This pipeline computes the correlation between significantly recurrent gene mutations and selected clinical features.
Testing the association between mutation status of 102 genes and 12 clinical features across 44 patients, 5 significant findings detected with Q value < 0.25.
-
FYN mutation correlated to 'Time to Death'.
-
FADS2 mutation correlated to 'NEOPLASM.DISEASESTAGE'.
-
CEL mutation correlated to 'NUMBER.OF.LYMPH.NODES'.
-
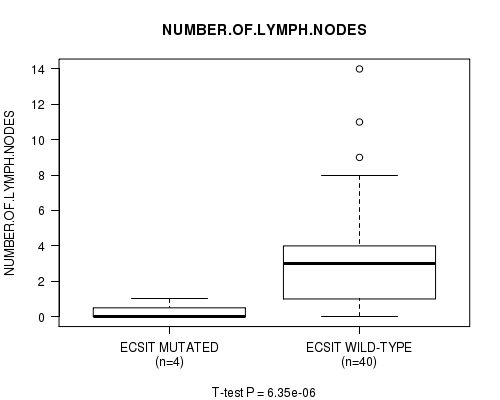
ECSIT mutation correlated to 'NUMBER.OF.LYMPH.NODES'.
-
PLCZ1 mutation correlated to 'NUMBER.OF.LYMPH.NODES'.
Table 1. Get Full Table Overview of the association between mutation status of 102 genes and 12 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 5 significant findings detected.
|
Clinical Features |
Time to Death |
AGE |
NEOPLASM DISEASESTAGE |
PATHOLOGY T STAGE |
PATHOLOGY N STAGE |
PATHOLOGY M STAGE |
GENDER |
HISTOLOGICAL TYPE |
NUMBERPACKYEARSSMOKED | YEAROFTOBACCOSMOKINGONSET |
COMPLETENESS OF RESECTION |
NUMBER OF LYMPH NODES |
||
| nMutated (%) | nWild-Type | logrank test | t-test | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | t-test | t-test | Fisher's exact test | t-test | |
| FYN | 5 (11%) | 39 |
0 (0) |
0.731 (1.00) |
0.0354 (1.00) |
0.394 (1.00) |
0.317 (1.00) |
0.0472 (1.00) |
0.345 (1.00) |
1 (1.00) |
0.0482 (1.00) |
0.0909 (1.00) |
||
| FADS2 | 5 (11%) | 39 |
0.627 (1.00) |
0.789 (1.00) |
7.36e-05 (0.0754) |
0.0575 (1.00) |
0.00681 (1.00) |
1 (1.00) |
0.345 (1.00) |
1 (1.00) |
0.178 (1.00) |
0.0974 (1.00) |
||
| CEL | 4 (9%) | 40 |
0.000604 (0.617) |
0.468 (1.00) |
0.00547 (1.00) |
0.327 (1.00) |
0.0316 (1.00) |
0.45 (1.00) |
0.607 (1.00) |
1 (1.00) |
0.306 (1.00) |
6.35e-06 (0.00651) |
||
| ECSIT | 4 (9%) | 40 |
0.000604 (0.617) |
0.468 (1.00) |
0.00547 (1.00) |
0.327 (1.00) |
0.0316 (1.00) |
0.45 (1.00) |
0.607 (1.00) |
1 (1.00) |
0.306 (1.00) |
6.35e-06 (0.00651) |
||
| PLCZ1 | 3 (7%) | 41 |
0.983 (1.00) |
0.264 (1.00) |
0.000858 (0.875) |
0.254 (1.00) |
0.125 (1.00) |
0.651 (1.00) |
1 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.000227 (0.232) |
||
| C15ORF24 | 6 (14%) | 38 |
0.908 (1.00) |
0.281 (1.00) |
0.081 (1.00) |
0.456 (1.00) |
0.12 (1.00) |
0.554 (1.00) |
0.0211 (1.00) |
1 (1.00) |
0.0702 (1.00) |
0.509 (1.00) |
||
| IRS1 | 4 (9%) | 40 |
0.418 (1.00) |
0.00806 (1.00) |
0.422 (1.00) |
1 (1.00) |
1 (1.00) |
0.265 (1.00) |
1 (1.00) |
1 (1.00) |
0.612 (1.00) |
0.568 (1.00) |
||
| C14ORF49 | 8 (18%) | 36 |
0.584 (1.00) |
0.332 (1.00) |
0.614 (1.00) |
0.566 (1.00) |
0.659 (1.00) |
0.8 (1.00) |
0.24 (1.00) |
1 (1.00) |
0.182 (1.00) |
0.166 (1.00) |
||
| FGF10 | 6 (14%) | 38 |
