This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 17 genes and 12 molecular subtypes across 177 patients, 7 significant findings detected with P value < 0.05 and Q value < 0.25.
-
TP53 mutation correlated to 'CN_CNMF'.
-
NFE2L2 mutation correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
KEAP1 mutation correlated to 'MRNA_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between mutation status of 17 genes and 12 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 7 significant findings detected.
|
Clinical Features |
MRNA CNMF |
MRNA CHIERARCHICAL |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| NFE2L2 | 27 (15%) | 150 |
0.000356 (0.0663) |
0.00115 (0.211) |
0.0185 (1.00) |
0.0334 (1.00) |
0.261 (1.00) |
0.149 (1.00) |
6.26e-06 (0.00118) |
2.46e-06 (0.000465) |
0.0296 (1.00) |
2.36e-06 (0.000449) |
0.589 (1.00) |
0.14 (1.00) |
| TP53 | 141 (80%) | 36 |
0.0846 (1.00) |
0.0524 (1.00) |
0.00018 (0.0338) |
0.00153 (0.28) |
1 (1.00) |
0.894 (1.00) |
0.0021 (0.378) |
0.0183 (1.00) |
0.119 (1.00) |
0.565 (1.00) |
0.824 (1.00) |
0.726 (1.00) |
| KEAP1 | 22 (12%) | 155 |
0.00196 (0.354) |
0.000403 (0.0746) |
0.17 (1.00) |
0.0758 (1.00) |
0.204 (1.00) |
0.0963 (1.00) |
0.159 (1.00) |
0.0428 (1.00) |
0.701 (1.00) |
0.824 (1.00) |
0.00504 (0.903) |
0.61 (1.00) |
| CDKN2A | 26 (15%) | 151 |
0.00177 (0.323) |
0.0627 (1.00) |
0.0183 (1.00) |
0.668 (1.00) |
0.34 (1.00) |
0.203 (1.00) |
0.331 (1.00) |
0.646 (1.00) |
0.751 (1.00) |
1 (1.00) |
1 (1.00) |
0.0993 (1.00) |
| PIK3CA | 27 (15%) | 150 |
0.64 (1.00) |
0.578 (1.00) |
0.448 (1.00) |
0.545 (1.00) |
1 (1.00) |
0.702 (1.00) |
0.39 (1.00) |
0.916 (1.00) |
0.615 (1.00) |
0.671 (1.00) |
0.456 (1.00) |
0.726 (1.00) |
| PTEN | 14 (8%) | 163 |
0.376 (1.00) |
0.427 (1.00) |
0.439 (1.00) |
0.134 (1.00) |
0.721 (1.00) |
0.473 (1.00) |
0.613 (1.00) |
0.158 (1.00) |
0.575 (1.00) |
0.431 (1.00) |
0.357 (1.00) |
1 (1.00) |
| MLL2 | 35 (20%) | 142 |
0.146 (1.00) |
0.809 (1.00) |
0.253 (1.00) |
0.315 (1.00) |
0.696 (1.00) |
0.221 (1.00) |
0.248 (1.00) |
0.611 (1.00) |
0.948 (1.00) |
0.638 (1.00) |
0.614 (1.00) |
1 (1.00) |
| CSMD3 | 81 (46%) | 96 |
0.24 (1.00) |
0.878 (1.00) |
0.746 (1.00) |
0.479 (1.00) |
0.275 (1.00) |
0.765 (1.00) |
0.429 (1.00) |
0.769 (1.00) |
0.642 (1.00) |
1 (1.00) |
0.389 (1.00) |
0.164 (1.00) |
| RB1 | 12 (7%) | 165 |
0.343 (1.00) |
0.198 (1.00) |
0.418 (1.00) |
0.108 (1.00) |
0.657 (1.00) |
0.433 (1.00) |
1 (1.00) |
0.355 (1.00) |
0.575 (1.00) |
0.911 (1.00) |
1 (1.00) |
|
| HS6ST1 | 5 (3%) | 172 |
0.269 (1.00) |
0.295 (1.00) |
0.53 (1.00) |
0.568 (1.00) |
0.101 (1.00) |
1 (1.00) |
1 (1.00) |
|||||
| FBXW7 | 10 (6%) | 167 |
0.458 (1.00) |
1 (1.00) |
0.508 (1.00) |
0.621 (1.00) |
0.785 (1.00) |
0.834 (1.00) |
0.574 (1.00) |
0.935 (1.00) |
0.772 (1.00) |
1 (1.00) |
||
| ZNF567 | 3 (2%) | 174 |
0.364 (1.00) |
0.359 (1.00) |
0.424 (1.00) |
0.389 (1.00) |
0.427 (1.00) |
0.253 (1.00) |
0.19 (1.00) |
1 (1.00) |
||||
| ELTD1 | 18 (10%) | 159 |
0.0295 (1.00) |
0.303 (1.00) |
0.0533 (1.00) |
0.6 (1.00) |
0.341 (1.00) |
0.648 (1.00) |
0.554 (1.00) |
0.42 (1.00) |
0.0887 (1.00) |
0.722 (1.00) |
0.305 (1.00) |
0.419 (1.00) |
| ASCL4 | 6 (3%) | 171 |
0.0924 (1.00) |
0.534 (1.00) |
0.882 (1.00) |
0.783 (1.00) |
0.817 (1.00) |
0.835 (1.00) |
0.348 (1.00) |
0.247 (1.00) |
0.91 (1.00) |
0.219 (1.00) |
||
| OR2G6 | 16 (9%) | 161 |
0.366 (1.00) |
0.211 (1.00) |
0.909 (1.00) |
1 (1.00) |
1 (1.00) |
0.717 (1.00) |
0.199 (1.00) |
0.0466 (1.00) |
0.428 (1.00) |
0.245 (1.00) |
0.668 (1.00) |
0.61 (1.00) |
| HLA-A | 6 (3%) | 171 |
0.0391 (1.00) |
0.33 (1.00) |
0.446 (1.00) |
0.178 (1.00) |
1 (1.00) |
0.513 (1.00) |
0.128 (1.00) |
0.114 (1.00) |
0.471 (1.00) |
0.73 (1.00) |
0.381 (1.00) |
1 (1.00) |
| NOTCH1 | 14 (8%) | 163 |
0.812 (1.00) |
0.921 (1.00) |
0.543 (1.00) |
0.0342 (1.00) |
0.591 (1.00) |
0.824 (1.00) |
0.686 (1.00) |
0.401 (1.00) |
0.185 (1.00) |
0.844 (1.00) |
0.483 (1.00) |
0.668 (1.00) |
P value = 0.00177 (Fisher's exact test), Q value = 0.32
Table S1. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| CDKN2A MUTATED | 0 | 13 | 3 | 2 |
| CDKN2A WILD-TYPE | 37 | 35 | 13 | 17 |
Figure S1. Get High-res Image Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
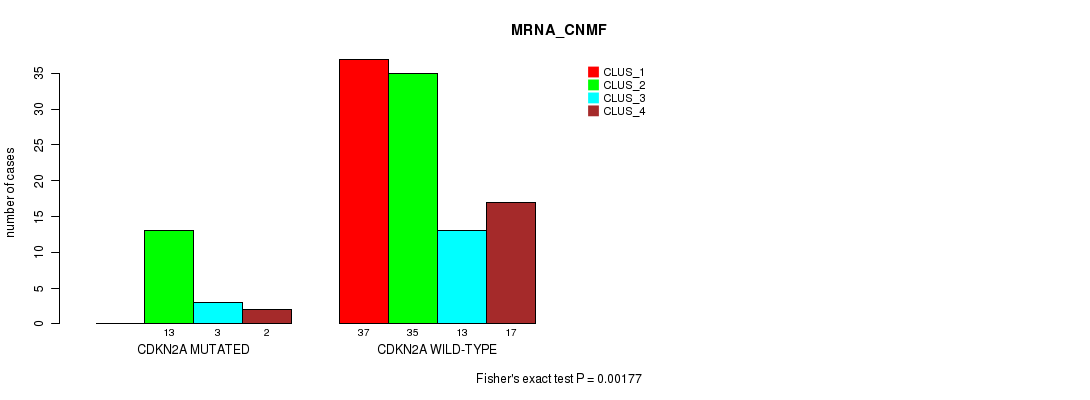
P value = 0.0627 (Fisher's exact test), Q value = 1
Table S2. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| CDKN2A MUTATED | 1 | 13 | 1 | 3 |
| CDKN2A WILD-TYPE | 6 | 39 | 19 | 38 |
P value = 0.0183 (Fisher's exact test), Q value = 1
Table S3. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| CDKN2A MUTATED | 3 | 14 | 9 |
| CDKN2A WILD-TYPE | 58 | 55 | 38 |
Figure S2. Get High-res Image Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
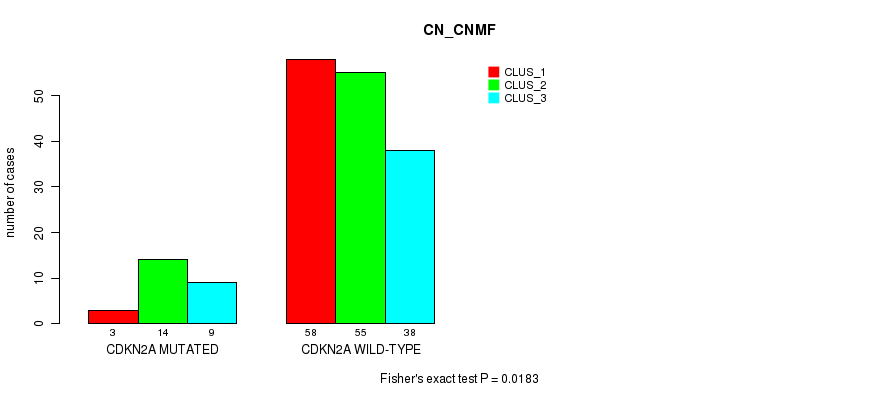
P value = 0.668 (Fisher's exact test), Q value = 1
Table S4. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| CDKN2A MUTATED | 2 | 4 | 3 |
| CDKN2A WILD-TYPE | 25 | 25 | 15 |
P value = 0.34 (Fisher's exact test), Q value = 1
Table S5. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| CDKN2A MUTATED | 2 | 8 | 3 |
| CDKN2A WILD-TYPE | 32 | 39 | 28 |
P value = 0.203 (Fisher's exact test), Q value = 1
Table S6. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| CDKN2A MUTATED | 3 | 1 | 4 | 5 |
| CDKN2A WILD-TYPE | 28 | 26 | 29 | 16 |
