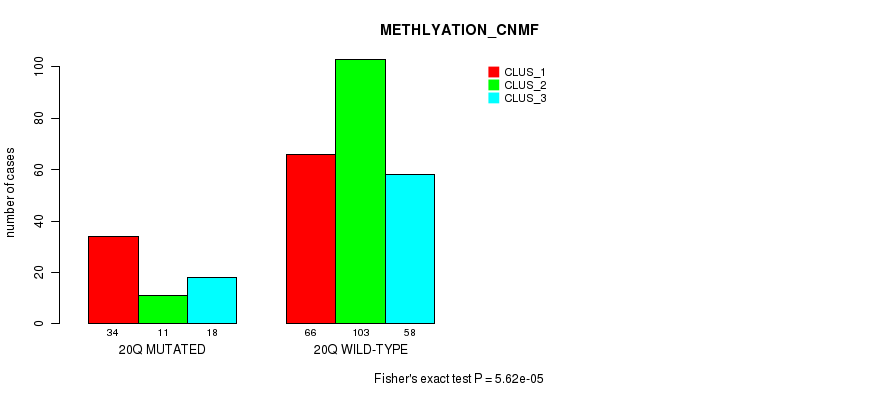
P value = 1.75e-09 (Chi-square test), Q value = 7.2e-07
Table S168. Gene #37: '20q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 20Q MUTATED | 7 | 21 | 21 | 18 | 5 | 31 |
| 20Q WILD-TYPE | 93 | 67 | 63 | 57 | 38 | 26 |
Figure S168. Get High-res Image Gene #37: '20q' versus Molecular Subtype #5: 'RPPA_CNMF'
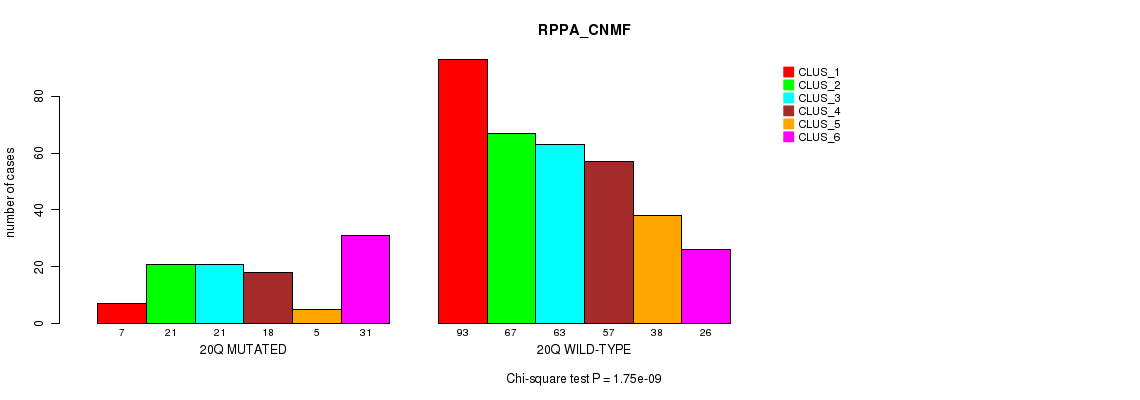
P value = 5.17e-16 (Fisher's exact test), Q value = 2.4e-13
Table S169. Gene #37: '20q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 20Q MUTATED | 18 | 82 | 16 |
| 20Q WILD-TYPE | 187 | 107 | 88 |
Figure S169. Get High-res Image Gene #37: '20q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
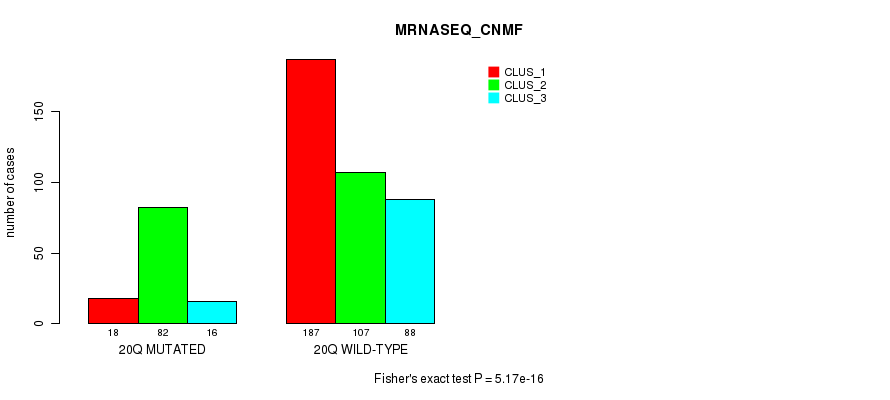
P value = 2.04e-13 (Fisher's exact test), Q value = 9.1e-11
Table S170. Gene #37: '20q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 20Q MUTATED | 21 | 83 | 12 |
