This pipeline computes the correlation between significant copy number variation (cnv focal) genes and molecular subtypes.
Testing the association between copy number variation 43 focal events and 8 molecular subtypes across 91 patients, 45 significant findings detected with P value < 0.05 and Q value < 0.25.
-
amp_7q22.1 cnv correlated to 'METHLYATION_CNMF'.
-
amp_8q24.21 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
amp_12p13.33 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
amp_12p11.21 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
amp_12q15 cnv correlated to 'CN_CNMF'.
-
amp_18q11.2 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MIRSEQ_CNMF', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
amp_19q13.2 cnv correlated to 'CN_CNMF'.
-
amp_20q13.2 cnv correlated to 'CN_CNMF'.
-
del_1p36.32 cnv correlated to 'CN_CNMF'.
-
del_1p36.11 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
del_3p24.1 cnv correlated to 'CN_CNMF'.
-
del_5q14.2 cnv correlated to 'CN_CNMF'.
-
del_6p25.2 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
del_8p21.3 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CHIERARCHICAL'.
-
del_9p21.3 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
del_10q23.2 cnv correlated to 'CN_CNMF'.
-
del_12p12.1 cnv correlated to 'CN_CNMF'.
-
del_12q12 cnv correlated to 'CN_CNMF'.
-
del_12q24.33 cnv correlated to 'CN_CNMF'.
-
del_17q22 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
del_18q21.2 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
del_19p13.3 cnv correlated to 'CN_CNMF'.
-
del_21q21.1 cnv correlated to 'CN_CNMF'.
-
del_22q13.31 cnv correlated to 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 43 focal events and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 45 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| del 9p21 3 | 58 (64%) | 33 |
1.17e-08 (3.98e-06) |
1.16e-09 (3.98e-07) |
0.000292 (0.0908) |
0.000152 (0.0488) |
1.09e-05 (0.00361) |
0.000183 (0.0579) |
2.56e-05 (0.00837) |
2.54e-05 (0.00834) |
| amp 18q11 2 | 30 (33%) | 61 |
2.8e-12 (9.64e-10) |
0.000255 (0.0796) |
0.0216 (1.00) |
0.00219 (0.649) |
0.000407 (0.124) |
0.00517 (1.00) |
0.000418 (0.126) |
2.03e-05 (0.00669) |
| del 6p25 2 | 38 (42%) | 53 |
4.43e-07 (0.00015) |
5.97e-06 (0.00199) |
0.0776 (1.00) |
0.000369 (0.113) |
0.0113 (1.00) |
0.0599 (1.00) |
0.0107 (1.00) |
0.00493 (1.00) |
| del 8p21 3 | 26 (29%) | 65 |
0.000218 (0.0683) |
0.000425 (0.128) |
0.0225 (1.00) |
0.000415 (0.126) |
0.19 (1.00) |
0.028 (1.00) |
0.0113 (1.00) |
0.0143 (1.00) |