0.607 (1.00) |
0.626 (1.00) |
0.0779 (1.00) |
0.456 (1.00) |
0.606 (1.00) |
0.751 (1.00) |
0.664 (1.00) |
1 (1.00) |
0.547 (1.00) |
0.541 (1.00) |
||
| CCR3 | 5 (11%) | 39 |
0.997 (1.00) |
0.875 (1.00) |
0.949 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.742 (1.00) |
||
| OR10A2 | 6 (14%) | 38 |
0.707 (1.00) |
0.352 (1.00) |
0.589 (1.00) |
0.456 (1.00) |
1 (1.00) |
0.751 (1.00) |
0.664 (1.00) |
1 (1.00) |
0.547 (1.00) |
0.139 (1.00) |
||
| OR10A7 | 9 (20%) | 35 |
0.431 (1.00) |
0.877 (1.00) |
0.0233 (1.00) |
0.18 (1.00) |
0.175 (1.00) |
0.82 (1.00) |
0.132 (1.00) |
1 (1.00) |
0.156 (1.00) |
0.612 (1.00) |
||
| POP5 | 5 (11%) | 39 |
0.344 (1.00) |
0.522 (1.00) |
0.0354 (1.00) |
0.394 (1.00) |
0.317 (1.00) |
0.353 (1.00) |
0.345 (1.00) |
1 (1.00) |
0.178 (1.00) |
0.696 (1.00) |
||
| SRP14 | 7 (16%) | 37 |
0.602 (1.00) |
0.29 (1.00) |
0.932 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.364 (1.00) |
0.841 (1.00) |
||
| WRN | 4 (9%) | 40 |
0.908 (1.00) |
0.387 (1.00) |
0.354 (1.00) |
0.327 (1.00) |
0.218 (1.00) |
1 (1.00) |
0.108 (1.00) |
1 (1.00) |
0.306 (1.00) |
0.916 (1.00) |
||
| TMEM40 | 8 (18%) | 36 |
0.54 (1.00) |
0.752 (1.00) |
0.614 (1.00) |
0.566 (1.00) |
0.659 (1.00) |
0.8 (1.00) |
0.698 (1.00) |
1 (1.00) |
0.785 (1.00) |
0.458 (1.00) |
||
| KRAS | 23 (52%) | 21 |
0.351 (1.00) |
0.731 (1.00) |
0.483 (1.00) |
0.335 (1.00) |
0.155 (1.00) |
0.674 (1.00) |
1 (1.00) |
0.41 (1.00) |
0.251 (1.00) |
0.981 (1.00) |
0.653 (1.00) |
0.465 (1.00) |
| TP53 | 27 (61%) | 17 |
0.32 (1.00) |
0.34 (1.00) |
0.464 (1.00) |
0.634 (1.00) |
0.473 (1.00) |
0.728 (1.00) |
1 (1.00) |
0.731 (1.00) |
0.604 (1.00) |
0.686 (1.00) |
0.0169 (1.00) |
0.441 (1.00) |
| QRICH1 | 7 (16%) | 37 |
0.743 (1.00) |
0.953 (1.00) |
0.932 (1.00) |
1 (1.00) |
1 (1.00) |
0.782 (1.00) |
1 (1.00) |
0.523 (1.00) |
0.181 (1.00) |
0.0241 (1.00) |
0.0482 (1.00) |
0.568 (1.00) |
| ST6GALNAC5 | 5 (11%) | 39 |
0.797 (1.00) |
0.517 (1.00) |
0.865 (1.00) |
1 (1.00) |
0.573 (1.00) |
0.731 (1.00) |
1 (1.00) |
1 (1.00) |
0.306 (1.00) |
0.646 (1.00) |
||
| TNFSF9 | 6 (14%) | 38 |
0.664 (1.00) |
0.0667 (1.00) |
0.555 (1.00) |
1 (1.00) |
0.606 (1.00) |
0.554 (1.00) |
0.664 (1.00) |
1 (1.00) |
0.227 (1.00) |
0.0737 (1.00) |
||
| CDKN2A | 10 (23%) | 34 |
0.155 (1.00) |
0.368 (1.00) |
0.505 (1.00) |
1 (1.00) |
0.0853 (1.00) |
0.835 (1.00) |
0.721 (1.00) |
1 (1.00) |
0.139 (1.00) |
0.479 (1.00) |
0.191 (1.00) |
0.0626 (1.00) |
| SEH1L | 8 (18%) | 36 |
0.993 (1.00) |
0.212 (1.00) |
0.812 (1.00) |
0.566 (1.00) |
1 (1.00) |
0.8 (1.00) |
0.698 (1.00) |
1 (1.00) |
0.0244 (1.00) |
0.794 (1.00) |
||
| SCD | 8 (18%) | 36 |
0.997 (1.00) |
0.483 (1.00) |
0.0104 (1.00) |