| 20Q WILD-TYPE | 194 | 123 | 65 |
Figure S170. Get High-res Image Gene #37: '20q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
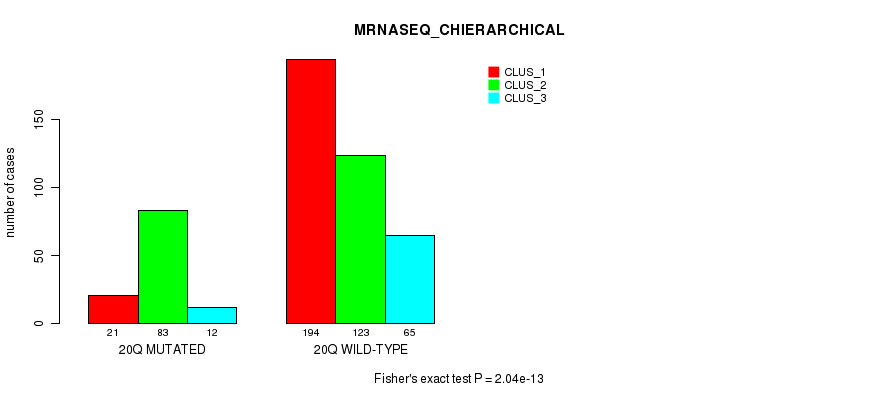
P value = 1.77e-12 (Fisher's exact test), Q value = 7.7e-10
Table S171. Gene #37: '20q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 20Q MUTATED | 27 | 19 | 69 |
| 20Q WILD-TYPE | 91 | 184 | 96 |
Figure S171. Get High-res Image Gene #37: '20q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
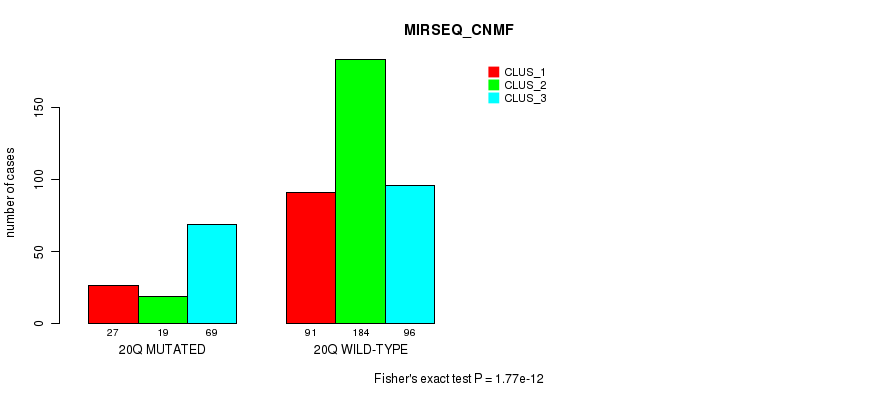
P value = 3.04e-05 (Fisher's exact test), Q value = 0.011
Table S172. Gene #37: '20q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 264 | 40 | 182 |
| 20Q MUTATED | 42 | 16 | 57 |
| 20Q WILD-TYPE | 222 | 24 | 125 |
Figure S172. Get High-res Image Gene #37: '20q' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
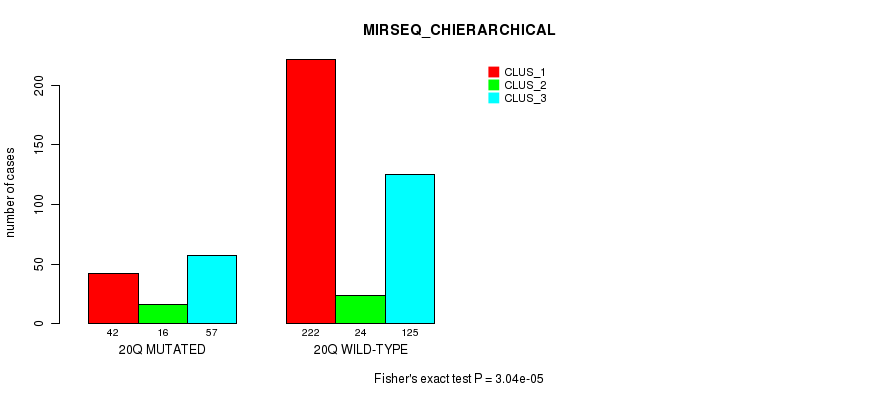
P value = 1.29e-07 (Fisher's exact test), Q value = 5.2e-05