P value = 0.331 (Fisher's exact test), Q value = 1
Table S7. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| CDKN2A MUTATED | 4 | 12 | 5 | 5 |
| CDKN2A WILD-TYPE | 38 | 49 | 44 | 20 |
P value = 0.646 (Fisher's exact test), Q value = 1
Table S8. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| CDKN2A MUTATED | 4 | 11 | 11 |
| CDKN2A WILD-TYPE | 30 | 49 | 72 |
P value = 0.751 (Fisher's exact test), Q value = 1
Table S9. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| CDKN2A MUTATED | 15 | 3 | 2 | 4 |
| CDKN2A WILD-TYPE | 84 | 13 | 21 | 19 |
P value = 1 (Fisher's exact test), Q value = 1
Table S10. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| CDKN2A MUTATED | 4 | 5 | 15 |
| CDKN2A WILD-TYPE | 26 | 27 | 84 |
P value = 1 (Fisher's exact test), Q value = 1
Table S11. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| CDKN2A MUTATED | 2 | 2 | 3 |
| CDKN2A WILD-TYPE | 13 | 16 | 16 |
P value = 0.0993 (Fisher's exact test), Q value = 1
Table S12. Gene #1: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| CDKN2A MUTATED | 1 | 6 |
| CDKN2A WILD-TYPE | 25 | 20 |
P value = 0.0846 (Fisher's exact test), Q value = 1
Table S13. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| TP53 MUTATED | 29 | 43 | 10 | 14 |
| TP53 WILD-TYPE | 8 | 5 | 6 | 5 |
P value = 0.0524 (Fisher's exact test), Q value = 1
Table S14. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| TP53 MUTATED | 6 | 47 | 14 | 29 |
| TP53 WILD-TYPE | 1 | 5 | 6 | 12 |
P value = 0.00018 (Fisher's exact test), Q value = 0.034
Table S15. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| TP53 MUTATED | 38 | 63 | 40 |
| TP53 WILD-TYPE | 23 | 6 | 7 |
Figure S3. Get High-res Image Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'

P value = 0.00153 (Fisher's exact test), Q value = 0.28
Table S16. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| TP53 MUTATED | 16 | 28 | 15 |
| TP53 WILD-TYPE | 11 | 1 | 3 |
Figure S4. Get High-res Image Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
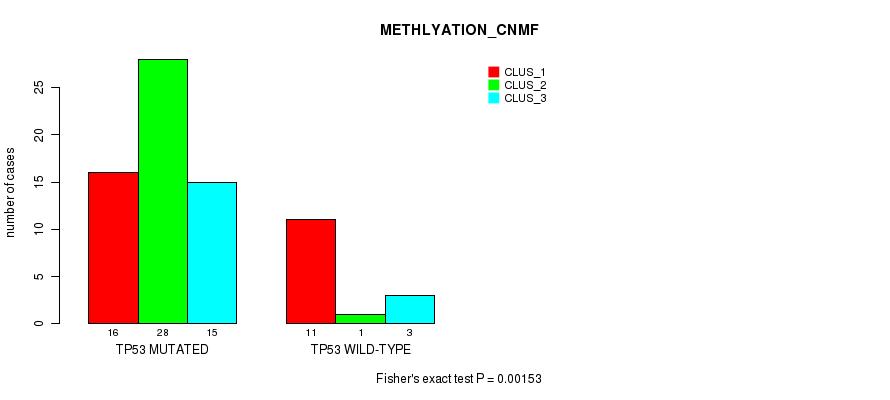
P value = 1 (Fisher's exact test), Q value = 1
Table S17. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| TP53 MUTATED | 26 | 37 | 24 |
| TP53 WILD-TYPE | 8 | 10 | 7 |
P value = 0.894 (Fisher's exact test), Q value = 1
Table S18. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| TP53 MUTATED | 23 | 21 | 27 | 16 |
| TP53 WILD-TYPE | 8 | 6 | 6 | 5 |
P value = 0.0021 (Fisher's exact test), Q value = 0.38
Table S19. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| TP53 MUTATED | 33 | 55 | 40 | 13 |
| TP53 WILD-TYPE | 9 | 6 | 9 | 12 |
Figure S5. Get High-res Image Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
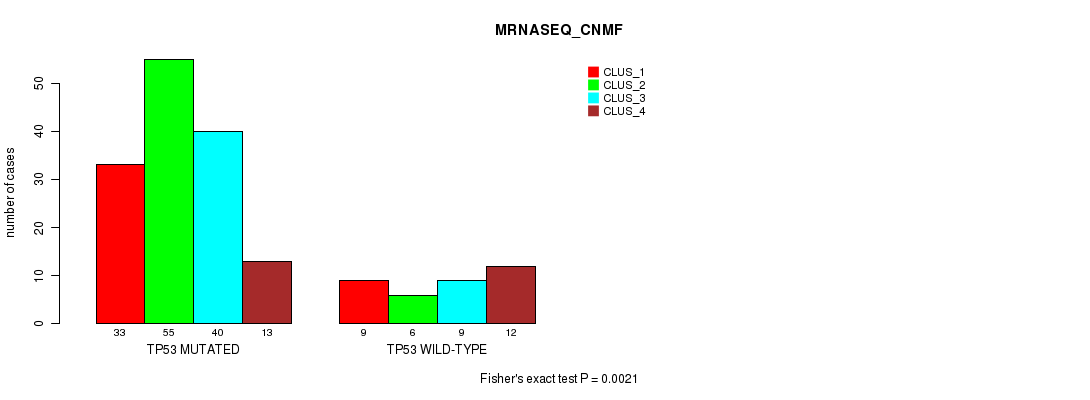
P value = 0.0183 (Fisher's exact test), Q value = 1
Table S20. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| TP53 MUTATED | 28 | 54 | 59 |
| TP53 WILD-TYPE | 6 | 6 | 24 |
Figure S6. Get High-res Image Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
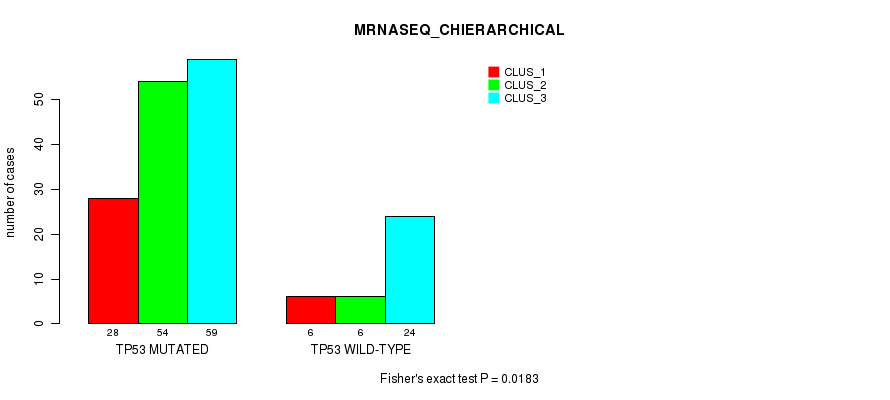
P value = 0.119 (Fisher's exact test), Q value = 1
Table S21. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| TP53 MUTATED | 83 | 14 | 15 | 21 |
| TP53 WILD-TYPE | 16 | 2 | 8 | 2 |
P value = 0.565 (Fisher's exact test), Q value = 1
Table S22. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| TP53 MUTATED | 26 | 28 | 79 |
| TP53 WILD-TYPE | 4 | 4 | 20 |
P value = 0.824 (Fisher's exact test), Q value = 1
Table S23. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| TP53 MUTATED | 13 | 14 | 15 |
| TP53 WILD-TYPE | 2 | 4 | 4 |
P value = 0.726 (Fisher's exact test), Q value = 1
Table S24. Gene #2: 'TP53 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| TP53 MUTATED | 20 | 22 |
| TP53 WILD-TYPE | 6 | 4 |
P value = 0.000356 (Fisher's exact test), Q value = 0.066
Table S25. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| NFE2L2 MUTATED | 0 | 14 | 1 | 1 |
| NFE2L2 WILD-TYPE | 37 | 34 | 15 | 18 |
Figure S7. Get High-res Image Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'

P value = 0.00115 (Fisher's exact test), Q value = 0.21
Table S26. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| NFE2L2 MUTATED | 1 | 14 | 0 | 1 |
| NFE2L2 WILD-TYPE | 6 | 38 | 20 | 40 |
Figure S8. Get High-res Image Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'

P value = 0.0185 (Fisher's exact test), Q value = 1
Table S27. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| NFE2L2 MUTATED | 7 | 17 | 3 |
| NFE2L2 WILD-TYPE | 54 | 52 | 44 |
Figure S9. Get High-res Image Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
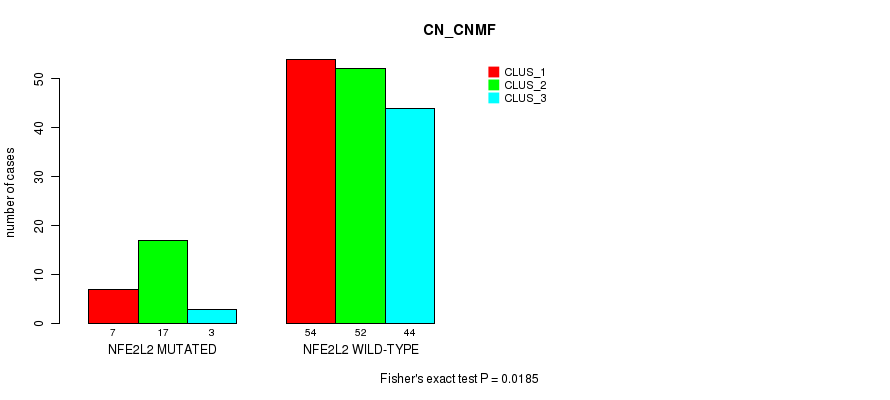
P value = 0.0334 (Fisher's exact test), Q value = 1
Table S28. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| NFE2L2 MUTATED | 4 | 8 | 0 |
| NFE2L2 WILD-TYPE | 23 | 21 | 18 |
Figure S10. Get High-res Image Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
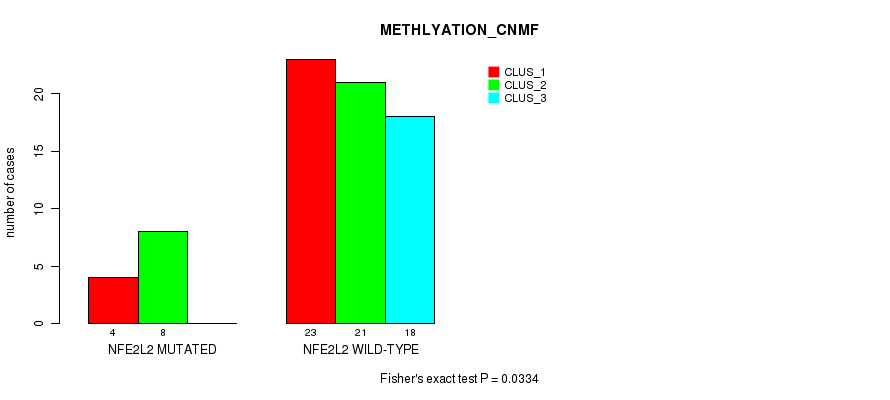
P value = 0.261 (Fisher's exact test), Q value = 1
Table S29. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| NFE2L2 MUTATED | 8 | 5 | 6 |
| NFE2L2 WILD-TYPE | 26 | 42 | 25 |
P value = 0.149 (Fisher's exact test), Q value = 1
Table S30. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| NFE2L2 MUTATED | 6 | 5 | 2 | 6 |
| NFE2L2 WILD-TYPE | 25 | 22 | 31 | 15 |