0.145 (1.00) |
0.0641 (1.00) |
0.8 (1.00) |
0.698 (1.00) |
1 (1.00) |
0.0321 (1.00) |
0.139 (1.00) |
||
| TMC4 | 7 (16%) | 37 |
0.976 (1.00) |
0.438 (1.00) |
0.932 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.227 (1.00) |
0.578 (1.00) |
||
| MED15 | 9 (20%) | 35 |
0.922 (1.00) |
0.658 (1.00) |
0.642 (1.00) |
1 (1.00) |
0.402 (1.00) |
1 (1.00) |
0.457 (1.00) |
1 (1.00) |
0.156 (1.00) |
0.068 (1.00) |
||
| TNFRSF9 | 6 (14%) | 38 |
0.784 (1.00) |
0.254 (1.00) |
0.946 (1.00) |
1 (1.00) |
1 (1.00) |
0.751 (1.00) |
1 (1.00) |
1 (1.00) |
0.547 (1.00) |
0.494 (1.00) |
||
| BRDT | 8 (18%) | 36 |
0.422 (1.00) |
0.23 (1.00) |
0.907 (1.00) |
1 (1.00) |
0.659 (1.00) |
0.318 (1.00) |
1 (1.00) |
1 (1.00) |
0.646 (1.00) |
0.378 (1.00) |
||
| CXXC4 | 4 (9%) | 40 |
0.233 (1.00) |
0.555 (1.00) |
0.933 (1.00) |
1 (1.00) |
1 (1.00) |
0.45 (1.00) |
1 (1.00) |
0.334 (1.00) |
0.502 (1.00) |
0.173 (1.00) |
||
| MBD3 | 6 (14%) | 38 |
0.762 (1.00) |
0.633 (1.00) |
0.946 (1.00) |
1 (1.00) |
1 (1.00) |
0.313 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.409 (1.00) |
||
| TULP1 | 6 (14%) | 38 |
0.812 (1.00) |
0.315 (1.00) |
0.0183 (1.00) |
0.456 (1.00) |
0.0179 (1.00) |
1 (1.00) |
0.185 (1.00) |
1 (1.00) |
0.1 (1.00) |
0.227 (1.00) |
||
| PHF13 | 4 (9%) | 40 |
0.919 (1.00) |
0.821 (1.00) |
0.933 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.607 (1.00) |
0.334 (1.00) |
0.855 (1.00) |
1 (1.00) |
0.85 (1.00) |
|
| BHLHB9 | 4 (9%) | 40 |
0.0191 (1.00) |
0.173 (1.00) |
0.422 (1.00) |
1 (1.00) |
1 (1.00) |
0.265 (1.00) |
1 (1.00) |
1 (1.00) |
0.0298 (1.00) |
0.632 (1.00) |
||
| PRDM8 | 6 (14%) | 38 |
0.63 (1.00) |
0.479 (1.00) |
0.946 (1.00) |
1 (1.00) |
1 (1.00) |
0.751 (1.00) |
1 (1.00) |
1 (1.00) |
0.1 (1.00) |
0.212 (1.00) |
||
| SMAD4 | 7 (16%) | 37 |
0.648 (1.00) |
0.902 (1.00) |
0.766 (1.00) |
1 (1.00) |
1 (1.00) |
0.272 (1.00) |
1 (1.00) |
1 (1.00) |
0.694 (1.00) |
0.149 (1.00) |
1 (1.00) |
0.798 (1.00) |
| C14ORF43 | 7 (16%) | 37 |
0.148 (1.00) |
0.374 (1.00) |
0.0709 (1.00) |
0.513 (1.00) |
0.322 (1.00) |
0.272 (1.00) |
1 (1.00) |
1 (1.00) |
0.193 (1.00) |
0.183 (1.00) |
||
| MEPCE | 7 (16%) | 37 |
0.794 (1.00) |
0.68 (1.00) |
0.766 (1.00) |
0.513 (1.00) |
0.649 (1.00) |
0.588 (1.00) |
0.412 (1.00) |
1 (1.00) |
0.0655 (1.00) |
0.224 (1.00) |
||
| HVCN1 | 4 (9%) | 40 |
0.222 (1.00) |
0.753 (1.00) |
0.00547 (1.00) |
0.327 (1.00) |
0.0316 (1.00) |
1 (1.00) |
0.607 (1.00) |
1 (1.00) |
0.306 (1.00) |
0.00295 (1.00) |
||
| FOXP2 | 7 (16%) | 37 |
0.547 (1.00) |
0.385 (1.00) |
0.766 (1.00) |
0.513 (1.00) |
0.649 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.785 (1.00) |
0.837 (1.00) |
||