Table S173. Gene #37: '20q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 20Q MUTATED | 10 | 6 | 32 |
| 20Q WILD-TYPE | 43 | 89 | 46 |
Figure S173. Get High-res Image Gene #37: '20q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
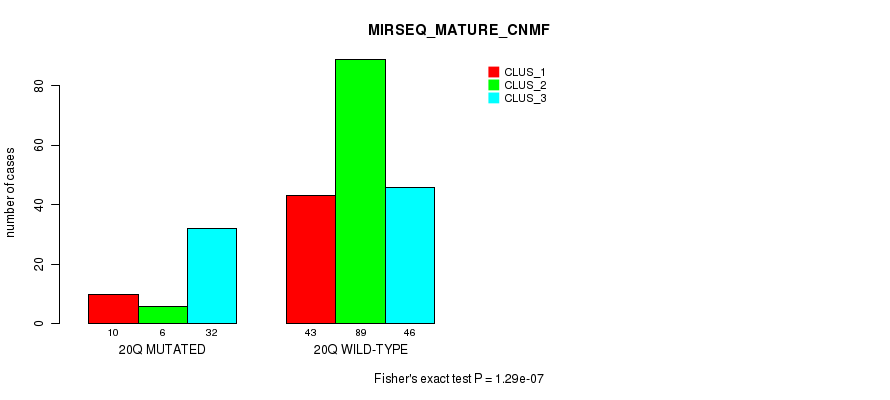
P value = 2.08e-11 (Fisher's exact test), Q value = 8.9e-09
Table S174. Gene #38: '21q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 21Q MUTATED | 59 | 15 | 31 |
| 21Q WILD-TYPE | 112 | 197 | 90 |
Figure S174. Get High-res Image Gene #38: '21q' versus Molecular Subtype #3: 'CN_CNMF'
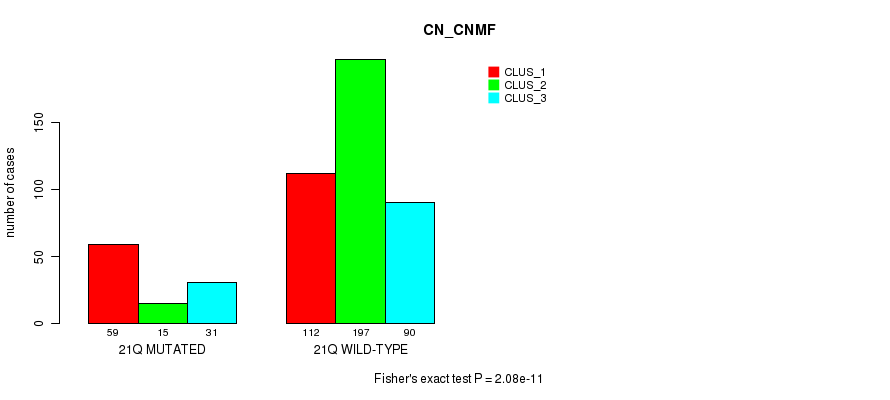
P value = 8.91e-05 (Fisher's exact test), Q value = 0.03
Table S175. Gene #38: '21q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 100 | 114 | 76 |
| 21Q MUTATED | 36 | 13 | 16 |
| 21Q WILD-TYPE | 64 | 101 | 60 |
Figure S175. Get High-res Image Gene #38: '21q' versus Molecular Subtype #4: 'METHLYATION_CNMF'