P value = 6.26e-06 (Fisher's exact test), Q value = 0.0012
Table S31. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| NFE2L2 MUTATED | 2 | 21 | 1 | 3 |
| NFE2L2 WILD-TYPE | 40 | 40 | 48 | 22 |
Figure S11. Get High-res Image Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
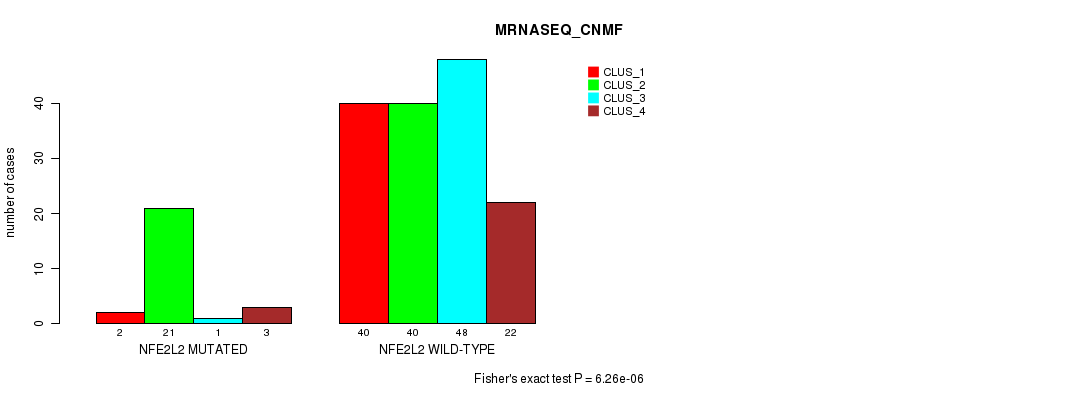
P value = 2.46e-06 (Fisher's exact test), Q value = 0.00046
Table S32. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| NFE2L2 MUTATED | 1 | 21 | 5 |
| NFE2L2 WILD-TYPE | 33 | 39 | 78 |
Figure S12. Get High-res Image Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
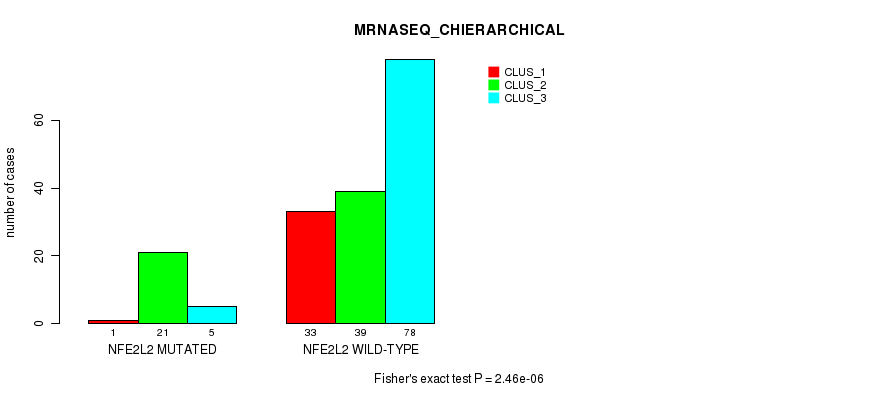
P value = 0.0296 (Fisher's exact test), Q value = 1
Table S33. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| NFE2L2 MUTATED | 15 | 0 | 3 | 8 |
| NFE2L2 WILD-TYPE | 84 | 16 | 20 | 15 |
Figure S13. Get High-res Image Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
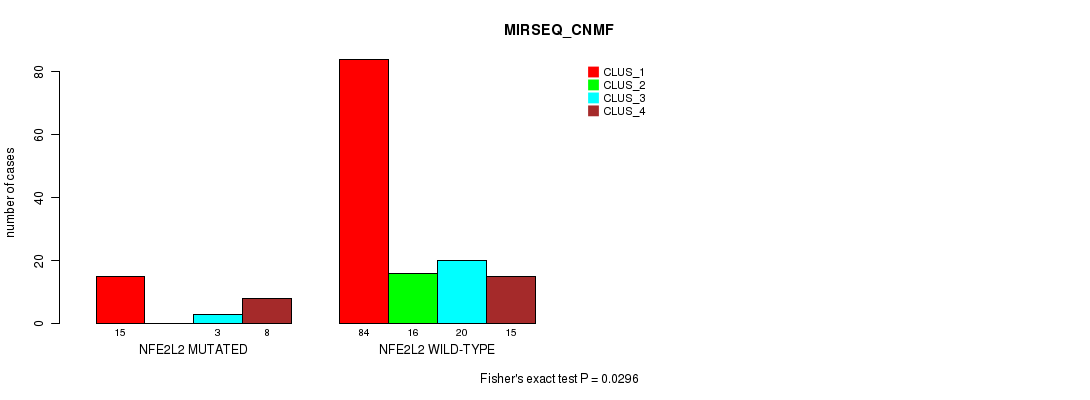
P value = 2.36e-06 (Fisher's exact test), Q value = 0.00045
Table S34. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| NFE2L2 MUTATED | 14 | 0 | 12 |
| NFE2L2 WILD-TYPE | 16 | 32 | 87 |
Figure S14. Get High-res Image Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
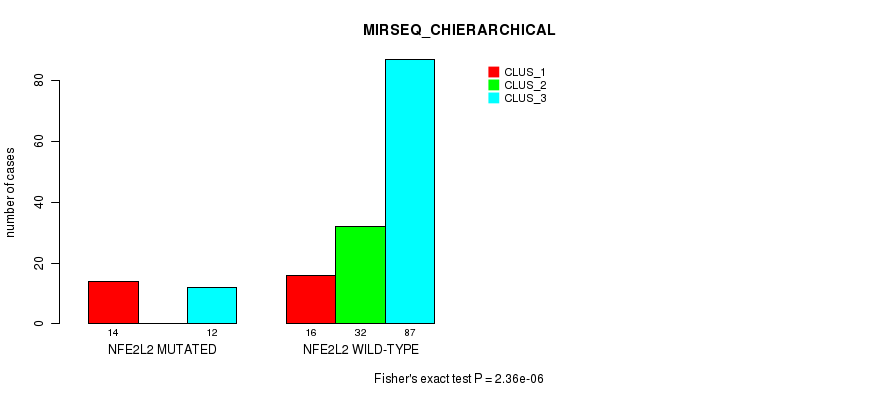
P value = 0.589 (Fisher's exact test), Q value = 1
Table S35. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| NFE2L2 MUTATED | 4 | 2 | 3 |
| NFE2L2 WILD-TYPE | 11 | 16 | 16 |
P value = 0.14 (Fisher's exact test), Q value = 1
Table S36. Gene #3: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| NFE2L2 MUTATED | 2 | 7 |
| NFE2L2 WILD-TYPE | 24 | 19 |
P value = 0.64 (Fisher's exact test), Q value = 1
Table S37. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| PIK3CA MUTATED | 4 | 6 | 3 | 4 |
| PIK3CA WILD-TYPE | 33 | 42 | 13 | 15 |
P value = 0.578 (Fisher's exact test), Q value = 1
Table S38. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| PIK3CA MUTATED | 0 | 6 | 4 | 7 |
| PIK3CA WILD-TYPE | 7 | 46 | 16 | 34 |
P value = 0.448 (Fisher's exact test), Q value = 1
Table S39. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| PIK3CA MUTATED | 12 | 8 | 7 |
| PIK3CA WILD-TYPE | 49 | 61 | 40 |
P value = 0.545 (Fisher's exact test), Q value = 1
Table S40. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| PIK3CA MUTATED | 4 | 3 | 4 |
| PIK3CA WILD-TYPE | 23 | 26 | 14 |
P value = 1 (Fisher's exact test), Q value = 1
Table S41. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| PIK3CA MUTATED | 5 | 7 | 5 |
| PIK3CA WILD-TYPE | 29 | 40 | 26 |
P value = 0.702 (Fisher's exact test), Q value = 1
Table S42. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| PIK3CA MUTATED | 4 | 4 | 7 | 2 |
| PIK3CA WILD-TYPE | 27 | 23 | 26 | 19 |
P value = 0.39 (Fisher's exact test), Q value = 1
Table S43. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| PIK3CA MUTATED | 8 | 7 | 10 | 2 |
| PIK3CA WILD-TYPE | 34 | 54 | 39 | 23 |
P value = 0.916 (Fisher's exact test), Q value = 1
Table S44. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| PIK3CA MUTATED | 6 | 9 | 12 |
| PIK3CA WILD-TYPE | 28 | 51 | 71 |
P value = 0.615 (Fisher's exact test), Q value = 1
Table S45. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| PIK3CA MUTATED | 18 | 2 | 2 | 2 |
| PIK3CA WILD-TYPE | 81 | 14 | 21 | 21 |
P value = 0.671 (Fisher's exact test), Q value = 1
Table S46. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| PIK3CA MUTATED | 3 | 4 | 17 |
| PIK3CA WILD-TYPE | 27 | 28 | 82 |
P value = 0.456 (Fisher's exact test), Q value = 1
Table S47. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| PIK3CA MUTATED | 4 | 4 | 2 |
| PIK3CA WILD-TYPE | 11 | 14 | 17 |
P value = 0.726 (Fisher's exact test), Q value = 1
Table S48. Gene #4: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| PIK3CA MUTATED | 6 | 4 |
| PIK3CA WILD-TYPE | 20 | 22 |
P value = 0.376 (Fisher's exact test), Q value = 1
Table S49. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| PTEN MUTATED | 4 | 2 | 0 | 2 |
| PTEN WILD-TYPE | 33 | 46 | 16 | 17 |
P value = 0.427 (Fisher's exact test), Q value = 1
Table S50. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| PTEN MUTATED | 1 | 2 | 1 | 4 |
| PTEN WILD-TYPE | 6 | 50 | 19 | 37 |
P value = 0.439 (Fisher's exact test), Q value = 1
Table S51. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| PTEN MUTATED | 7 | 4 | 3 |
| PTEN WILD-TYPE | 54 | 65 | 44 |
P value = 0.134 (Fisher's exact test), Q value = 1
Table S52. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| PTEN MUTATED | 5 | 3 | 0 |
| PTEN WILD-TYPE | 22 | 26 | 18 |
P value = 0.721 (Fisher's exact test), Q value = 1
Table S53. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| PTEN MUTATED | 3 | 4 | 1 |
| PTEN WILD-TYPE | 31 | 43 | 30 |
P value = 0.473 (Fisher's exact test), Q value = 1
Table S54. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| PTEN MUTATED | 2 | 2 | 1 | 3 |
| PTEN WILD-TYPE | 29 | 25 | 32 | 18 |
P value = 0.613 (Fisher's exact test), Q value = 1
Table S55. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| PTEN MUTATED | 4 | 3 | 4 | 3 |
| PTEN WILD-TYPE | 38 | 58 | 45 | 22 |
P value = 0.158 (Fisher's exact test), Q value = 1
Table S56. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| PTEN MUTATED | 5 | 2 | 7 |
| PTEN WILD-TYPE | 29 | 58 | 76 |