| GRIN1 | 5 (11%) | 39 |
0.544 (1.00) |
0.631 (1.00) |
0.0292 (1.00) |
0.394 (1.00) |
0.0688 (1.00) |
0.731 (1.00) |
1 (1.00) |
1 (1.00) |
0.178 (1.00) |
0.00922 (1.00) |
||
| MAGEA10 | 4 (9%) | 40 |
0.583 (1.00) |
0.155 (1.00) |
0.0514 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.607 (1.00) |
1 (1.00) |
0.306 (1.00) |
0.836 (1.00) |
||
| C16ORF79 | 4 (9%) | 40 |
0.36 (1.00) |
0.957 (1.00) |
0.354 (1.00) |
0.327 (1.00) |
0.218 (1.00) |
1 (1.00) |
0.607 (1.00) |
1 (1.00) |
0.306 (1.00) |
0.146 (1.00) |
||
| ERF | 6 (14%) | 38 |
0.357 (1.00) |
0.825 (1.00) |
0.589 (1.00) |
0.456 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.93 (1.00) |
||
| IFNGR2 | 4 (9%) | 40 |
0.417 (1.00) |
0.544 (1.00) |
0.917 (1.00) |
1 (1.00) |
0.559 (1.00) |
0.677 (1.00) |
1 (1.00) |
1 (1.00) |
0.201 (1.00) |
0.292 (1.00) |
||
| OLIG3 | 5 (11%) | 39 |
0.673 (1.00) |
0.737 (1.00) |
0.415 (1.00) |
1 (1.00) |
0.317 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.716 (1.00) |
0.216 (1.00) |
||
| NDEL1 | 4 (9%) | 40 |
0.222 (1.00) |
0.456 (1.00) |
0.0409 (1.00) |
1 (1.00) |
0.218 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.306 (1.00) |
0.00461 (1.00) |
||
| EGR1 | 8 (18%) | 36 |
0.62 (1.00) |
0.725 (1.00) |
0.812 (1.00) |
0.566 (1.00) |
1 (1.00) |
0.318 (1.00) |
0.698 (1.00) |
1 (1.00) |
0.646 (1.00) |
0.541 (1.00) |
||
| CDH3 | 4 (9%) | 40 |
0.288 (1.00) |
0.263 (1.00) |
0.933 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.607 (1.00) |
1 (1.00) |
1 (1.00) |
0.632 (1.00) |
||
| PCDHAC2 | 7 (16%) | 37 |
0.908 (1.00) |
0.561 (1.00) |
0.766 (1.00) |
1 (1.00) |
1 (1.00) |
0.272 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.841 (1.00) |
||
| ZFP36 | 4 (9%) | 40 |
0.00545 (1.00) |
0.649 (1.00) |
0.917 (1.00) |
1 (1.00) |
0.559 (1.00) |
0.45 (1.00) |
0.607 (1.00) |
1 (1.00) |
1 (1.00) |
0.225 (1.00) |
||
| NR4A3 | 5 (11%) | 39 |
0.775 (1.00) |
0.541 (1.00) |
0.0354 (1.00) |
1 (1.00) |
0.317 (1.00) |
0.255 (1.00) |
1 (1.00) |
1 (1.00) |
0.716 (1.00) |
0.109 (1.00) |
||
| RAB11FIP5 | 5 (11%) | 39 |
0.411 (1.00) |
0.833 (1.00) |
0.53 (1.00) |
0.394 (1.00) |
1 (1.00) |
0.731 (1.00) |
1 (1.00) |
1 (1.00) |
0.716 (1.00) |
0.961 (1.00) |
||
| OTUD4 | 7 (16%) | 37 |
0.594 (1.00) |
0.776 (1.00) |
0.277 (1.00) |
1 (1.00) |
0.649 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.364 (1.00) |
0.156 (1.00) |
||
| PPARGC1B | 6 (14%) | 38 |
0.552 (1.00) |
0.578 (1.00) |
0.195 (1.00) |
1 (1.00) |
0.606 (1.00) |
0.554 (1.00) |
1 (1.00) |
1 (1.00) |
0.178 (1.00) |
0.87 (1.00) |
||
| PDZD2 | 9 (20%) | 35 |
0.288 (1.00) |
0.679 (1.00) |
0.394 (1.00) |
0.566 (1.00) |
1 (1.00) |
0.204 (1.00) |
1 (1.00) |
1 (1.00) |