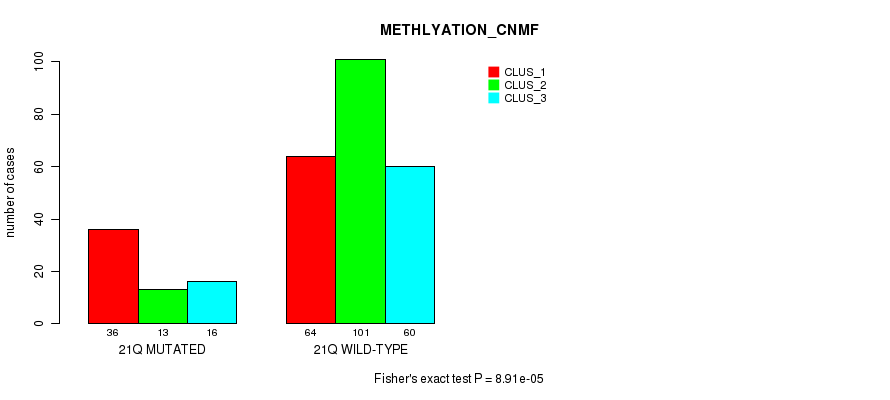
P value = 0.000125 (Chi-square test), Q value = 0.041
Table S176. Gene #38: '21q' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 100 | 88 | 84 | 75 | 43 | 57 |
| 21Q MUTATED | 10 | 10 | 21 | 19 | 8 | 22 |
| 21Q WILD-TYPE | 90 | 78 | 63 | 56 | 35 | 35 |
Figure S176. Get High-res Image Gene #38: '21q' versus Molecular Subtype #5: 'RPPA_CNMF'
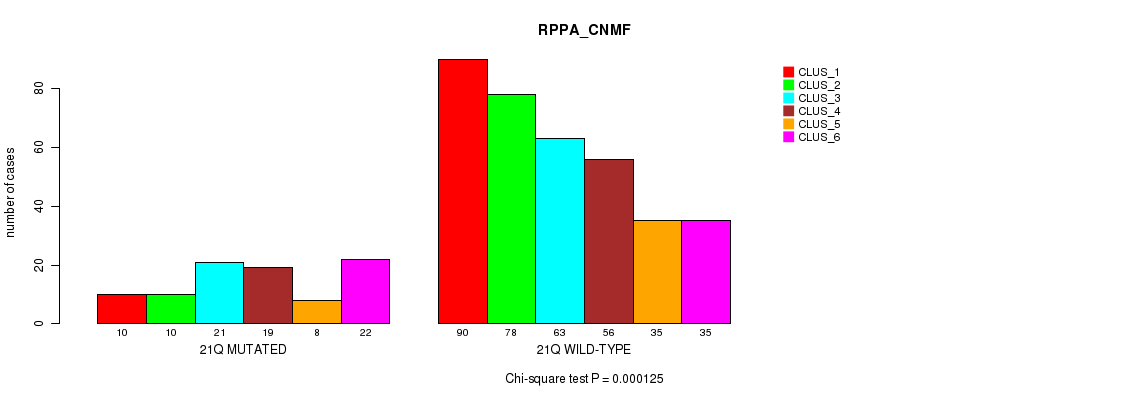
P value = 0.000154 (Fisher's exact test), Q value = 0.05
Table S177. Gene #38: '21q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 21Q MUTATED | 24 | 52 | 26 |
| 21Q WILD-TYPE | 181 | 137 | 78 |
Figure S177. Get High-res Image Gene #38: '21q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
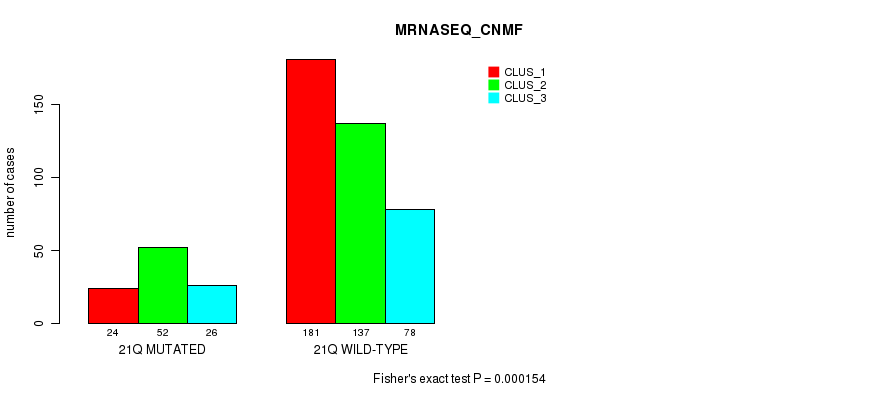
P value = 2.21e-05 (Fisher's exact test), Q value = 0.008
Table S178. Gene #38: '21q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 21Q MUTATED | 24 | 58 | 20 |