P value = 0.575 (Fisher's exact test), Q value = 1
Table S57. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| PTEN MUTATED | 9 | 1 | 2 | 0 |
| PTEN WILD-TYPE | 90 | 15 | 21 | 23 |
P value = 0.431 (Fisher's exact test), Q value = 1
Table S58. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| PTEN MUTATED | 1 | 4 | 7 |
| PTEN WILD-TYPE | 29 | 28 | 92 |
P value = 0.357 (Fisher's exact test), Q value = 1
Table S59. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| PTEN MUTATED | 0 | 2 | 3 |
| PTEN WILD-TYPE | 15 | 16 | 16 |
P value = 1 (Fisher's exact test), Q value = 1
Table S60. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| PTEN MUTATED | 3 | 2 |
| PTEN WILD-TYPE | 23 | 24 |
P value = 0.00196 (Fisher's exact test), Q value = 0.35
Table S61. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| KEAP1 MUTATED | 0 | 12 | 1 | 4 |
| KEAP1 WILD-TYPE | 37 | 36 | 15 | 15 |
Figure S15. Get High-res Image Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
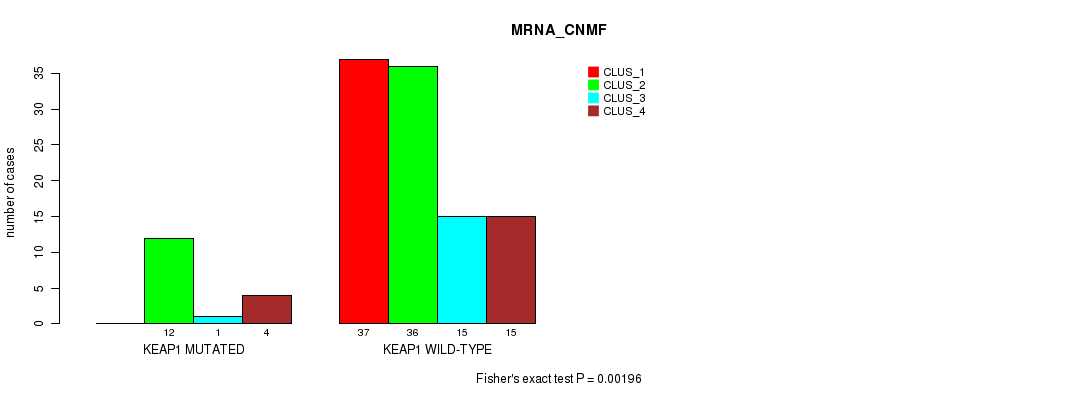
P value = 0.000403 (Fisher's exact test), Q value = 0.075
Table S62. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| KEAP1 MUTATED | 3 | 13 | 0 | 1 |
| KEAP1 WILD-TYPE | 4 | 39 | 20 | 40 |
Figure S16. Get High-res Image Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
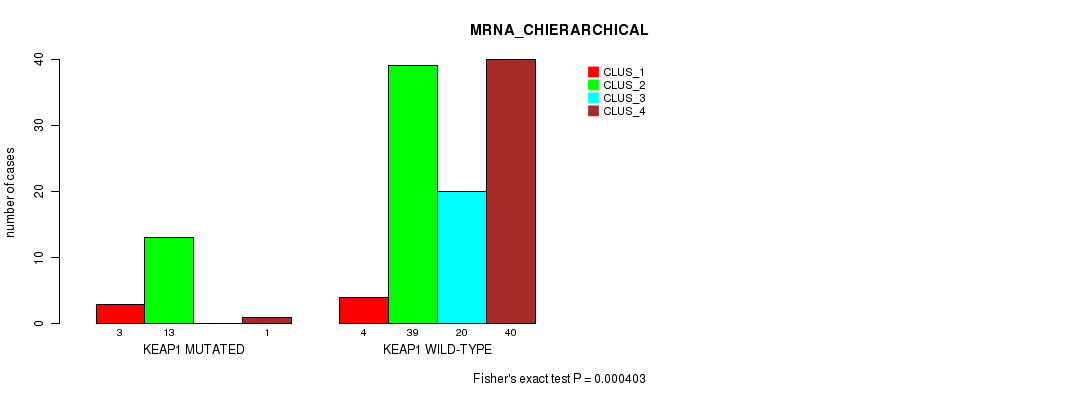
P value = 0.17 (Fisher's exact test), Q value = 1
Table S63. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| KEAP1 MUTATED | 4 | 12 | 6 |
| KEAP1 WILD-TYPE | 57 | 57 | 41 |
P value = 0.0758 (Fisher's exact test), Q value = 1
Table S64. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| KEAP1 MUTATED | 0 | 4 | 3 |
| KEAP1 WILD-TYPE | 27 | 25 | 15 |
P value = 0.204 (Fisher's exact test), Q value = 1
Table S65. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| KEAP1 MUTATED | 5 | 2 | 4 |
| KEAP1 WILD-TYPE | 29 | 45 | 27 |
P value = 0.0963 (Fisher's exact test), Q value = 1
Table S66. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| KEAP1 MUTATED | 3 | 6 | 1 | 1 |
| KEAP1 WILD-TYPE | 28 | 21 | 32 | 20 |
P value = 0.159 (Fisher's exact test), Q value = 1
Table S67. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| KEAP1 MUTATED | 2 | 12 | 5 | 3 |
| KEAP1 WILD-TYPE | 40 | 49 | 44 | 22 |
P value = 0.0428 (Fisher's exact test), Q value = 1
Table S68. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| KEAP1 MUTATED | 6 | 11 | 5 |
| KEAP1 WILD-TYPE | 28 | 49 | 78 |
Figure S17. Get High-res Image Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
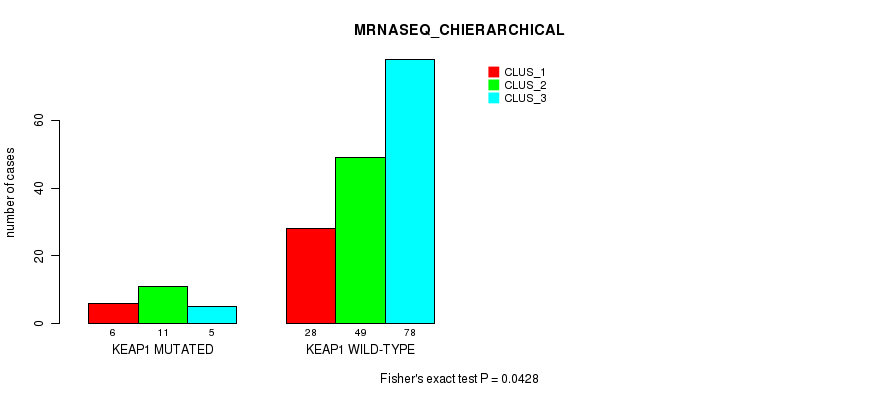
P value = 0.701 (Fisher's exact test), Q value = 1
Table S69. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| KEAP1 MUTATED | 13 | 1 | 1 | 3 |
| KEAP1 WILD-TYPE | 86 | 15 | 22 | 20 |
P value = 0.824 (Fisher's exact test), Q value = 1
Table S70. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| KEAP1 MUTATED | 4 | 4 | 10 |
| KEAP1 WILD-TYPE | 26 | 28 | 89 |
P value = 0.00504 (Fisher's exact test), Q value = 0.9
Table S71. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| KEAP1 MUTATED | 4 | 0 | 0 |
| KEAP1 WILD-TYPE | 11 | 18 | 19 |
Figure S18. Get High-res Image Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
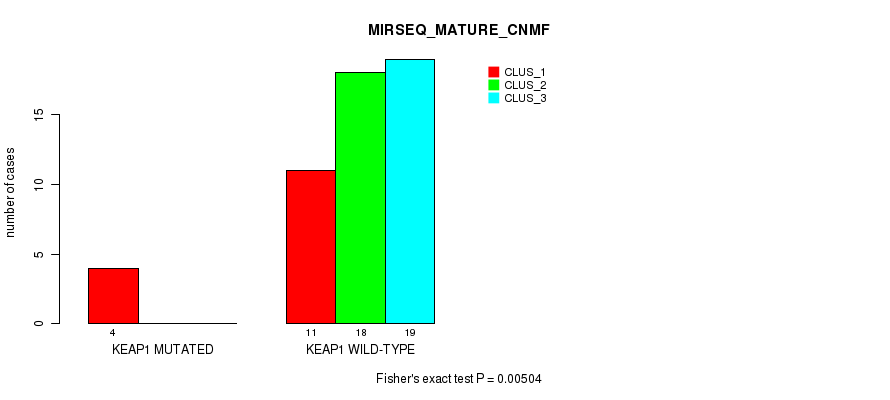
P value = 0.61 (Fisher's exact test), Q value = 1
Table S72. Gene #6: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| KEAP1 MUTATED | 1 | 3 |
| KEAP1 WILD-TYPE | 25 | 23 |
P value = 0.146 (Fisher's exact test), Q value = 1
Table S73. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| MLL2 MUTATED | 4 | 10 | 6 | 5 |
| MLL2 WILD-TYPE | 33 | 38 | 10 | 14 |
P value = 0.809 (Fisher's exact test), Q value = 1
Table S74. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| MLL2 MUTATED | 1 | 13 | 3 | 8 |
| MLL2 WILD-TYPE | 6 | 39 | 17 | 33 |
P value = 0.253 (Fisher's exact test), Q value = 1
Table S75. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| MLL2 MUTATED | 8 | 16 | 11 |
| MLL2 WILD-TYPE | 53 | 53 | 36 |
P value = 0.315 (Fisher's exact test), Q value = 1
Table S76. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| MLL2 MUTATED | 6 | 6 | 1 |
| MLL2 WILD-TYPE | 21 | 23 | 17 |
P value = 0.696 (Fisher's exact test), Q value = 1
Table S77. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| MLL2 MUTATED | 6 | 9 | 8 |
| MLL2 WILD-TYPE | 28 | 38 | 23 |
P value = 0.221 (Fisher's exact test), Q value = 1
Table S78. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| MLL2 MUTATED | 5 | 4 | 6 | 8 |
| MLL2 WILD-TYPE | 26 | 23 | 27 | 13 |
P value = 0.248 (Fisher's exact test), Q value = 1
Table S79. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| MLL2 MUTATED | 4 | 15 | 11 | 5 |
| MLL2 WILD-TYPE | 38 | 46 | 38 | 20 |
P value = 0.611 (Fisher's exact test), Q value = 1
Table S80. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| MLL2 MUTATED | 5 | 14 | 16 |
| MLL2 WILD-TYPE | 29 | 46 | 67 |
P value = 0.948 (Fisher's exact test), Q value = 1
Table S81. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| MLL2 MUTATED | 17 | 3 | 4 | 5 |
| MLL2 WILD-TYPE | 82 | 13 | 19 | 18 |
P value = 0.638 (Fisher's exact test), Q value = 1
Table S82. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| MLL2 MUTATED | 5 | 4 | 20 |
| MLL2 WILD-TYPE | 25 | 28 | 79 |
P value = 0.614 (Fisher's exact test), Q value = 1