| amp 8q24 21 | 36 (40%) | 55 |
2.62e-06 (0.000879) |
2.24e-05 (0.00739) |
0.171 (1.00) |
0.0102 (1.00) |
0.149 (1.00) |
0.0599 (1.00) |
0.0262 (1.00) |
0.0109 (1.00) |
| amp 12p13 33 | 15 (16%) | 76 |
2.52e-09 (8.61e-07) |
0.000591 (0.177) |
0.244 (1.00) |
0.0112 (1.00) |
0.821 (1.00) |
0.0629 (1.00) |
0.117 (1.00) |
0.0387 (1.00) |
| amp 12p11 21 | 16 (18%) | 75 |
6.1e-09 (2.08e-06) |
0.000146 (0.0471) |
0.483 (1.00) |
0.0112 (1.00) |
0.975 (1.00) |
0.207 (1.00) |
0.089 (1.00) |
0.1 (1.00) |
| del 1p36 11 | 30 (33%) | 61 |
0.000164 (0.0522) |
9.12e-06 (0.00303) |
0.0104 (1.00) |
0.00453 (1.00) |
0.239 (1.00) |
0.00306 (0.89) |
0.0191 (1.00) |
0.0105 (1.00) |
| del 17q22 | 25 (27%) | 66 |
1.17e-07 (3.95e-05) |
0.000359 (0.11) |
0.0125 (1.00) |
0.00583 (1.00) |
0.05 (1.00) |
0.0544 (1.00) |
0.0169 (1.00) |
0.0197 (1.00) |
| del 18q21 2 | 65 (71%) | 26 |
3.56e-06 (0.00119) |
0.000175 (0.0556) |
0.0888 (1.00) |
0.00743 (1.00) |
0.0124 (1.00) |
0.166 (1.00) |
0.00579 (1.00) |
0.0201 (1.00) |
| amp 7q22 1 | 26 (29%) | 65 |
0.00201 (0.596) |
0.000359 (0.11) |
0.153 (1.00) |
0.00583 (1.00) |
0.13 (1.00) |
0.0544 (1.00) |
0.0169 (1.00) |
0.0197 (1.00) |
| amp 12q15 | 10 (11%) | 81 |
0.000154 (0.0494) |
0.013 (1.00) |
0.122 (1.00) |
0.0257 (1.00) |
0.463 (1.00) |
0.0782 (1.00) |
0.0223 (1.00) |
0.0199 (1.00) |
| amp 19q13 2 | 23 (25%) | 68 |
0.0003 (0.0929) |
0.00819 (1.00) |
0.0884 (1.00) |
0.187 (1.00) |
0.11 (1.00) |
0.181 (1.00) |
0.241 (1.00) |
0.143 (1.00) |
| amp 20q13 2 | 24 (26%) | 67 |
9.64e-05 (0.0313) |
0.00895 (1.00) |
0.493 (1.00) |
0.187 (1.00) |
0.533 (1.00) |
0.484 (1.00) |
0.325 (1.00) |
0.143 (1.00) |
| del 1p36 32 | 21 (23%) | 70 |
0.00022 (0.0689) |
0.00115 (0.344) |
0.02 (1.00) |
0.00788 (1.00) |
0.713 (1.00) |
0.0277 (1.00) |
0.122 (1.00) |
0.131 (1.00) |
| del 3p24 1 | 19 (21%) | 72 |
0.000344 (0.106) |
0.0475 (1.00) |
0.698 (1.00) |
0.497 (1.00) |
0.775 (1.00) |
0.606 (1.00) |
0.562 (1.00) |
0.262 (1.00) |
| del 5q14 2 | 19 (21%) | 72 |
0.000131 (0.0424) |
0.0475 (1.00) |
0.478 (1.00) |
0.325 (1.00) |
0.633 (1.00) |
0.277 (1.00) |
0.134 (1.00) |
0.262 (1.00) |
| del 10q23 2 | 16 (18%) | 75 |
7.66e-05 (0.025) |
0.0158 (1.00) |
0.483 (1.00) |
0.102 (1.00) |
0.804 (1.00) |
0.00817 (1.00) |
0.701 (1.00) |
0.1 (1.00) |
| del 12p12 1 | 11 (12%) | 80 |
0.00034 (0.105) |
0.00513 (1.00) |
0.312 (1.00) |
0.191 (1.00) |
0.0945 (1.00) |
0.336 (1.00) |
0.309 (1.00) |
0.0707 (1.00) |
| del 12q12 | 11 (12%) | 80 |
0.000179 (0.0566) |
0.00221 (0.651) |
0.312 (1.00) |
0.191 (1.00) |
0.2 (1.00) |