0.272 (1.00) |
0.861 (1.00) |
||
| MED9 | 3 (7%) | 41 |
0.337 (1.00) |
0.0312 (1.00) |
0.256 (1.00) |
0.254 (1.00) |
0.548 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.77 (1.00) |
||
| BCL2L1 | 3 (7%) | 41 |
0.502 (1.00) |
0.0651 (1.00) |
0.0796 (1.00) |
1 (1.00) |
0.125 (1.00) |
0.651 (1.00) |
1 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.326 (1.00) |
||
| CPLX3 | 3 (7%) | 41 |
0.684 (1.00) |
0.567 (1.00) |
0.956 (1.00) |
1 (1.00) |
1 (1.00) |
0.651 (1.00) |
1 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.795 (1.00) |
||
| NFAT5 | 8 (18%) | 36 |
0.237 (1.00) |
0.146 (1.00) |
0.0402 (1.00) |
0.145 (1.00) |
0.0641 (1.00) |
0.8 (1.00) |
1 (1.00) |
0.258 (1.00) |
0.182 (1.00) |
0.526 (1.00) |
||
| WASF3 | 5 (11%) | 39 |
0.444 (1.00) |
0.871 (1.00) |
0.547 (1.00) |
0.394 (1.00) |
0.317 (1.00) |
0.731 (1.00) |
1 (1.00) |
1 (1.00) |
0.716 (1.00) |
0.933 (1.00) |
||
| ABCC2 | 5 (11%) | 39 |
0.945 (1.00) |
0.32 (1.00) |
0.53 (1.00) |
0.0575 (1.00) |
1 (1.00) |
0.731 (1.00) |
0.345 (1.00) |
1 (1.00) |
0.716 (1.00) |
0.127 (1.00) |
||
| ATP13A3 | 4 (9%) | 40 |
0.972 (1.00) |
0.838 (1.00) |
0.422 (1.00) |
0.327 (1.00) |
1 (1.00) |
1 (1.00) |
0.607 (1.00) |
1 (1.00) |
1 (1.00) |
0.611 (1.00) |
||
| ARHGAP18 | 4 (9%) | 40 |
0.797 (1.00) |
0.0982 (1.00) |
0.0514 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.108 (1.00) |
1 (1.00) |
1 (1.00) |
0.909 (1.00) |
||
| CIR1 | 3 (7%) | 41 |
0.893 (1.00) |
0.952 (1.00) |
0.871 (1.00) |
1 (1.00) |
0.548 (1.00) |
1 (1.00) |
0.233 (1.00) |
1 (1.00) |
1 (1.00) |
0.472 (1.00) |
||
| KIAA0020 | 4 (9%) | 40 |
0.00795 (1.00) |
0.466 (1.00) |
0.0514 (1.00) |
1 (1.00) |
1 (1.00) |
0.0815 (1.00) |
0.607 (1.00) |
1 (1.00) |
1 (1.00) |
0.861 (1.00) |
||
| ZNF608 | 6 (14%) | 38 |
0.213 (1.00) |
0.408 (1.00) |
0.158 (1.00) |
1 (1.00) |
1 (1.00) |
0.313 (1.00) |
0.664 (1.00) |
1 (1.00) |
0.1 (1.00) |
0.688 (1.00) |
||
| NEDD4L | 5 (11%) | 39 |
0.615 (1.00) |
0.502 (1.00) |
0.53 (1.00) |
0.0575 (1.00) |
1 (1.00) |
0.488 (1.00) |
1 (1.00) |
1 (1.00) |
0.716 (1.00) |
0.648 (1.00) |
||
| THBS4 | 6 (14%) | 38 |
0.702 (1.00) |
0.859 (1.00) |
0.0779 (1.00) |
0.456 (1.00) |
0.606 (1.00) |
1 (1.00) |
0.185 (1.00) |
1 (1.00) |
0.547 (1.00) |
0.331 (1.00) |
||
| MMRN1 | 6 (14%) | 38 |
0.555 (1.00) |
0.424 (1.00) |
0.946 (1.00) |
1 (1.00) |
1 (1.00) |
0.751 (1.00) |
0.664 (1.00) |
1 (1.00) |
0.547 (1.00) |
0.386 (1.00) |
||
| GPR25 | 3 (7%) | 41 |
0.796 (1.00) |
0.272 (1.00) |
0.956 (1.00) |
1 (1.00) |
1 (1.00) |
0.651 (1.00) |
1 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.795 (1.00) |
||
| SORBS2 | 5 (11%) | 39 |
0.199 (1.00) |
0.269 (1.00) |
0.865 (1.00) |