Table S83. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| MLL2 MUTATED | 2 | 5 | 3 |
| MLL2 WILD-TYPE | 13 | 13 | 16 |
P value = 1 (Fisher's exact test), Q value = 1
Table S84. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| MLL2 MUTATED | 5 | 5 |
| MLL2 WILD-TYPE | 21 | 21 |
P value = 0.24 (Fisher's exact test), Q value = 1
Table S85. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| CSMD3 MUTATED | 16 | 17 | 10 | 10 |
| CSMD3 WILD-TYPE | 21 | 31 | 6 | 9 |
P value = 0.878 (Fisher's exact test), Q value = 1
Table S86. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| CSMD3 MUTATED | 3 | 21 | 9 | 20 |
| CSMD3 WILD-TYPE | 4 | 31 | 11 | 21 |
P value = 0.746 (Fisher's exact test), Q value = 1
Table S87. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| CSMD3 MUTATED | 26 | 34 | 21 |
| CSMD3 WILD-TYPE | 35 | 35 | 26 |
P value = 0.479 (Fisher's exact test), Q value = 1
Table S88. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| CSMD3 MUTATED | 14 | 14 | 6 |
| CSMD3 WILD-TYPE | 13 | 15 | 12 |
P value = 0.275 (Fisher's exact test), Q value = 1
Table S89. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| CSMD3 MUTATED | 17 | 23 | 10 |
| CSMD3 WILD-TYPE | 17 | 24 | 21 |
P value = 0.765 (Fisher's exact test), Q value = 1
Table S90. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| CSMD3 MUTATED | 16 | 11 | 15 | 8 |
| CSMD3 WILD-TYPE | 15 | 16 | 18 | 13 |
P value = 0.429 (Fisher's exact test), Q value = 1
Table S91. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| CSMD3 MUTATED | 17 | 26 | 23 | 15 |
| CSMD3 WILD-TYPE | 25 | 35 | 26 | 10 |
P value = 0.769 (Fisher's exact test), Q value = 1
Table S92. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| CSMD3 MUTATED | 14 | 27 | 40 |
| CSMD3 WILD-TYPE | 20 | 33 | 43 |
P value = 0.642 (Fisher's exact test), Q value = 1
Table S93. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| CSMD3 MUTATED | 44 | 6 | 12 | 8 |
| CSMD3 WILD-TYPE | 55 | 10 | 11 | 15 |
P value = 1 (Fisher's exact test), Q value = 1
Table S94. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| CSMD3 MUTATED | 13 | 14 | 43 |
| CSMD3 WILD-TYPE | 17 | 18 | 56 |
P value = 0.389 (Fisher's exact test), Q value = 1
Table S95. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| CSMD3 MUTATED | 9 | 8 | 7 |
| CSMD3 WILD-TYPE | 6 | 10 | 12 |
P value = 0.164 (Fisher's exact test), Q value = 1
Table S96. Gene #8: 'CSMD3 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| CSMD3 MUTATED | 9 | 15 |
| CSMD3 WILD-TYPE | 17 | 11 |
P value = 0.343 (Fisher's exact test), Q value = 1
Table S97. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| RB1 MUTATED | 2 | 5 | 0 | 3 |
| RB1 WILD-TYPE | 35 | 43 | 16 | 16 |
P value = 0.198 (Fisher's exact test), Q value = 1
Table S98. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| RB1 MUTATED | 2 | 5 | 1 | 2 |
| RB1 WILD-TYPE | 5 | 47 | 19 | 39 |
P value = 0.418 (Fisher's exact test), Q value = 1
Table S99. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| RB1 MUTATED | 4 | 3 | 5 |
| RB1 WILD-TYPE | 57 | 66 | 42 |
P value = 0.108 (Fisher's exact test), Q value = 1
Table S100. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| RB1 MUTATED | 0 | 4 | 1 |
| RB1 WILD-TYPE | 27 | 25 | 17 |
P value = 0.657 (Fisher's exact test), Q value = 1
Table S101. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| RB1 MUTATED | 3 | 2 | 1 |
| RB1 WILD-TYPE | 31 | 45 | 30 |
P value = 0.433 (Fisher's exact test), Q value = 1
Table S102. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| RB1 MUTATED | 1 | 3 | 2 | 0 |
| RB1 WILD-TYPE | 30 | 24 | 31 | 21 |
P value = 1 (Fisher's exact test), Q value = 1
Table S103. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| RB1 MUTATED | 3 | 4 | 3 | 2 |
| RB1 WILD-TYPE | 39 | 57 | 46 | 23 |
P value = 0.355 (Fisher's exact test), Q value = 1
Table S104. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| RB1 MUTATED | 4 | 4 | 4 |
| RB1 WILD-TYPE | 30 | 56 | 79 |
P value = 0.575 (Fisher's exact test), Q value = 1
Table S105. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| RB1 MUTATED | 9 | 1 | 0 | 2 |
| RB1 WILD-TYPE | 90 | 15 | 23 | 21 |
P value = 0.911 (Fisher's exact test), Q value = 1
Table S106. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| RB1 MUTATED | 2 | 3 | 7 |
| RB1 WILD-TYPE | 28 | 29 | 92 |
P value = 1 (Fisher's exact test), Q value = 1
Table S107. Gene #9: 'RB1 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| RB1 MUTATED | 0 | 1 |
| RB1 WILD-TYPE | 26 | 25 |
P value = 0.269 (Fisher's exact test), Q value = 1
Table S108. Gene #10: 'HS6ST1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| HS6ST1 MUTATED | 1 | 1 | 2 | 1 |
| HS6ST1 WILD-TYPE | 36 | 47 | 14 | 18 |
P value = 0.295 (Fisher's exact test), Q value = 1
Table S109. Gene #10: 'HS6ST1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| HS6ST1 MUTATED | 1 | 1 | 1 | 2 |
| HS6ST1 WILD-TYPE | 6 | 51 | 19 | 39 |
P value = 0.53 (Fisher's exact test), Q value = 1
Table S110. Gene #10: 'HS6ST1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| HS6ST1 MUTATED | 3 | 1 | 1 |
| HS6ST1 WILD-TYPE | 58 | 68 | 46 |
P value = 0.568 (Fisher's exact test), Q value = 1
Table S111. Gene #10: 'HS6ST1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| HS6ST1 MUTATED | 1 | 1 | 3 | 0 |
| HS6ST1 WILD-TYPE | 41 | 60 | 46 | 25 |