0.0103 (1.00) |
0.309 (1.00) |
0.00883 (1.00) |
| del 12q24 33 | 19 (21%) | 72 |
0.000131 (0.0424) |
0.00102 (0.304) |
0.112 (1.00) |
0.00277 (0.811) |
0.651 (1.00) |
0.013 (1.00) |
0.487 (1.00) |
0.213 (1.00) |
| del 19p13 3 | 17 (19%) | 74 |
4.19e-06 (0.0014) |
0.00294 (0.859) |
0.108 (1.00) |
0.00496 (1.00) |
0.568 (1.00) |
0.196 (1.00) |
0.125 (1.00) |
0.492 (1.00) |
| del 21q21 1 | 35 (38%) | 56 |
1.42e-06 (0.000479) |
0.00381 (1.00) |
0.00731 (1.00) |
0.0022 (0.649) |
0.00785 (1.00) |
0.0438 (1.00) |
0.00703 (1.00) |
0.0462 (1.00) |
| del 22q13 31 | 27 (30%) | 64 |
0.000211 (0.0664) |
0.00553 (1.00) |
0.0201 (1.00) |
0.0764 (1.00) |
0.0703 (1.00) |
0.0507 (1.00) |
0.00955 (1.00) |
0.087 (1.00) |
| amp 1p34 2 | 7 (8%) | 84 |
0.883 (1.00) |
0.122 (1.00) |
0.755 (1.00) |
1 (1.00) |
1 (1.00) |
0.649 (1.00) |
0.566 (1.00) |
0.656 (1.00) |
| amp 1p12 | 15 (16%) | 76 |
0.533 (1.00) |
0.182 (1.00) |
0.922 (1.00) |
0.589 (1.00) |
0.946 (1.00) |
1 (1.00) |
0.721 (1.00) |
0.852 (1.00) |
| amp 9p13 3 | 9 (10%) | 82 |
0.138 (1.00) |
0.0786 (1.00) |
0.389 (1.00) |
0.319 (1.00) |
0.155 (1.00) |
0.0381 (1.00) |
0.309 (1.00) |
0.0442 (1.00) |
| amp 11p11 2 | 10 (11%) | 81 |
0.0037 (1.00) |
0.0116 (1.00) |
0.385 (1.00) |
0.235 (1.00) |
0.357 (1.00) |
0.424 (1.00) |
0.473 (1.00) |
0.141 (1.00) |
| amp 17p11 2 | 4 (4%) | 87 |
0.113 (1.00) |
0.131 (1.00) |
0.806 (1.00) |
1 (1.00) |
0.418 (1.00) |
0.824 (1.00) |
0.345 (1.00) |
0.564 (1.00) |
| amp 17q21 32 | 9 (10%) | 82 |
0.0115 (1.00) |
0.0786 (1.00) |
0.385 (1.00) |
0.529 (1.00) |
0.959 (1.00) |
0.424 (1.00) |
1 (1.00) |
0.479 (1.00) |
| amp xp22 11 | 5 (5%) | 86 |
0.346 (1.00) |
0.365 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.712 (1.00) |
0.848 (1.00) |
0.597 (1.00) |
| amp xq27 1 | 8 (9%) | 83 |
0.12 (1.00) |
0.586 (1.00) |
0.306 (1.00) |
0.515 (1.00) |
0.781 (1.00) |
0.55 (1.00) |
1 (1.00) |
0.353 (1.00) |
| del 1q42 2 | 3 (3%) | 88 |
0.796 (1.00) |
0.184 (1.00) |
0.322 (1.00) |
0.315 (1.00) |
0.864 (1.00) |
1 (1.00) |
0.596 (1.00) |
0.324 (1.00) |
| del 2p23 3 | 11 (12%) | 80 |
0.00548 (1.00) |
0.0391 (1.00) |
0.191 (1.00) |
0.302 (1.00) |
0.602 (1.00) |
0.0322 (1.00) |
0.418 (1.00) |
0.0438 (1.00) |
| del 4q35 2 | 16 (18%) | 75 |
0.211 (1.00) |
0.0349 (1.00) |
0.265 (1.00) |
0.773 (1.00) |
0.898 (1.00) |
1 (1.00) |
0.875 (1.00) |
0.667 (1.00) |
| del 7q36 1 | 9 (10%) | 82 |
0.0115 (1.00) |
0.193 (1.00) |
0.563 (1.00) |
0.319 (1.00) |
0.642 (1.00) |
0.624 (1.00) |
0.802 (1.00) |
0.712 (1.00) |
| del 8q12 1 | 7 (8%) | 84 |
0.587 (1.00) |
0.0536 (1.00) |