| 21Q WILD-TYPE | 191 | 148 | 57 |
Figure S178. Get High-res Image Gene #38: '21q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
P value = 0.000114 (Fisher's exact test), Q value = 0.037
Table S179. Gene #38: '21q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 21Q MUTATED | 23 | 28 | 53 |
| 21Q WILD-TYPE | 95 | 175 | 112 |
Figure S179. Get High-res Image Gene #38: '21q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
P value = 0.000485 (Fisher's exact test), Q value = 0.15
Table S180. Gene #39: '22q' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 14 | 32 | 24 |
| 22Q MUTATED | 4 | 0 | 8 |
| 22Q WILD-TYPE | 10 | 32 | 16 |
Figure S180. Get High-res Image Gene #39: '22q' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
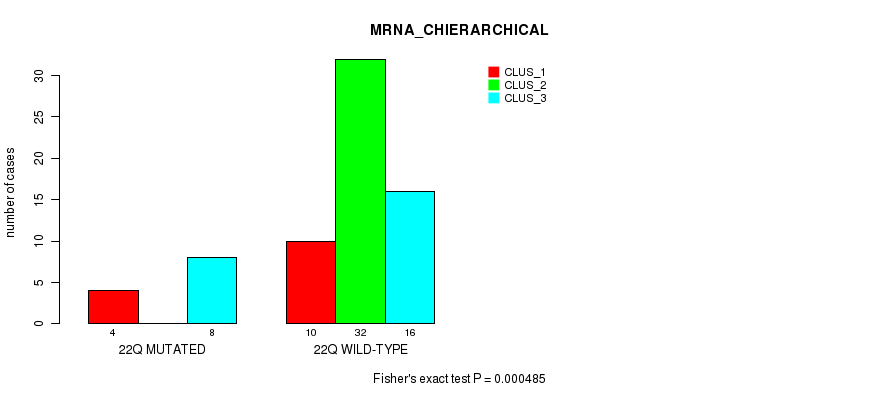
P value = 5.67e-14 (Fisher's exact test), Q value = 2.6e-11