1 (1.00) |
0.573 (1.00) |
0.488 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.34 (1.00) |
||
| ZNF326 | 4 (9%) | 40 |
0.554 (1.00) |
0.978 (1.00) |
0.917 (1.00) |
1 (1.00) |
0.559 (1.00) |
1 (1.00) |
0.607 (1.00) |
1 (1.00) |
1 (1.00) |
0.723 (1.00) |
||
| C15ORF52 | 3 (7%) | 41 |
0.249 (1.00) |
0.947 (1.00) |
0.956 (1.00) |
1 (1.00) |
1 (1.00) |
0.651 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.403 (1.00) |
||
| SLC39A5 | 3 (7%) | 41 |
0.648 (1.00) |
0.358 (1.00) |
0.956 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.233 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.251 (1.00) |
||
| SLC24A1 | 4 (9%) | 40 |
0.796 (1.00) |
0.895 (1.00) |
0.422 (1.00) |
0.327 (1.00) |
1 (1.00) |
0.45 (1.00) |
1 (1.00) |
1 (1.00) |
0.306 (1.00) |
0.37 (1.00) |
||
| PUM2 | 5 (11%) | 39 |
0.993 (1.00) |
0.167 (1.00) |
0.11 (1.00) |
1 (1.00) |
0.317 (1.00) |
0.488 (1.00) |
1 (1.00) |
1 (1.00) |
0.178 (1.00) |
0.681 (1.00) |
||
| SYT2 | 5 (11%) | 39 |
0.147 (1.00) |
0.0255 (1.00) |
0.865 (1.00) |
1 (1.00) |
0.573 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.178 (1.00) |
0.439 (1.00) |
||
| SLITRK5 | 6 (14%) | 38 |
0.993 (1.00) |
0.156 (1.00) |
0.195 (1.00) |
1 (1.00) |
0.606 (1.00) |
0.751 (1.00) |
1 (1.00) |
1 (1.00) |
0.1 (1.00) |
0.48 (1.00) |
||
| NAP1L3 | 3 (7%) | 41 |
0.897 (1.00) |
0.256 (1.00) |
0.254 (1.00) |
0.548 (1.00) |
0.651 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.177 (1.00) |
|||
| PHF14 | 3 (7%) | 41 |
0.222 (1.00) |
0.161 (1.00) |
0.871 (1.00) |
1 (1.00) |
0.548 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.488 (1.00) |
||
| CCDC28B | 4 (9%) | 40 |
0.715 (1.00) |
0.887 (1.00) |
0.00547 (1.00) |
0.327 (1.00) |
0.0316 (1.00) |
0.45 (1.00) |
0.607 (1.00) |
1 (1.00) |
0.306 (1.00) |
0.764 (1.00) |
||
| PDE12 | 3 (7%) | 41 |
0.00795 (1.00) |
0.898 (1.00) |
0.956 (1.00) |
1 (1.00) |
1 (1.00) |
0.219 (1.00) |
1 (1.00) |
1 (1.00) |
0.872 (1.00) |
|||
| FLAD1 | 3 (7%) | 41 |
0.982 (1.00) |
0.256 (1.00) |
0.254 (1.00) |
0.548 (1.00) |
0.219 (1.00) |
1 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.38 (1.00) |
|||
| ENG | 3 (7%) | 41 |
0.335 (1.00) |
0.256 (1.00) |
0.254 (1.00) |
0.548 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.976 (1.00) |
|||
| PABPC1 | 4 (9%) | 40 |
0.158 (1.00) |
0.0459 (1.00) |
0.917 (1.00) |
1 (1.00) |
0.559 (1.00) |
1 (1.00) |
0.607 (1.00) |
1 (1.00) |
1 (1.00) |
0.494 (1.00) |
||
| ATG16L2 | 3 (7%) | 41 |
0.278 (1.00) |
0.836 (1.00) |
0.0131 (1.00) |
1 (1.00) |
0.548 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.488 (1.00) |
||
| LRCH1 | 7 (16%) | 37 |
0.592 (1.00) |
0.829 (1.00) |
0.00344 (1.00) |
0.113 (1.00) |