P value = 0.101 (Fisher's exact test), Q value = 1
Table S112. Gene #10: 'HS6ST1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| HS6ST1 MUTATED | 3 | 1 | 1 |
| HS6ST1 WILD-TYPE | 31 | 59 | 82 |
P value = 1 (Fisher's exact test), Q value = 1
Table S113. Gene #10: 'HS6ST1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| HS6ST1 MUTATED | 3 | 0 | 0 | 0 |
| HS6ST1 WILD-TYPE | 96 | 16 | 23 | 23 |
P value = 1 (Fisher's exact test), Q value = 1
Table S114. Gene #10: 'HS6ST1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| HS6ST1 MUTATED | 0 | 0 | 3 |
| HS6ST1 WILD-TYPE | 30 | 32 | 96 |
P value = 0.458 (Fisher's exact test), Q value = 1
Table S115. Gene #11: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| FBXW7 MUTATED | 1 | 4 | 2 | 2 |
| FBXW7 WILD-TYPE | 36 | 44 | 14 | 17 |
P value = 1 (Fisher's exact test), Q value = 1
Table S116. Gene #11: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| FBXW7 MUTATED | 0 | 5 | 1 | 3 |
| FBXW7 WILD-TYPE | 7 | 47 | 19 | 38 |
P value = 0.508 (Fisher's exact test), Q value = 1
Table S117. Gene #11: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| FBXW7 MUTATED | 4 | 5 | 1 |
| FBXW7 WILD-TYPE | 57 | 64 | 46 |
P value = 0.621 (Fisher's exact test), Q value = 1
Table S118. Gene #11: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| FBXW7 MUTATED | 0 | 2 | 1 |
| FBXW7 WILD-TYPE | 34 | 45 | 30 |
P value = 0.785 (Fisher's exact test), Q value = 1
Table S119. Gene #11: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| FBXW7 MUTATED | 0 | 1 | 1 | 1 |
| FBXW7 WILD-TYPE | 31 | 26 | 32 | 20 |
P value = 0.834 (Fisher's exact test), Q value = 1
Table S120. Gene #11: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| FBXW7 MUTATED | 2 | 5 | 2 | 1 |
| FBXW7 WILD-TYPE | 40 | 56 | 47 | 24 |
P value = 0.574 (Fisher's exact test), Q value = 1
Table S121. Gene #11: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| FBXW7 MUTATED | 1 | 5 | 4 |
| FBXW7 WILD-TYPE | 33 | 55 | 79 |
P value = 0.935 (Fisher's exact test), Q value = 1
Table S122. Gene #11: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| FBXW7 MUTATED | 6 | 0 | 1 | 1 |
| FBXW7 WILD-TYPE | 93 | 16 | 22 | 22 |
P value = 0.772 (Fisher's exact test), Q value = 1
Table S123. Gene #11: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| FBXW7 MUTATED | 2 | 1 | 5 |
| FBXW7 WILD-TYPE | 28 | 31 | 94 |
P value = 1 (Fisher's exact test), Q value = 1
Table S124. Gene #11: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| FBXW7 MUTATED | 0 | 1 |
| FBXW7 WILD-TYPE | 26 | 25 |
P value = 0.364 (Fisher's exact test), Q value = 1
Table S125. Gene #12: 'ZNF567 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| ZNF567 MUTATED | 2 | 0 | 1 |
| ZNF567 WILD-TYPE | 59 | 69 | 46 |
P value = 0.359 (Fisher's exact test), Q value = 1
Table S126. Gene #12: 'ZNF567 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| ZNF567 MUTATED | 0 | 1 | 2 |
| ZNF567 WILD-TYPE | 34 | 46 | 29 |
P value = 0.424 (Fisher's exact test), Q value = 1
Table S127. Gene #12: 'ZNF567 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| ZNF567 MUTATED | 2 | 1 | 0 | 0 |
| ZNF567 WILD-TYPE | 29 | 26 | 33 | 21 |
P value = 0.389 (Fisher's exact test), Q value = 1
Table S128. Gene #12: 'ZNF567 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| ZNF567 MUTATED | 1 | 0 | 1 | 1 |
| ZNF567 WILD-TYPE | 41 | 61 | 48 | 24 |
P value = 0.427 (Fisher's exact test), Q value = 1
Table S129. Gene #12: 'ZNF567 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| ZNF567 MUTATED | 1 | 0 | 2 |
| ZNF567 WILD-TYPE | 33 | 60 | 81 |
P value = 0.253 (Fisher's exact test), Q value = 1
Table S130. Gene #12: 'ZNF567 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| ZNF567 MUTATED | 1 | 1 | 1 | 0 |
| ZNF567 WILD-TYPE | 98 | 15 | 22 | 23 |
P value = 0.19 (Fisher's exact test), Q value = 1
Table S131. Gene #12: 'ZNF567 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| ZNF567 MUTATED | 0 | 2 | 1 |
| ZNF567 WILD-TYPE | 30 | 30 | 98 |
P value = 1 (Fisher's exact test), Q value = 1
Table S132. Gene #12: 'ZNF567 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| ZNF567 MUTATED | 0 | 1 |
| ZNF567 WILD-TYPE | 26 | 25 |
P value = 0.0295 (Fisher's exact test), Q value = 1
Table S133. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| ELTD1 MUTATED | 3 | 2 | 5 | 2 |
| ELTD1 WILD-TYPE | 34 | 46 | 11 | 17 |
Figure S19. Get High-res Image Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'

P value = 0.303 (Fisher's exact test), Q value = 1
Table S134. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| ELTD1 MUTATED | 0 | 3 | 2 | 7 |
| ELTD1 WILD-TYPE | 7 | 49 | 18 | 34 |
P value = 0.0533 (Fisher's exact test), Q value = 1
Table S135. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| ELTD1 MUTATED | 11 | 4 | 3 |
| ELTD1 WILD-TYPE | 50 | 65 | 44 |
P value = 0.6 (Fisher's exact test), Q value = 1
Table S136. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| ELTD1 MUTATED | 4 | 2 | 1 |
| ELTD1 WILD-TYPE | 23 | 27 | 17 |
P value = 0.341 (Fisher's exact test), Q value = 1
Table S137. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| ELTD1 MUTATED | 4 | 6 | 1 |
| ELTD1 WILD-TYPE | 30 | 41 | 30 |
P value = 0.648 (Fisher's exact test), Q value = 1
Table S138. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| ELTD1 MUTATED | 4 | 1 | 4 | 2 |
| ELTD1 WILD-TYPE | 27 | 26 | 29 | 19 |