0.594 (1.00) |
0.311 (1.00) |
0.457 (1.00) |
0.882 (1.00) |
0.265 (1.00) |
0.153 (1.00) |
| del 9q22 33 | 24 (26%) | 67 |
0.144 (1.00) |
0.0849 (1.00) |
0.952 (1.00) |
1 (1.00) |
0.51 (1.00) |
0.908 (1.00) |
0.423 (1.00) |
0.681 (1.00) |
| del 9q32 | 23 (25%) | 68 |
0.0889 (1.00) |
0.4 (1.00) |
1 (1.00) |
1 (1.00) |
0.931 (1.00) |
0.827 (1.00) |
0.905 (1.00) |
0.614 (1.00) |
| del 11p15 4 | 15 (16%) | 76 |
0.0147 (1.00) |
0.38 (1.00) |
0.857 (1.00) |
1 (1.00) |
0.624 (1.00) |
0.791 (1.00) |
0.741 (1.00) |
1 (1.00) |
| del 16q23 1 | 6 (7%) | 85 |
0.136 (1.00) |
0.118 (1.00) |
0.503 (1.00) |
0.667 (1.00) |
0.924 (1.00) |
0.712 (1.00) |
0.848 (1.00) |
0.852 (1.00) |
| del 18p11 32 | 34 (37%) | 57 |
0.827 (1.00) |
0.0613 (1.00) |
0.382 (1.00) |
1 (1.00) |
0.574 (1.00) |
0.498 (1.00) |
1 (1.00) |
0.835 (1.00) |
| del 19q13 41 | 10 (11%) | 81 |
0.0141 (1.00) |
0.0325 (1.00) |
0.598 (1.00) |
0.529 (1.00) |
0.583 (1.00) |
0.817 (1.00) |
0.742 (1.00) |
0.817 (1.00) |
P value = 0.000359 (Fisher's exact test), Q value = 0.11
Table S1. Gene #3: 'amp_7q22.1' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| AMP PEAK 3(7Q22.1) MUTATED | 14 | 2 | 9 |
| AMP PEAK 3(7Q22.1) WILD-TYPE | 17 | 33 | 15 |
Figure S1. Get High-res Image Gene #3: 'amp_7q22.1' versus Molecular Subtype #2: 'METHLYATION_CNMF'
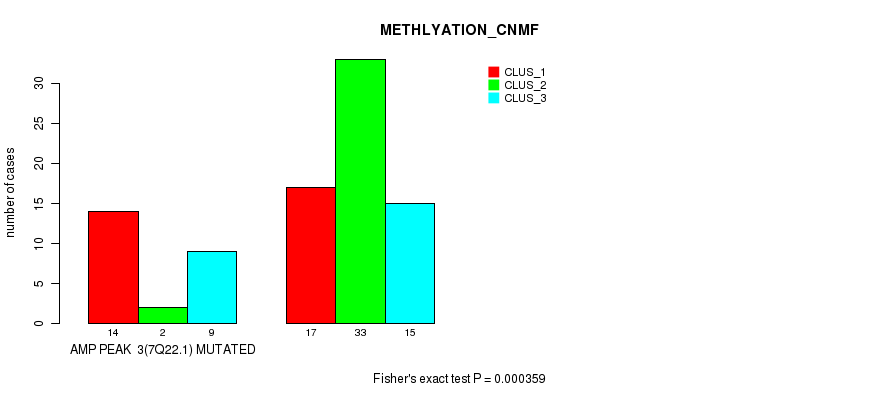
P value = 2.62e-06 (Fisher's exact test), Q value = 0.00088
Table S2. Gene #4: 'amp_8q24.21' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| AMP PEAK 4(8Q24.21) MUTATED | 11 | 8 | 17 |
| AMP PEAK 4(8Q24.21) WILD-TYPE | 13 | 38 | 4 |
Figure S2. Get High-res Image Gene #4: 'amp_8q24.21' versus Molecular Subtype #1: 'CN_CNMF'
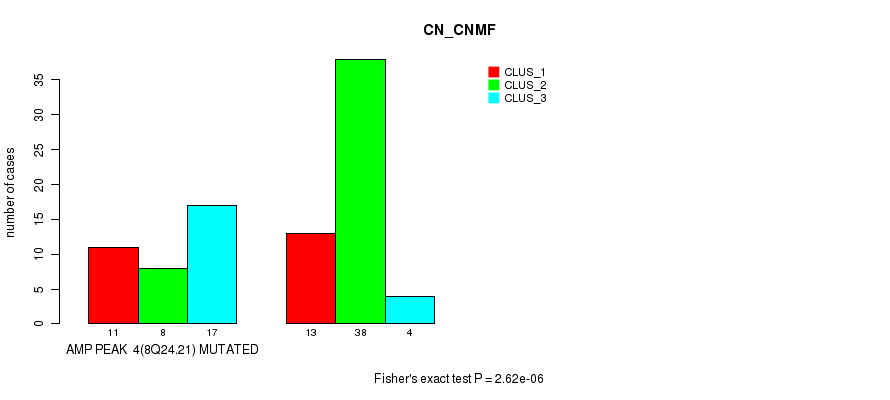
P value = 2.24e-05 (Fisher's exact test), Q value = 0.0074
Table S3. Gene #4: 'amp_8q24.21' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| AMP PEAK 4(8Q24.21) MUTATED | 20 | 4 | 11 |
| AMP PEAK 4(8Q24.21) WILD-TYPE | 11 | 31 | 13 |
Figure S3. Get High-res Image Gene #4: 'amp_8q24.21' versus Molecular Subtype #2: 'METHLYATION_CNMF'
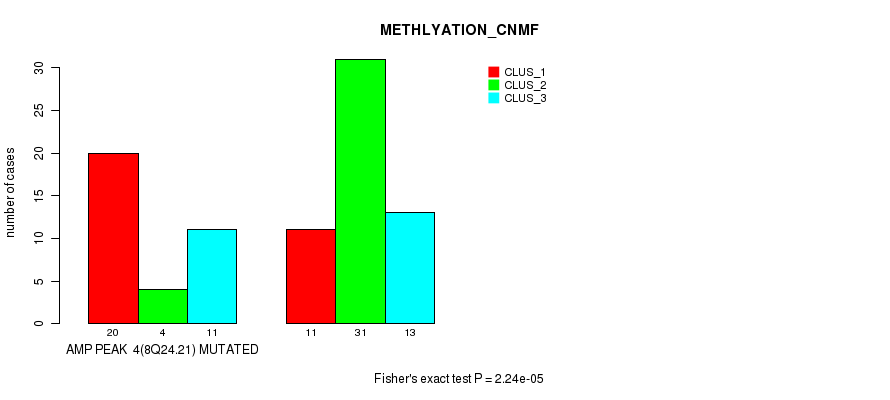
P value = 2.52e-09 (Fisher's exact test), Q value = 8.6e-07
Table S4. Gene #7: 'amp_12p13.33' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| AMP PEAK 7(12P13.33) MUTATED | 2 | 0 | 13 |
| AMP PEAK 7(12P13.33) WILD-TYPE | 22 | 46 | 8 |
Figure S4. Get High-res Image Gene #7: 'amp_12p13.33' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.000591 (Fisher's exact test), Q value = 0.18