0.0367 (1.00) |
1 (1.00) |
0.412 (1.00) |
1 (1.00) |
0.0655 (1.00) |
0.032 (1.00) |
||
| TAOK2 | 4 (9%) | 40 |
0.841 (1.00) |
0.676 (1.00) |
0.354 (1.00) |
0.327 (1.00) |
0.218 (1.00) |
1 (1.00) |
0.607 (1.00) |
1 (1.00) |
1 (1.00) |
0.481 (1.00) |
||
| ZMIZ1 | 5 (11%) | 39 |
0.774 (1.00) |
0.293 (1.00) |
0.949 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.345 (1.00) |
1 (1.00) |
0.0482 (1.00) |
0.0145 (1.00) |
||
| SYT15 | 6 (14%) | 38 |
0.316 (1.00) |
0.569 (1.00) |
0.0779 (1.00) |
1 (1.00) |
0.606 (1.00) |
0.115 (1.00) |
1 (1.00) |
1 (1.00) |
0.752 (1.00) |
0.333 (1.00) |
||
| LRTM1 | 3 (7%) | 41 |
0.0182 (1.00) |
0.646 (1.00) |
0.871 (1.00) |
1 (1.00) |
0.548 (1.00) |
1 (1.00) |
0.233 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.867 (1.00) |
||
| TIMM50 | 3 (7%) | 41 |
0.531 (1.00) |
0.259 (1.00) |
0.871 (1.00) |
1 (1.00) |
0.548 (1.00) |
0.336 (1.00) |
1 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.261 (1.00) |
||
| CCNK | 4 (9%) | 40 |
0.531 (1.00) |
0.23 (1.00) |
0.933 (1.00) |
1 (1.00) |
1 (1.00) |
0.677 (1.00) |
0.607 (1.00) |
1 (1.00) |
0.306 (1.00) |
0.0926 (1.00) |
||
| TTK | 5 (11%) | 39 |
0.495 (1.00) |
0.0517 (1.00) |
0.949 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.345 (1.00) |
1 (1.00) |
0.716 (1.00) |
0.488 (1.00) |
||
| TMEM175 | 4 (9%) | 40 |
0.484 (1.00) |
0.976 (1.00) |
0.917 (1.00) |
1 (1.00) |
0.559 (1.00) |
0.677 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.145 (1.00) |
||
| CD93 | 5 (11%) | 39 |
0.641 (1.00) |
0.949 (1.00) |
0.0292 (1.00) |
0.394 (1.00) |
0.0688 (1.00) |
1 (1.00) |
0.345 (1.00) |
1 (1.00) |
0.0482 (1.00) |
0.0221 (1.00) |
||
| C19ORF55 | 4 (9%) | 40 |
0.15 (1.00) |
0.26 (1.00) |
0.917 (1.00) |
1 (1.00) |
0.559 (1.00) |
0.677 (1.00) |
0.607 (1.00) |
1 (1.00) |
0.7 (1.00) |
0.341 (1.00) |
||
| MRPL48 | 3 (7%) | 41 |
0.796 (1.00) |
0.646 (1.00) |
0.956 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.375 (1.00) |
0.544 (1.00) |
||
| PVRL1 | 5 (11%) | 39 |
0.985 (1.00) |
0.84 (1.00) |
0.53 (1.00) |
0.394 (1.00) |
1 (1.00) |
0.731 (1.00) |
1 (1.00) |
1 (1.00) |
0.178 (1.00) |
0.965 (1.00) |
||
| ACIN1 | 3 (7%) | 41 |
0.575 (1.00) |
0.654 (1.00) |
0.956 (1.00) |
1 (1.00) |
1 (1.00) |
0.651 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.28 (1.00) |
||
| MAML3 | 6 (14%) | 38 |
0.229 (1.00) |
0.497 (1.00) |
0.158 (1.00) |
1 (1.00) |
1 (1.00) |
0.313 (1.00) |
0.664 (1.00) |
1 (1.00) |
0.752 (1.00) |
0.535 (1.00) |
||
| EDC4 | 4 (9%) | 40 |
0.402 (1.00) |
0.914 (1.00) |
0.917 (1.00) |
1 (1.00) |
0.559 (1.00) |
0.677 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.413 (1.00) |
P value = 0 (logrank test), Q value = 0