Table S181. Gene #39: '22q' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| 22Q MUTATED | 48 | 5 | 26 |
| 22Q WILD-TYPE | 123 | 207 | 95 |
Figure S181. Get High-res Image Gene #39: '22q' versus Molecular Subtype #3: 'CN_CNMF'
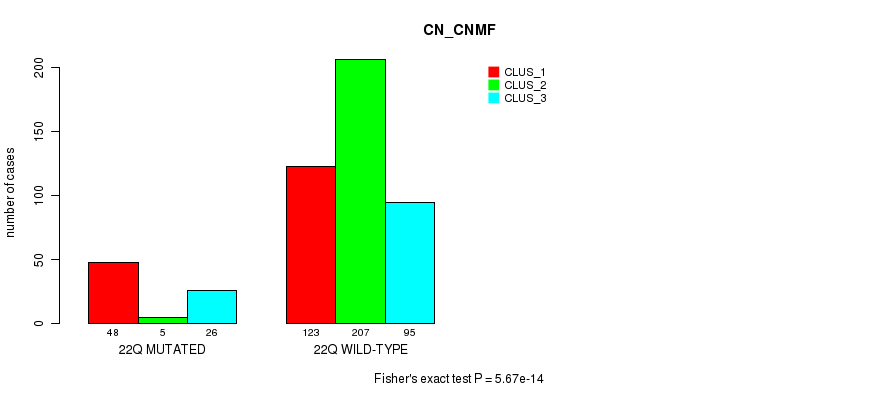
P value = 9.26e-06 (Fisher's exact test), Q value = 0.0035
Table S182. Gene #39: '22q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 205 | 189 | 104 |
| 22Q MUTATED | 16 | 48 | 15 |
| 22Q WILD-TYPE | 189 | 141 | 89 |
Figure S182. Get High-res Image Gene #39: '22q' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
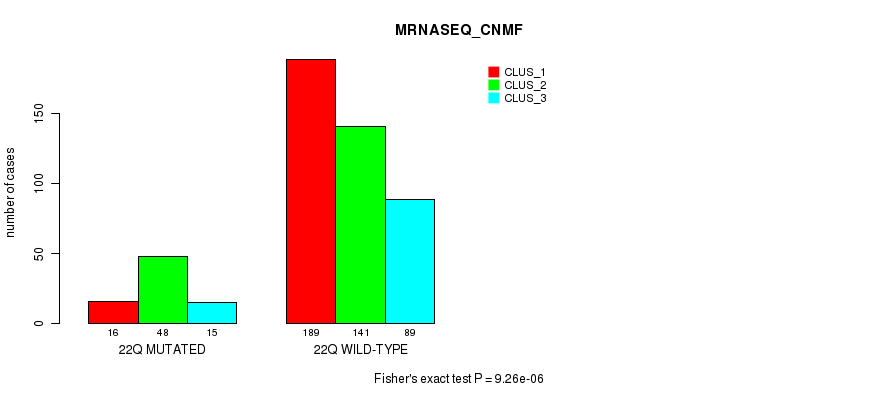
P value = 1.52e-05 (Fisher's exact test), Q value = 0.0056
Table S183. Gene #39: '22q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 215 | 206 | 77 |
| 22Q MUTATED | 16 | 48 | 15 |
| 22Q WILD-TYPE | 199 | 158 | 62 |
Figure S183. Get High-res Image Gene #39: '22q' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