Table S5. Gene #7: 'amp_12p13.33' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| AMP PEAK 7(12P13.33) MUTATED | 9 | 0 | 6 |
| AMP PEAK 7(12P13.33) WILD-TYPE | 22 | 35 | 18 |
Figure S5. Get High-res Image Gene #7: 'amp_12p13.33' versus Molecular Subtype #2: 'METHLYATION_CNMF'
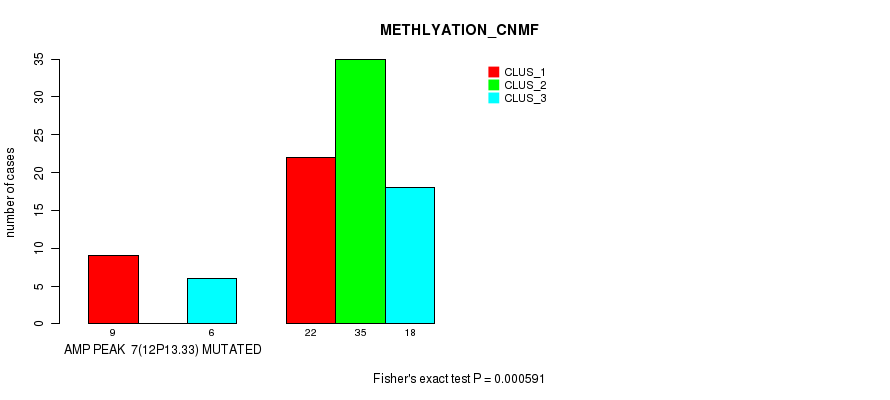
P value = 6.1e-09 (Fisher's exact test), Q value = 2.1e-06
Table S6. Gene #8: 'amp_12p11.21' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| AMP PEAK 8(12P11.21) MUTATED | 3 | 0 | 13 |
| AMP PEAK 8(12P11.21) WILD-TYPE | 21 | 46 | 8 |
Figure S6. Get High-res Image Gene #8: 'amp_12p11.21' versus Molecular Subtype #1: 'CN_CNMF'
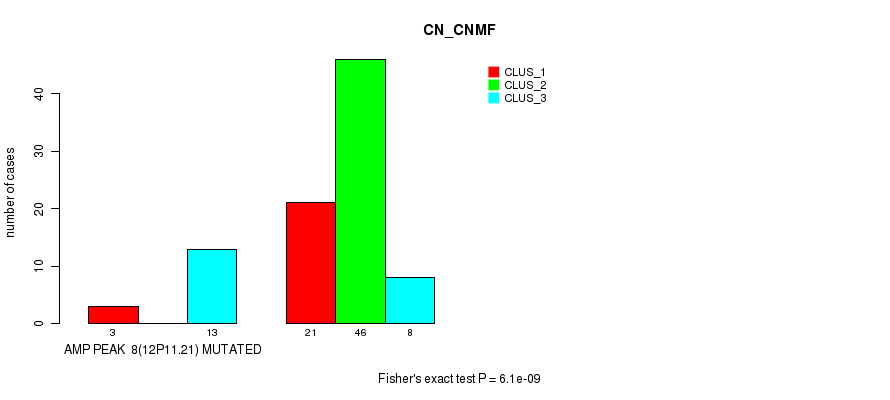
P value = 0.000146 (Fisher's exact test), Q value = 0.047
Table S7. Gene #8: 'amp_12p11.21' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| AMP PEAK 8(12P11.21) MUTATED | 7 | 0 | 9 |
| AMP PEAK 8(12P11.21) WILD-TYPE | 24 | 35 | 15 |
Figure S7. Get High-res Image Gene #8: 'amp_12p11.21' versus Molecular Subtype #2: 'METHLYATION_CNMF'
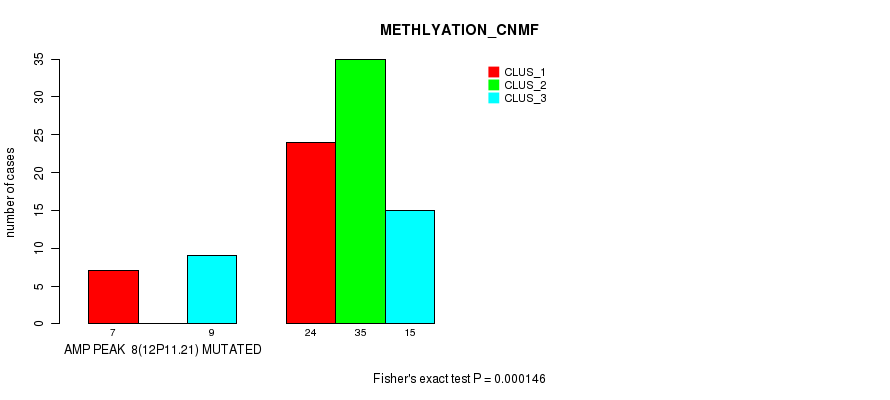
P value = 0.000154 (Fisher's exact test), Q value = 0.049
Table S8. Gene #9: 'amp_12q15' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| AMP PEAK 9(12Q15) MUTATED | 3 | 0 | 7 |
| AMP PEAK 9(12Q15) WILD-TYPE | 21 | 46 | 14 |
Figure S8. Get High-res Image Gene #9: 'amp_12q15' versus Molecular Subtype #1: 'CN_CNMF'
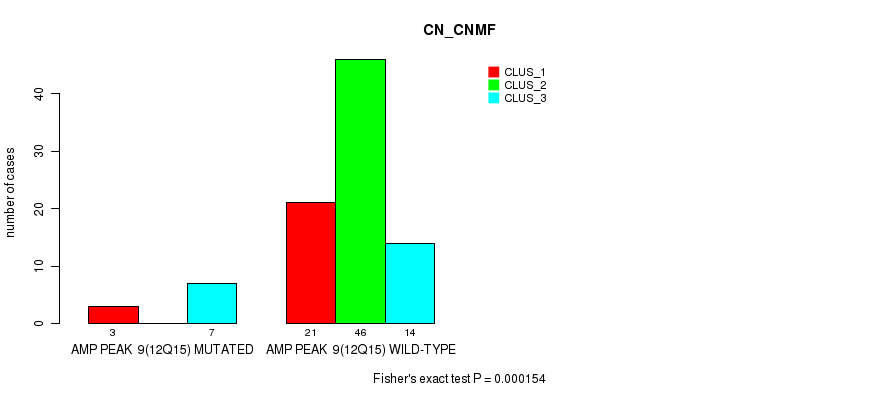
P value = 2.8e-12 (Fisher's exact test), Q value = 9.6e-10
Table S9. Gene #12: 'amp_18q11.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| AMP PEAK 12(18Q11.2) MUTATED | 22 | 4 | 4 |
| AMP PEAK 12(18Q11.2) WILD-TYPE | 2 | 42 | 17 |
Figure S9. Get High-res Image Gene #12: 'amp_18q11.2' versus Molecular Subtype #1: 'CN_CNMF'
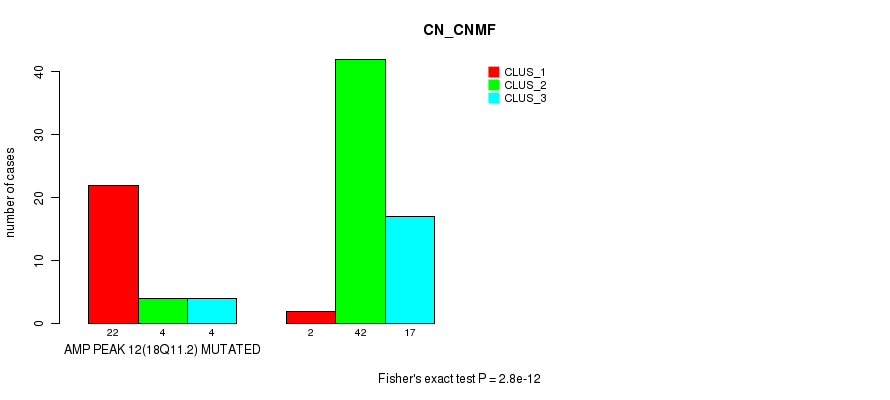
P value = 0.000255 (Fisher's exact test), Q value = 0.08
Table S10. Gene #12: 'amp_18q11.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| AMP PEAK 12(18Q11.2) MUTATED | 16 | 3 | 10 |
| AMP PEAK 12(18Q11.2) WILD-TYPE | 15 | 32 | 14 |
Figure S10. Get High-res Image Gene #12: 'amp_18q11.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
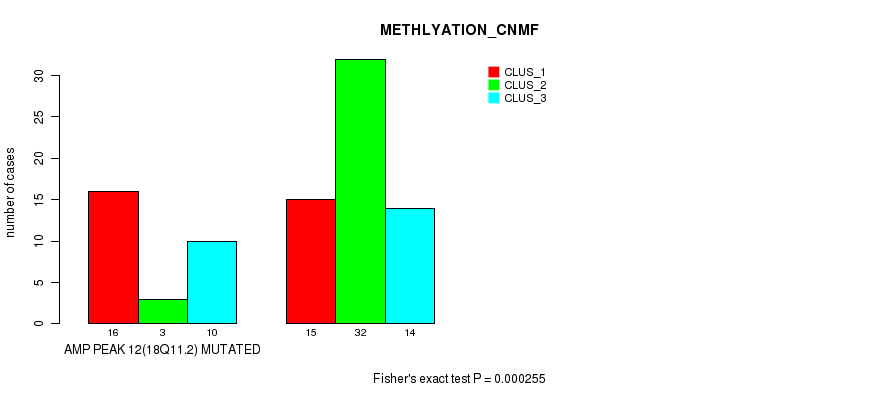
P value = 0.000407 (Fisher's exact test), Q value = 0.12
Table S11. Gene #12: 'amp_18q11.2' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 19 | 12 | 22 |
| AMP PEAK 12(18Q11.2) MUTATED | 19 | 5 | 1 | 3 |
| AMP PEAK 12(18Q11.2) WILD-TYPE | 12 | 14 | 11 | 19 |
Figure S11. Get High-res Image Gene #12: 'amp_18q11.2' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
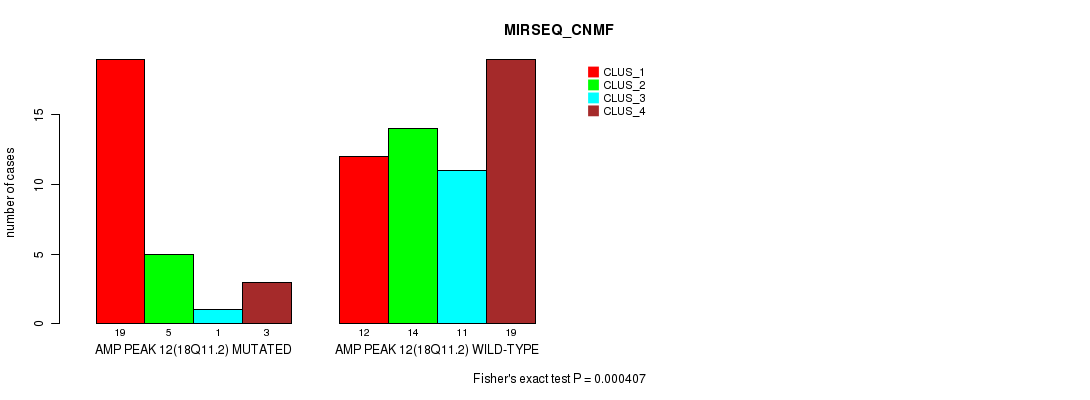