Table S1. Gene #3: 'FYN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 43 | 16 | 0.0 - 49.4 (5.0) |
| FYN MUTATED | 4 | 0 | 0.3 - 1.0 (0.7) |
| FYN WILD-TYPE | 39 | 16 | 0.0 - 49.4 (6.0) |
Figure S1. Get High-res Image Gene #3: 'FYN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
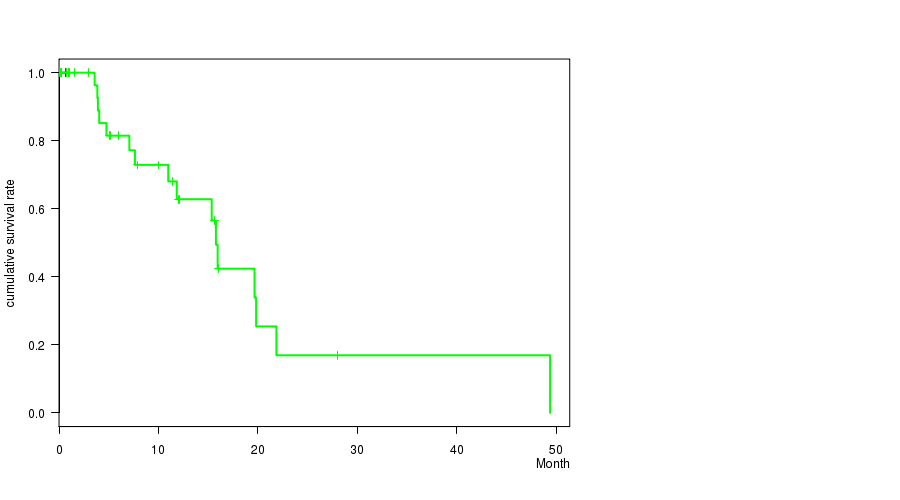
P value = 7.36e-05 (Chi-square test), Q value = 0.075
Table S2. Gene #4: 'FADS2 MUTATION STATUS' versus Clinical Feature #3: 'NEOPLASM.DISEASESTAGE'
| nPatients | STAGE IA | STAGE IB | STAGE IIA | STAGE IIB | STAGE III | STAGE IV |
|---|---|---|---|---|---|---|
| ALL | 1 | 2 | 5 | 33 | 1 | 2 |
| FADS2 MUTATED | 0 | 2 | 1 | 1 | 1 | 0 |
| FADS2 WILD-TYPE | 1 | 0 | 4 | 32 | 0 | 2 |
Figure S2. Get High-res Image Gene #4: 'FADS2 MUTATION STATUS' versus Clinical Feature #3: 'NEOPLASM.DISEASESTAGE'
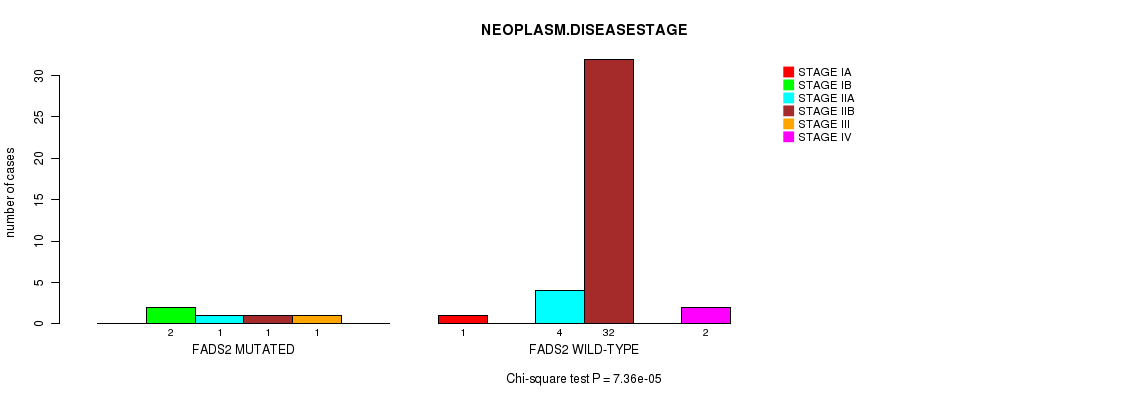
P value = 6.35e-06 (t-test), Q value = 0.0065
Table S3. Gene #7: 'CEL MUTATION STATUS' versus Clinical Feature #12: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 44 | 2.9 (3.1) |
| CEL MUTATED | 4 | 0.2 (0.5) |
| CEL WILD-TYPE | 40 | 3.2 (3.1) |
Figure S3. Get High-res Image Gene #7: 'CEL MUTATION STATUS' versus Clinical Feature #12: 'NUMBER.OF.LYMPH.NODES'
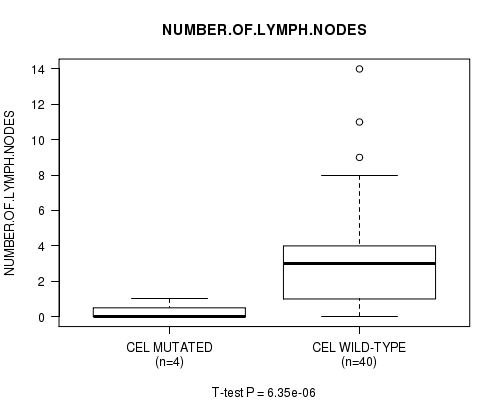
P value = 6.35e-06 (t-test), Q value = 0.0065
Table S4. Gene #70: 'ECSIT MUTATION STATUS' versus Clinical Feature #12: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 44 | 2.9 (3.1) |
| ECSIT MUTATED | 4 | 0.2 (0.5) |
| ECSIT WILD-TYPE | 40 | 3.2 (3.1) |
Figure S4. Get High-res Image Gene #70: 'ECSIT MUTATION STATUS' versus Clinical Feature #12: 'NUMBER.OF.LYMPH.NODES'
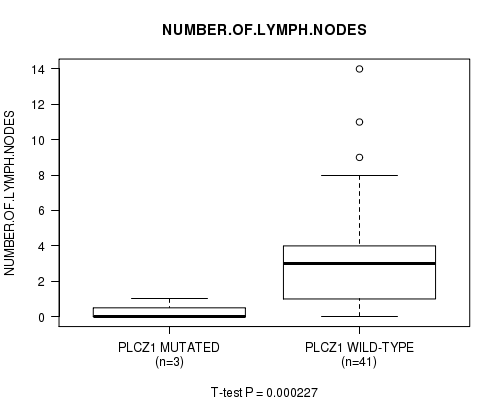
P value = 0.000227 (t-test), Q value = 0.23
Table S5. Gene #87: 'PLCZ1 MUTATION STATUS' versus Clinical Feature #12: 'NUMBER.OF.LYMPH.NODES'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 44 | 2.9 (3.1) |
| PLCZ1 MUTATED | 3 | 0.3 (0.6) |
| PLCZ1 WILD-TYPE | 41 | 3.1 (3.1) |
Figure S5. Get High-res Image Gene #87: 'PLCZ1 MUTATION STATUS' versus Clinical Feature #12: 'NUMBER.OF.LYMPH.NODES'
-
Mutation data file = transformed.cor.cli.txt
-
Clinical data file = PAAD-TP.merged_data.txt
-
Number of patients = 44
-
Number of significantly mutated genes = 102
-
Number of selected clinical features = 12
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For continuous numerical clinical features, two-tailed Student's t test with unequal variance (Lehmann and Romano 2005) was applied to compare the clinical values between tumors with and without gene mutations using 't.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.