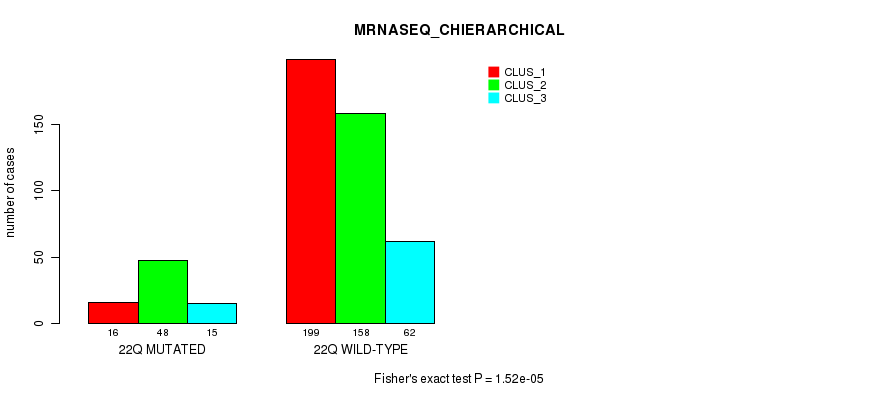
P value = 2.35e-06 (Fisher's exact test), Q value = 0.00091
Table S184. Gene #39: '22q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 118 | 203 | 165 |
| 22Q MUTATED | 15 | 17 | 46 |
| 22Q WILD-TYPE | 103 | 186 | 119 |
Figure S184. Get High-res Image Gene #39: '22q' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
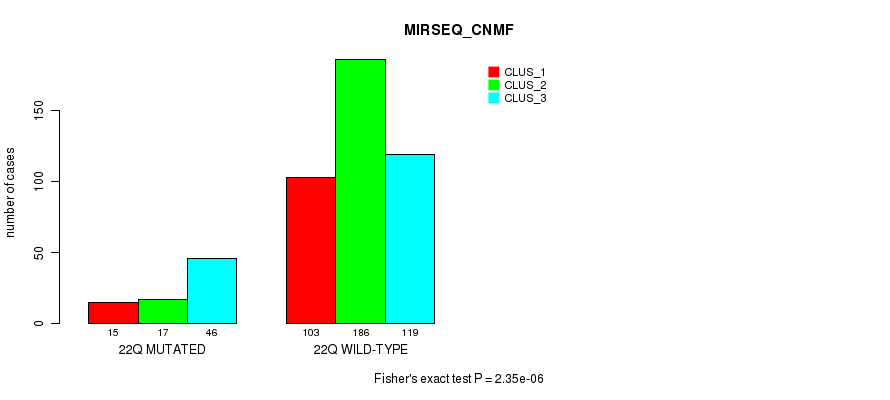
P value = 0.000659 (Fisher's exact test), Q value = 0.2
Table S185. Gene #39: '22q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 53 | 95 | 78 |
| 22Q MUTATED | 9 | 4 | 18 |
| 22Q WILD-TYPE | 44 | 91 | 60 |
Figure S185. Get High-res Image Gene #39: '22q' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
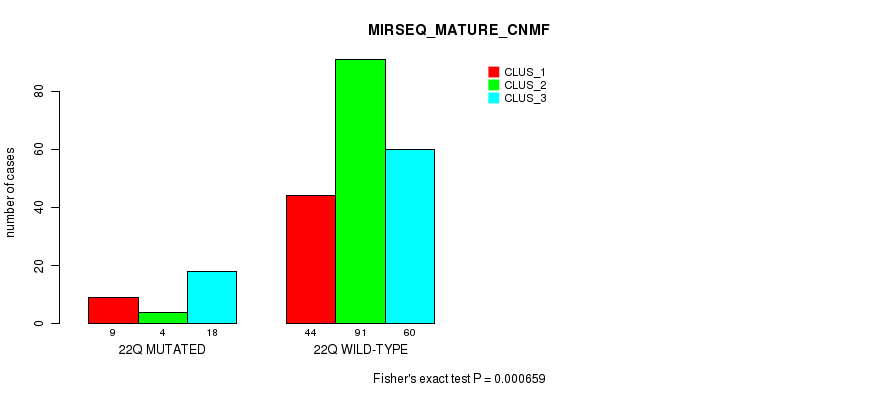
P value = 7.68e-05 (Fisher's exact test), Q value = 0.026
Table S186. Gene #40: 'xq' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 171 | 212 | 121 |