P value = 0.554 (Fisher's exact test), Q value = 1
Table S139. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| ELTD1 MUTATED | 4 | 4 | 7 | 3 |
| ELTD1 WILD-TYPE | 38 | 57 | 42 | 22 |
P value = 0.42 (Fisher's exact test), Q value = 1
Table S140. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| ELTD1 MUTATED | 5 | 4 | 9 |
| ELTD1 WILD-TYPE | 29 | 56 | 74 |
P value = 0.0887 (Fisher's exact test), Q value = 1
Table S141. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| ELTD1 MUTATED | 7 | 0 | 5 | 2 |
| ELTD1 WILD-TYPE | 92 | 16 | 18 | 21 |
P value = 0.722 (Fisher's exact test), Q value = 1
Table S142. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| ELTD1 MUTATED | 2 | 4 | 8 |
| ELTD1 WILD-TYPE | 28 | 28 | 91 |
P value = 0.305 (Fisher's exact test), Q value = 1
Table S143. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| ELTD1 MUTATED | 2 | 4 | 1 |
| ELTD1 WILD-TYPE | 13 | 14 | 18 |
P value = 0.419 (Fisher's exact test), Q value = 1
Table S144. Gene #13: 'ELTD1 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| ELTD1 MUTATED | 5 | 2 |
| ELTD1 WILD-TYPE | 21 | 24 |
P value = 0.0924 (Fisher's exact test), Q value = 1
Table S145. Gene #14: 'ASCL4 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| ASCL4 MUTATED | 0 | 2 | 2 | 2 |
| ASCL4 WILD-TYPE | 37 | 46 | 14 | 17 |
P value = 0.534 (Fisher's exact test), Q value = 1
Table S146. Gene #14: 'ASCL4 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| ASCL4 MUTATED | 1 | 2 | 1 | 2 |
| ASCL4 WILD-TYPE | 6 | 50 | 19 | 39 |
P value = 0.882 (Fisher's exact test), Q value = 1
Table S147. Gene #14: 'ASCL4 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| ASCL4 MUTATED | 2 | 3 | 1 |
| ASCL4 WILD-TYPE | 59 | 66 | 46 |
P value = 0.783 (Fisher's exact test), Q value = 1
Table S148. Gene #14: 'ASCL4 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| ASCL4 MUTATED | 1 | 2 | 0 |
| ASCL4 WILD-TYPE | 26 | 27 | 18 |
P value = 0.817 (Fisher's exact test), Q value = 1
Table S149. Gene #14: 'ASCL4 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| ASCL4 MUTATED | 2 | 1 | 1 |
| ASCL4 WILD-TYPE | 32 | 46 | 30 |
P value = 0.835 (Fisher's exact test), Q value = 1
Table S150. Gene #14: 'ASCL4 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| ASCL4 MUTATED | 2 | 1 | 1 | 0 |
| ASCL4 WILD-TYPE | 29 | 26 | 32 | 21 |
P value = 0.348 (Fisher's exact test), Q value = 1
Table S151. Gene #14: 'ASCL4 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| ASCL4 MUTATED | 0 | 3 | 3 | 0 |
| ASCL4 WILD-TYPE | 42 | 58 | 46 | 25 |
P value = 0.247 (Fisher's exact test), Q value = 1
Table S152. Gene #14: 'ASCL4 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| ASCL4 MUTATED | 2 | 3 | 1 |
| ASCL4 WILD-TYPE | 32 | 57 | 82 |
P value = 0.91 (Fisher's exact test), Q value = 1
Table S153. Gene #14: 'ASCL4 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| ASCL4 MUTATED | 5 | 0 | 0 | 1 |
| ASCL4 WILD-TYPE | 94 | 16 | 23 | 22 |
P value = 0.219 (Fisher's exact test), Q value = 1
Table S154. Gene #14: 'ASCL4 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| ASCL4 MUTATED | 2 | 2 | 2 |
| ASCL4 WILD-TYPE | 28 | 30 | 97 |
P value = 0.366 (Fisher's exact test), Q value = 1
Table S155. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| OR2G6 MUTATED | 5 | 2 | 2 | 2 |
| OR2G6 WILD-TYPE | 32 | 46 | 14 | 17 |
P value = 0.211 (Fisher's exact test), Q value = 1
Table S156. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| OR2G6 MUTATED | 1 | 2 | 2 | 6 |
| OR2G6 WILD-TYPE | 6 | 50 | 18 | 35 |
P value = 0.909 (Fisher's exact test), Q value = 1
Table S157. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| OR2G6 MUTATED | 5 | 6 | 5 |
| OR2G6 WILD-TYPE | 56 | 63 | 42 |
P value = 1 (Fisher's exact test), Q value = 1
Table S158. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| OR2G6 MUTATED | 2 | 3 | 1 |
| OR2G6 WILD-TYPE | 25 | 26 | 17 |
P value = 1 (Fisher's exact test), Q value = 1
Table S159. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| OR2G6 MUTATED | 2 | 4 | 2 |
| OR2G6 WILD-TYPE | 32 | 43 | 29 |
P value = 0.717 (Fisher's exact test), Q value = 1
Table S160. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| OR2G6 MUTATED | 2 | 1 | 4 | 1 |
| OR2G6 WILD-TYPE | 29 | 26 | 29 | 20 |
P value = 0.199 (Fisher's exact test), Q value = 1
Table S161. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| OR2G6 MUTATED | 4 | 3 | 8 | 1 |
| OR2G6 WILD-TYPE | 38 | 58 | 41 | 24 |
P value = 0.0466 (Fisher's exact test), Q value = 1
Table S162. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| OR2G6 MUTATED | 7 | 3 | 6 |
| OR2G6 WILD-TYPE | 27 | 57 | 77 |
Figure S20. Get High-res Image Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
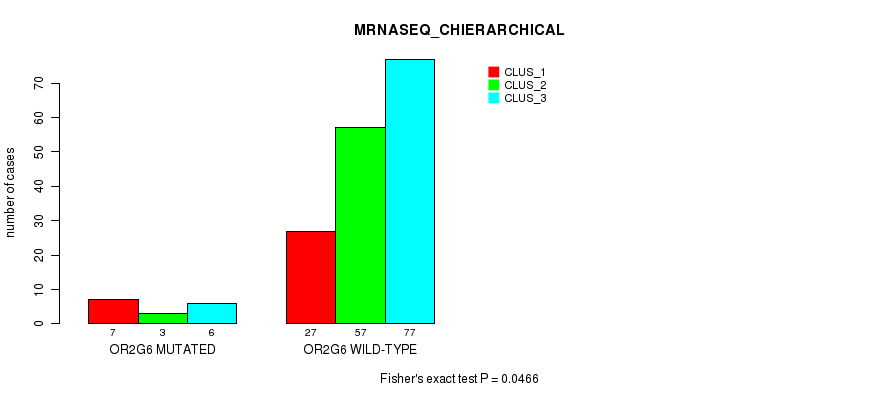
P value = 0.428 (Fisher's exact test), Q value = 1
Table S163. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| OR2G6 MUTATED | 9 | 3 | 1 | 1 |
| OR2G6 WILD-TYPE | 90 | 13 | 22 | 22 |
P value = 0.245 (Fisher's exact test), Q value = 1
Table S164. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| OR2G6 MUTATED | 1 | 5 | 8 |
| OR2G6 WILD-TYPE | 29 | 27 | 91 |
P value = 0.668 (Fisher's exact test), Q value = 1
Table S165. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| OR2G6 MUTATED | 2 | 1 | 1 |
| OR2G6 WILD-TYPE | 13 | 17 | 18 |