P value = 0.000418 (Fisher's exact test), Q value = 0.13
Table S12. Gene #12: 'amp_18q11.2' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 38 | 19 | 27 |
| AMP PEAK 12(18Q11.2) MUTATED | 21 | 4 | 3 |
| AMP PEAK 12(18Q11.2) WILD-TYPE | 17 | 15 | 24 |
Figure S12. Get High-res Image Gene #12: 'amp_18q11.2' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
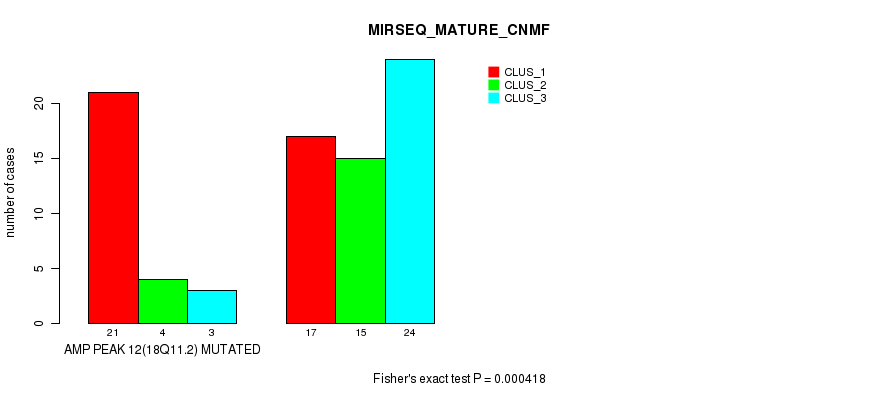
P value = 2.03e-05 (Fisher's exact test), Q value = 0.0067
Table S13. Gene #12: 'amp_18q11.2' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 26 | 20 | 38 |
| AMP PEAK 12(18Q11.2) MUTATED | 7 | 0 | 21 |
| AMP PEAK 12(18Q11.2) WILD-TYPE | 19 | 20 | 17 |
Figure S13. Get High-res Image Gene #12: 'amp_18q11.2' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
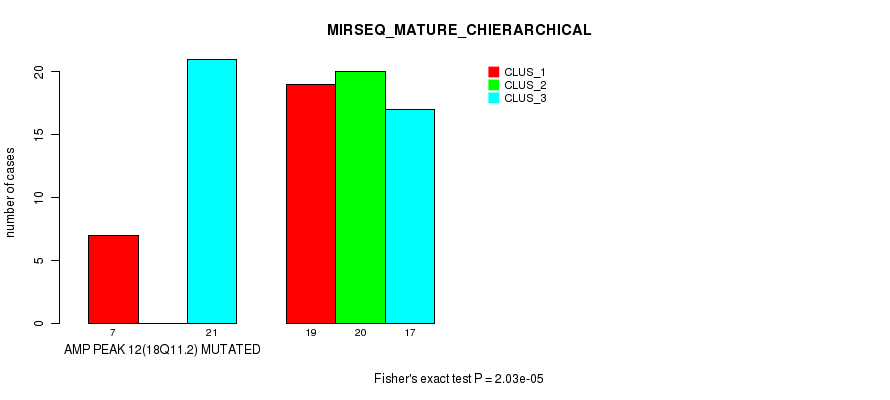
P value = 3e-04 (Fisher's exact test), Q value = 0.093
Table S14. Gene #13: 'amp_19q13.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| AMP PEAK 13(19Q13.2) MUTATED | 8 | 4 | 11 |
| AMP PEAK 13(19Q13.2) WILD-TYPE | 16 | 42 | 10 |
Figure S14. Get High-res Image Gene #13: 'amp_19q13.2' versus Molecular Subtype #1: 'CN_CNMF'
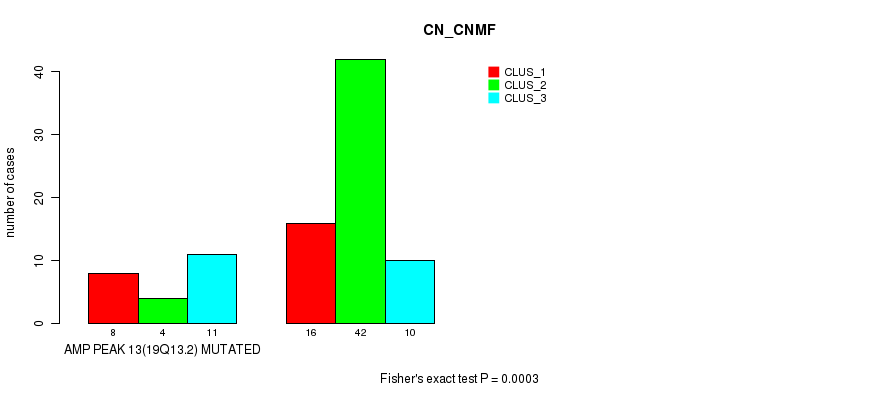
P value = 9.64e-05 (Fisher's exact test), Q value = 0.031
Table S15. Gene #14: 'amp_20q13.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| AMP PEAK 14(20Q13.2) MUTATED | 6 | 5 | 13 |
| AMP PEAK 14(20Q13.2) WILD-TYPE | 18 | 41 | 8 |
Figure S15. Get High-res Image Gene #14: 'amp_20q13.2' versus Molecular Subtype #1: 'CN_CNMF'
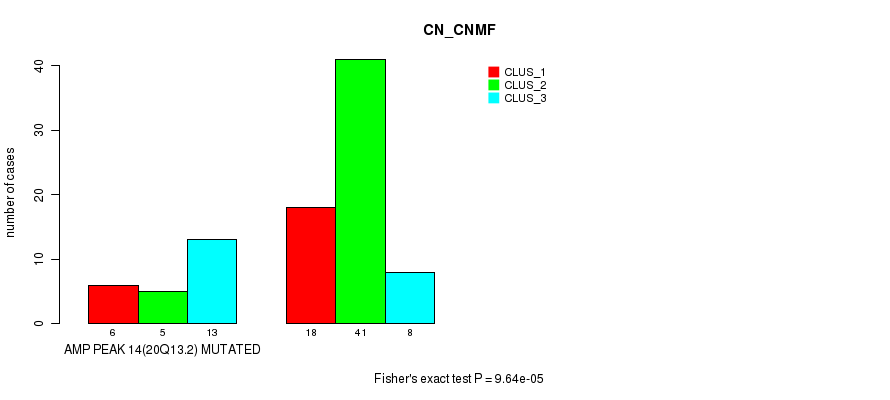
P value = 0.00022 (Fisher's exact test), Q value = 0.069
Table S16. Gene #17: 'del_1p36.32' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 1(1P36.32) MUTATED | 4 | 5 | 12 |
| DEL PEAK 1(1P36.32) WILD-TYPE | 20 | 41 | 9 |
Figure S16. Get High-res Image Gene #17: 'del_1p36.32' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.000164 (Fisher's exact test), Q value = 0.052
Table S17. Gene #18: 'del_1p36.11' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 2(1P36.11) MUTATED | 9 | 7 | 14 |
| DEL PEAK 2(1P36.11) WILD-TYPE | 15 | 39 | 7 |
Figure S17. Get High-res Image Gene #18: 'del_1p36.11' versus Molecular Subtype #1: 'CN_CNMF'
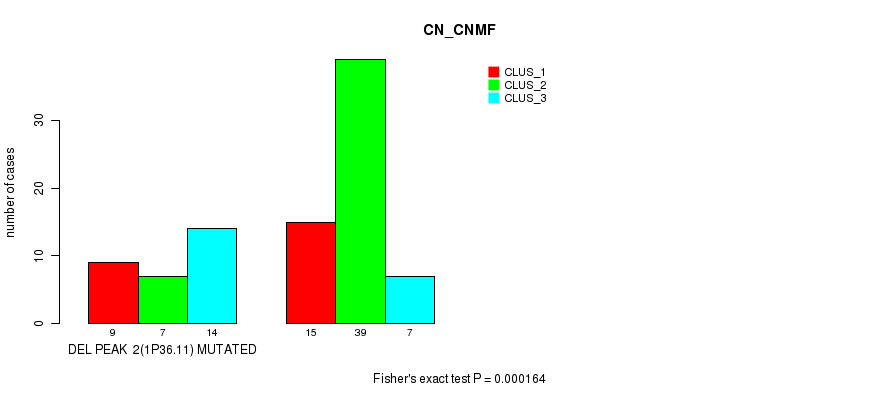
P value = 9.12e-06 (Fisher's exact test), Q value = 0.003
Table S18. Gene #18: 'del_1p36.11' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| DEL PEAK 2(1P36.11) MUTATED | 14 | 2 | 14 |
| DEL PEAK 2(1P36.11) WILD-TYPE | 17 | 33 | 10 |