| XQ MUTATED | 41 | 17 | 17 |
| XQ WILD-TYPE | 130 | 195 | 104 |
Figure S186. Get High-res Image Gene #40: 'xq' versus Molecular Subtype #3: 'CN_CNMF'
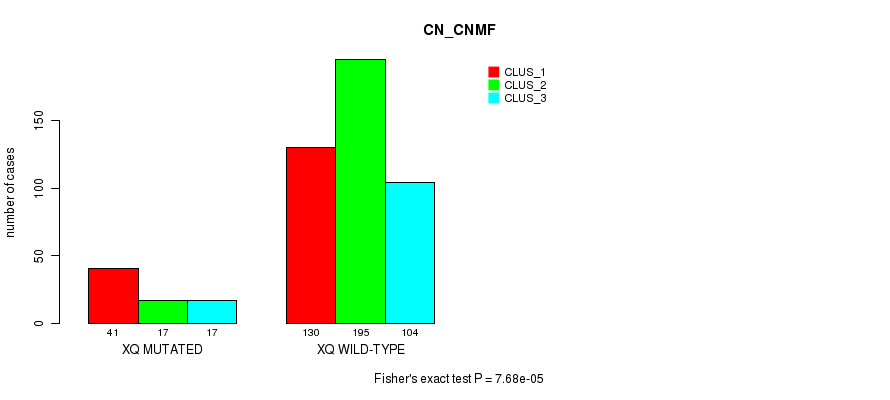
P value = 2.25e-05 (Fisher's exact test), Q value = 0.0081
Table S187. Gene #40: 'xq' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 186 | 152 | 109 |
| XQ MUTATED | 14 | 19 | 30 |
| XQ WILD-TYPE | 172 | 133 | 79 |
Figure S187. Get High-res Image Gene #40: 'xq' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
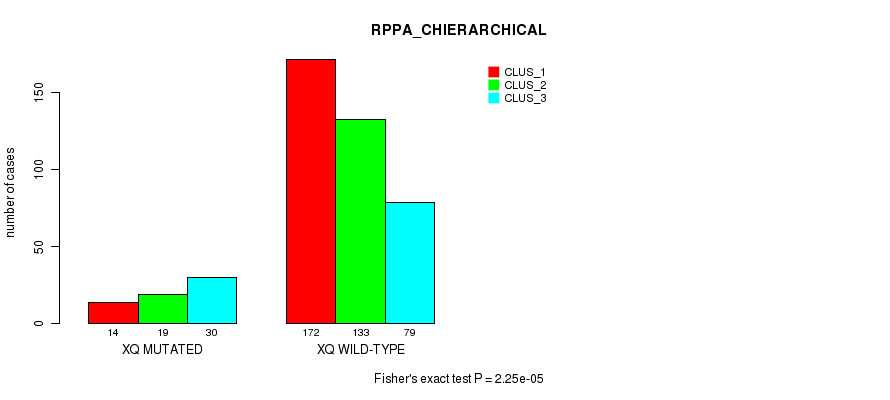
-
Copy number data file = transformed.cor.cli.txt
-
Molecular subtype file = KIRC-TP.transferedmergedcluster.txt
-
Number of patients = 504
-
Number of significantly focal cnvs = 40
-
Number of molecular subtypes = 12
-
Exclude genes that fewer than K tumors have alterations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.