P value = 0.61 (Fisher's exact test), Q value = 1
Table S166. Gene #15: 'OR2G6 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| OR2G6 MUTATED | 1 | 3 |
| OR2G6 WILD-TYPE | 25 | 23 |
P value = 0.0391 (Fisher's exact test), Q value = 1
Table S167. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| HLA-A MUTATED | 1 | 0 | 2 | 0 |
| HLA-A WILD-TYPE | 36 | 48 | 14 | 19 |
Figure S21. Get High-res Image Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
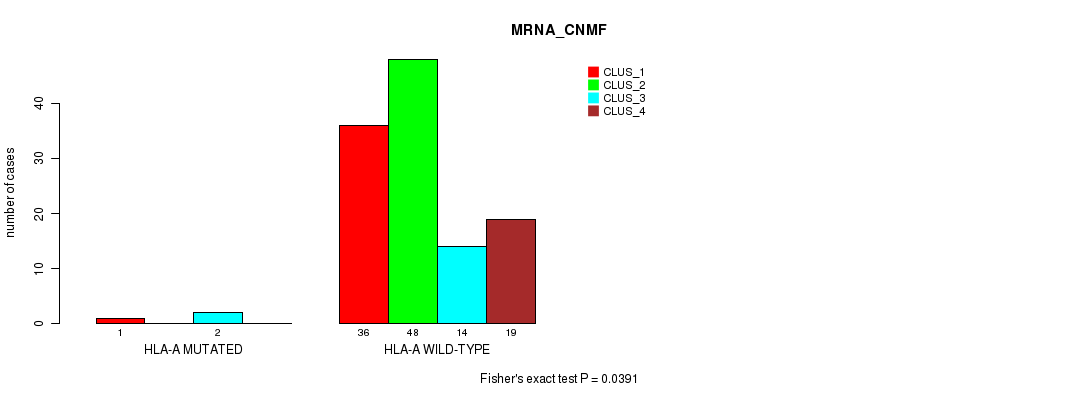
P value = 0.33 (Fisher's exact test), Q value = 1
Table S168. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| HLA-A MUTATED | 0 | 0 | 1 | 2 |
| HLA-A WILD-TYPE | 7 | 52 | 19 | 39 |
P value = 0.446 (Fisher's exact test), Q value = 1
Table S169. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| HLA-A MUTATED | 1 | 2 | 3 |
| HLA-A WILD-TYPE | 60 | 67 | 44 |
P value = 0.178 (Fisher's exact test), Q value = 1
Table S170. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| HLA-A MUTATED | 1 | 0 | 2 |
| HLA-A WILD-TYPE | 26 | 29 | 16 |
P value = 1 (Fisher's exact test), Q value = 1
Table S171. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| HLA-A MUTATED | 1 | 2 | 1 |
| HLA-A WILD-TYPE | 33 | 45 | 30 |
P value = 0.513 (Fisher's exact test), Q value = 1
Table S172. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| HLA-A MUTATED | 2 | 0 | 2 | 0 |
| HLA-A WILD-TYPE | 29 | 27 | 31 | 21 |
P value = 0.128 (Fisher's exact test), Q value = 1
Table S173. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| HLA-A MUTATED | 2 | 0 | 2 | 2 |
| HLA-A WILD-TYPE | 40 | 61 | 47 | 23 |
P value = 0.114 (Fisher's exact test), Q value = 1
Table S174. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| HLA-A MUTATED | 1 | 0 | 5 |
| HLA-A WILD-TYPE | 33 | 60 | 78 |
P value = 0.471 (Fisher's exact test), Q value = 1
Table S175. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| HLA-A MUTATED | 4 | 0 | 2 | 0 |
| HLA-A WILD-TYPE | 95 | 16 | 21 | 23 |
P value = 0.73 (Fisher's exact test), Q value = 1
Table S176. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| HLA-A MUTATED | 0 | 1 | 5 |
| HLA-A WILD-TYPE | 30 | 31 | 94 |
P value = 0.381 (Fisher's exact test), Q value = 1
Table S177. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| HLA-A MUTATED | 1 | 2 | 0 |
| HLA-A WILD-TYPE | 14 | 16 | 19 |
P value = 1 (Fisher's exact test), Q value = 1
Table S178. Gene #16: 'HLA-A MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| HLA-A MUTATED | 1 | 2 |
| HLA-A WILD-TYPE | 25 | 24 |
P value = 0.812 (Fisher's exact test), Q value = 1
Table S179. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 16 | 19 |
| NOTCH1 MUTATED | 1 | 4 | 1 | 1 |
| NOTCH1 WILD-TYPE | 36 | 44 | 15 | 18 |
P value = 0.921 (Fisher's exact test), Q value = 1
Table S180. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 7 | 52 | 20 | 41 |
| NOTCH1 MUTATED | 0 | 4 | 1 | 2 |
| NOTCH1 WILD-TYPE | 7 | 48 | 19 | 39 |
P value = 0.543 (Fisher's exact test), Q value = 1
Table S181. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 69 | 47 |
| NOTCH1 MUTATED | 3 | 7 | 4 |
| NOTCH1 WILD-TYPE | 58 | 62 | 43 |
P value = 0.0342 (Fisher's exact test), Q value = 1
Table S182. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| NOTCH1 MUTATED | 0 | 4 | 4 |
| NOTCH1 WILD-TYPE | 27 | 25 | 14 |
Figure S22. Get High-res Image Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
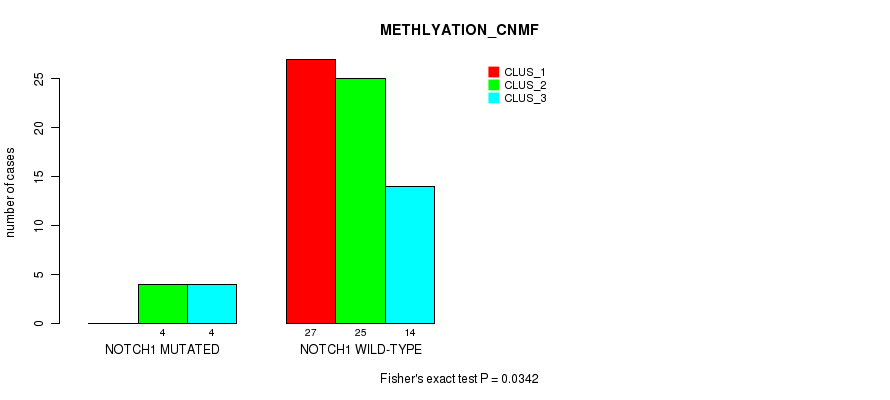
P value = 0.591 (Fisher's exact test), Q value = 1
Table S183. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| NOTCH1 MUTATED | 1 | 4 | 3 |
| NOTCH1 WILD-TYPE | 33 | 43 | 28 |
P value = 0.824 (Fisher's exact test), Q value = 1
Table S184. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 27 | 33 | 21 |
| NOTCH1 MUTATED | 3 | 1 | 3 | 1 |
| NOTCH1 WILD-TYPE | 28 | 26 | 30 | 20 |
P value = 0.686 (Fisher's exact test), Q value = 1
Table S185. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 42 | 61 | 49 | 25 |
| NOTCH1 MUTATED | 2 | 6 | 5 | 1 |
| NOTCH1 WILD-TYPE | 40 | 55 | 44 | 24 |
P value = 0.401 (Fisher's exact test), Q value = 1
Table S186. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 60 | 83 |
| NOTCH1 MUTATED | 2 | 7 | 5 |
| NOTCH1 WILD-TYPE | 32 | 53 | 78 |
P value = 0.185 (Fisher's exact test), Q value = 1
Table S187. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 99 | 16 | 23 | 23 |
| NOTCH1 MUTATED | 8 | 3 | 0 | 2 |
| NOTCH1 WILD-TYPE | 91 | 13 | 23 | 21 |
P value = 0.844 (Fisher's exact test), Q value = 1
Table S188. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 30 | 32 | 99 |
| NOTCH1 MUTATED | 3 | 2 | 8 |
| NOTCH1 WILD-TYPE | 27 | 30 | 91 |
P value = 0.483 (Fisher's exact test), Q value = 1
Table S189. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 15 | 18 | 19 |
| NOTCH1 MUTATED | 3 | 1 | 2 |
| NOTCH1 WILD-TYPE | 12 | 17 | 17 |
P value = 0.668 (Fisher's exact test), Q value = 1
Table S190. Gene #17: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 |
|---|---|---|
| ALL | 26 | 26 |
| NOTCH1 MUTATED | 2 | 4 |
| NOTCH1 WILD-TYPE | 24 | 22 |
-
Mutation data file = transformed.cor.cli.txt
-
Molecular subtypes file = LUSC-TP.transferedmergedcluster.txt
-
Number of patients = 177
-
Number of significantly mutated genes = 17
-
Number of Molecular subtypes = 12
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.