Figure S18. Get High-res Image Gene #18: 'del_1p36.11' versus Molecular Subtype #2: 'METHLYATION_CNMF'
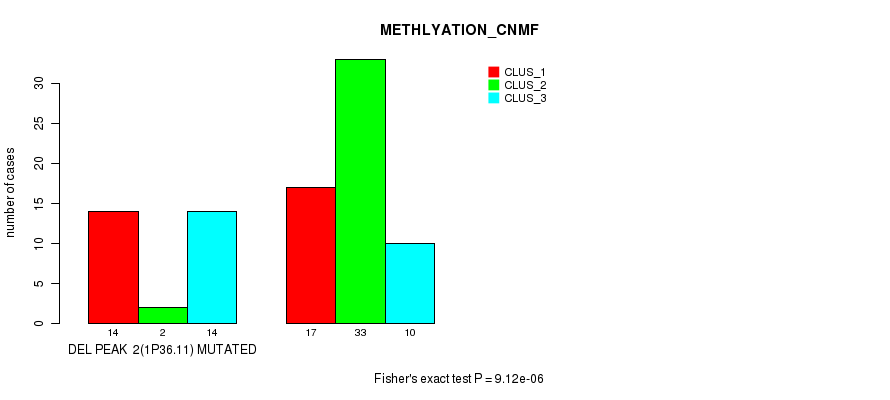
P value = 0.000344 (Fisher's exact test), Q value = 0.11
Table S19. Gene #21: 'del_3p24.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 5(3P24.1) MUTATED | 4 | 4 | 11 |
| DEL PEAK 5(3P24.1) WILD-TYPE | 20 | 42 | 10 |
Figure S19. Get High-res Image Gene #21: 'del_3p24.1' versus Molecular Subtype #1: 'CN_CNMF'
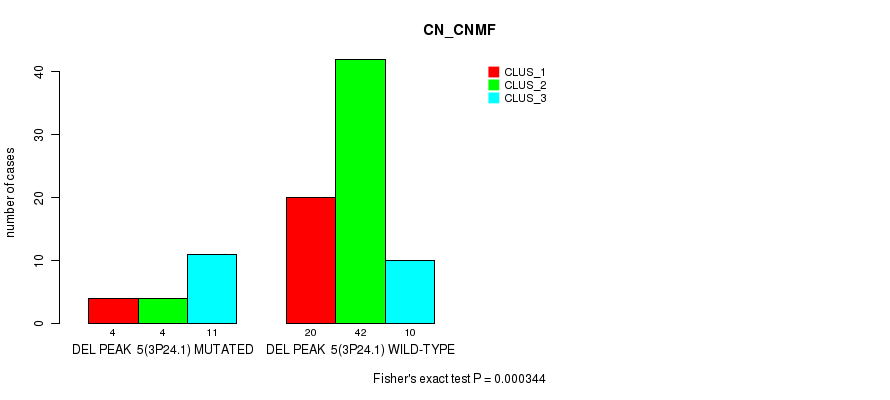
P value = 0.000131 (Fisher's exact test), Q value = 0.042
Table S20. Gene #23: 'del_5q14.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 7(5Q14.2) MUTATED | 5 | 3 | 11 |
| DEL PEAK 7(5Q14.2) WILD-TYPE | 19 | 43 | 10 |
Figure S20. Get High-res Image Gene #23: 'del_5q14.2' versus Molecular Subtype #1: 'CN_CNMF'
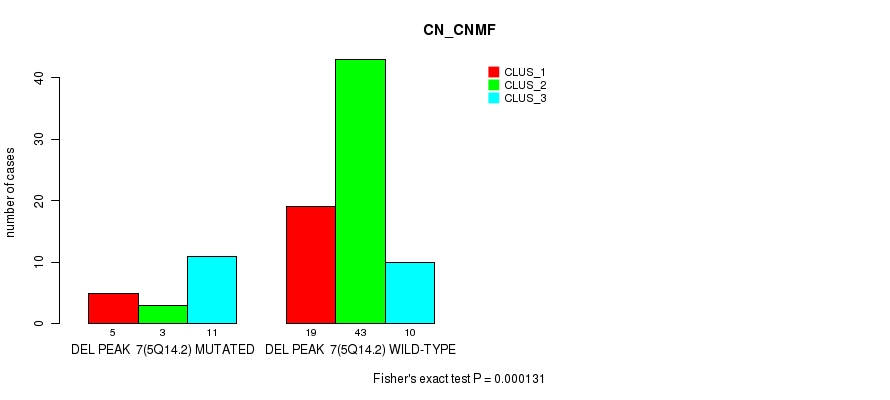
P value = 4.43e-07 (Fisher's exact test), Q value = 0.00015
Table S21. Gene #24: 'del_6p25.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 8(6P25.2) MUTATED | 15 | 7 | 16 |
| DEL PEAK 8(6P25.2) WILD-TYPE | 9 | 39 | 5 |
Figure S21. Get High-res Image Gene #24: 'del_6p25.2' versus Molecular Subtype #1: 'CN_CNMF'
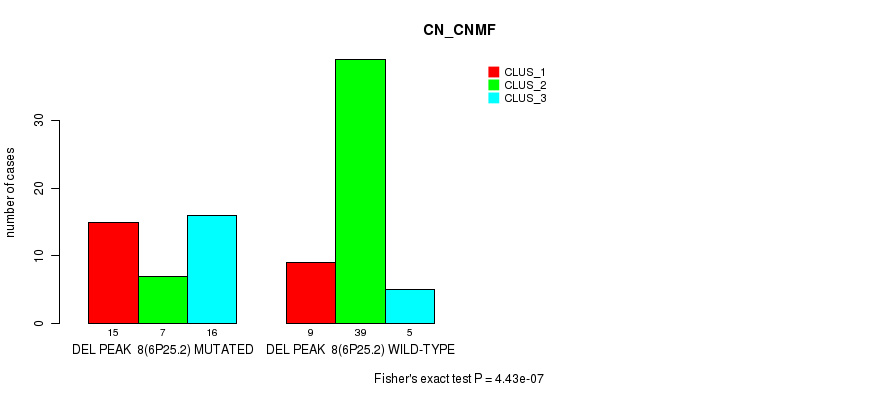
P value = 5.97e-06 (Fisher's exact test), Q value = 0.002
Table S22. Gene #24: 'del_6p25.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| DEL PEAK 8(6P25.2) MUTATED | 21 | 4 | 12 |
| DEL PEAK 8(6P25.2) WILD-TYPE | 10 | 31 | 12 |
Figure S22. Get High-res Image Gene #24: 'del_6p25.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
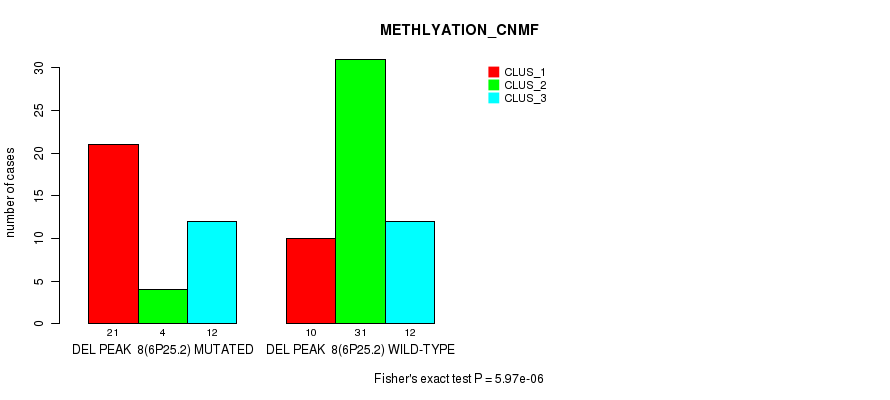
P value = 0.000369 (Fisher's exact test), Q value = 0.11
Table S23. Gene #24: 'del_6p25.2' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 31 | 52 |
| DEL PEAK 8(6P25.2) MUTATED | 0 | 5 | 29 |
| DEL PEAK 8(6P25.2) WILD-TYPE | 1 | 26 | 23 |
Figure S23. Get High-res Image Gene #24: 'del_6p25.2' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
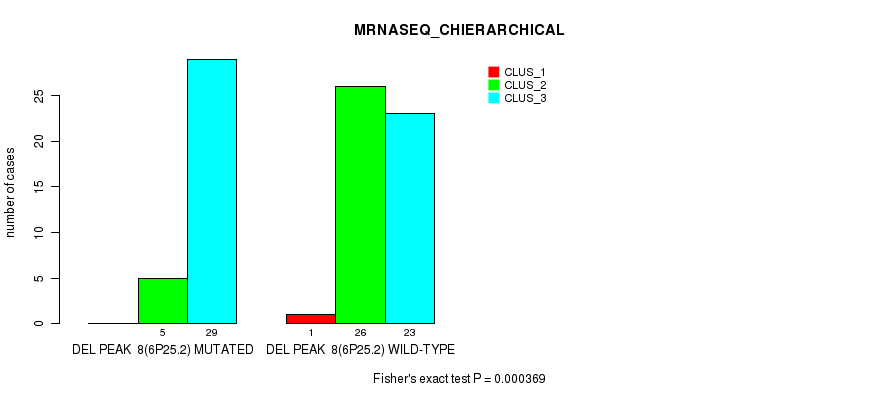
P value = 0.000218 (Fisher's exact test), Q value = 0.068
Table S24. Gene #26: 'del_8p21.3' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 10(8P21.3) MUTATED | 9 | 5 | 12 |
| DEL PEAK 10(8P21.3) WILD-TYPE | 15 | 41 | 9 |
Figure S24. Get High-res Image Gene #26: 'del_8p21.3' versus Molecular Subtype #1: 'CN_CNMF'
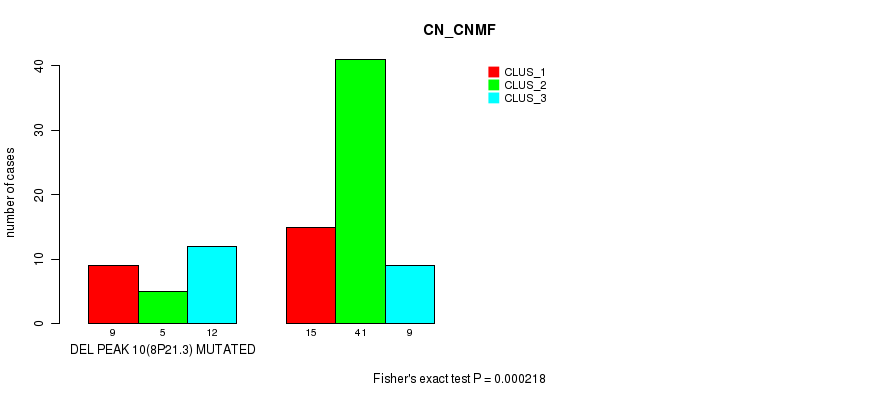
P value = 0.000425 (Fisher's exact test), Q value = 0.13
Table S25. Gene #26: 'del_8p21.3' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| DEL PEAK 10(8P21.3) MUTATED | 13 | 2 | 10 |
| DEL PEAK 10(8P21.3) WILD-TYPE | 18 | 33 | 14 |
Figure S25. Get High-res Image Gene #26: 'del_8p21.3' versus Molecular Subtype #2: 'METHLYATION_CNMF'
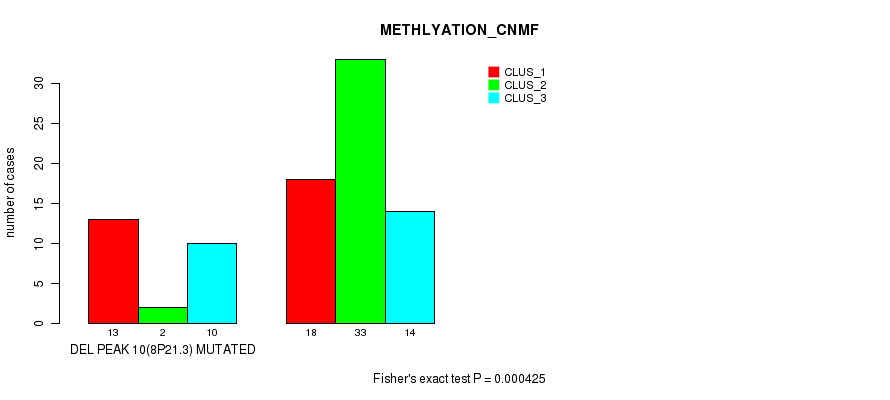
P value = 0.000415 (Fisher's exact test), Q value = 0.13
Table S26. Gene #26: 'del_8p21.3' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 31 | 52 |
| DEL PEAK 10(8P21.3) MUTATED | 0 | 2 | 22 |
| DEL PEAK 10(8P21.3) WILD-TYPE | 1 | 29 | 30 |
Figure S26. Get High-res Image Gene #26: 'del_8p21.3' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
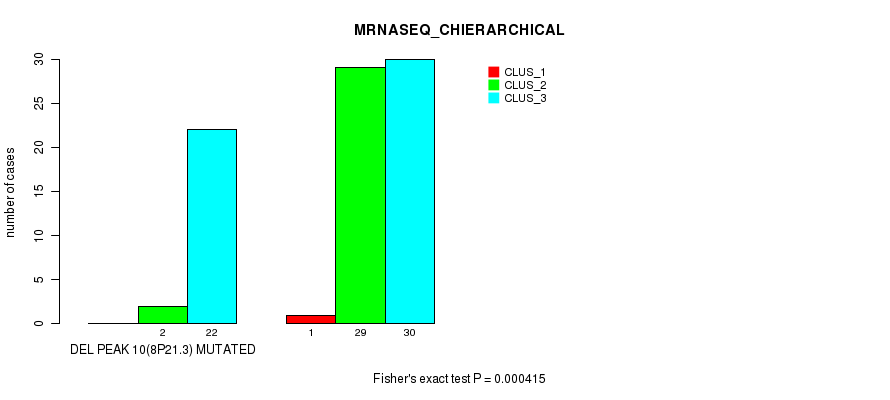
P value = 1.17e-08 (Fisher's exact test), Q value = 4e-06
Table S27. Gene #28: 'del_9p21.3' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 12(9P21.3) MUTATED | 23 | 16 | 19 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 1 | 30 | 2 |
Figure S27. Get High-res Image Gene #28: 'del_9p21.3' versus Molecular Subtype #1: 'CN_CNMF'
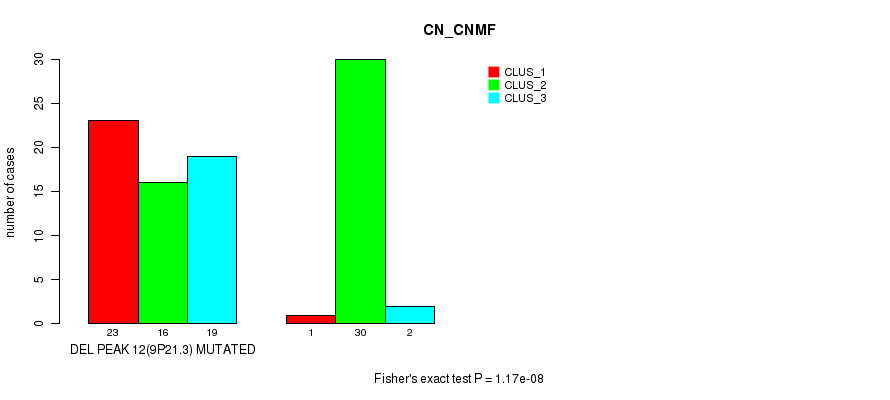
P value = 1.16e-09 (Fisher's exact test), Q value = 4e-07
Table S28. Gene #28: 'del_9p21.3' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| DEL PEAK 12(9P21.3) MUTATED | 30 | 9 | 18 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 1 | 26 | 6 |
Figure S28. Get High-res Image Gene #28: 'del_9p21.3' versus Molecular Subtype #2: 'METHLYATION_CNMF'
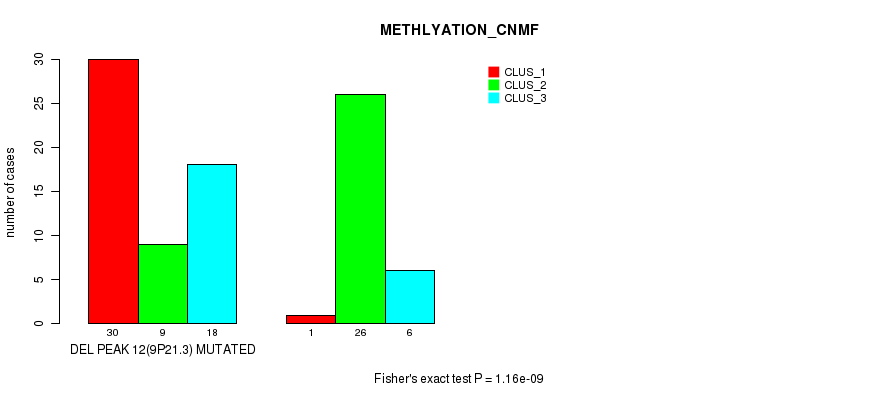
P value = 0.000292 (Fisher's exact test), Q value = 0.091
Table S29. Gene #28: 'del_9p21.3' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 40 | 24 | 20 |
| DEL PEAK 12(9P21.3) MUTATED | 33 | 11 | 7 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 7 | 13 | 13 |
Figure S29. Get High-res Image Gene #28: 'del_9p21.3' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
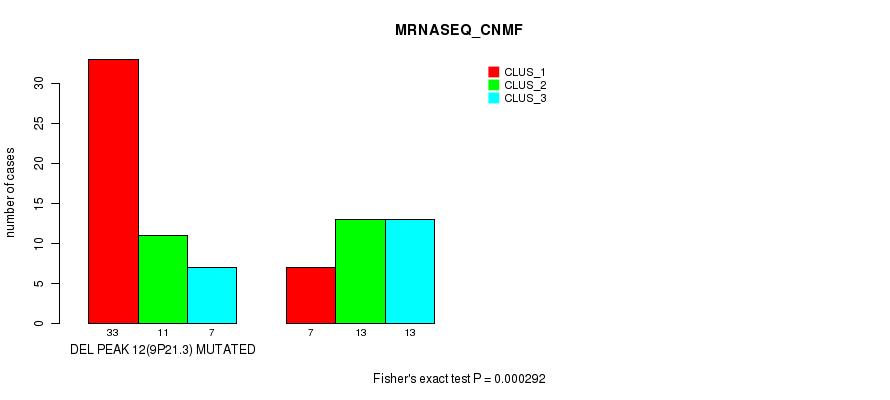
P value = 0.000152 (Fisher's exact test), Q value = 0.049
Table S30. Gene #28: 'del_9p21.3' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 1 | 31 | 52 |
| DEL PEAK 12(9P21.3) MUTATED | 0 | 11 | 40 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 1 | 20 | 12 |
Figure S30. Get High-res Image Gene #28: 'del_9p21.3' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
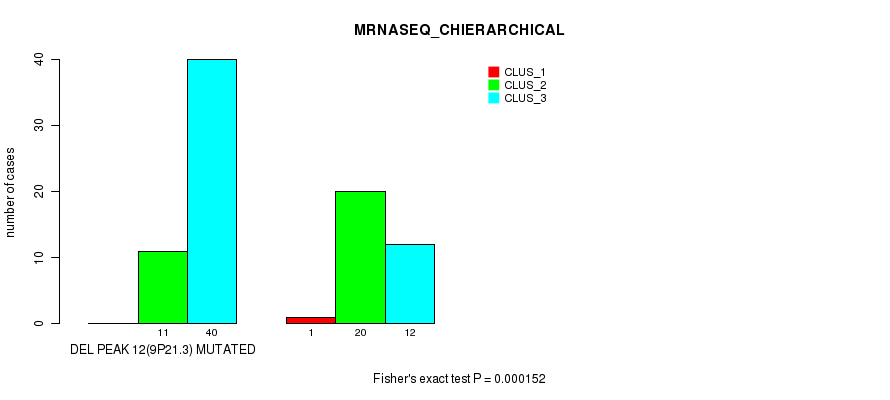
P value = 1.09e-05 (Fisher's exact test), Q value = 0.0036
Table S31. Gene #28: 'del_9p21.3' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 31 | 19 | 12 | 22 |
| DEL PEAK 12(9P21.3) MUTATED | 29 | 9 | 4 | 9 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 2 | 10 | 8 | 13 |
Figure S31. Get High-res Image Gene #28: 'del_9p21.3' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
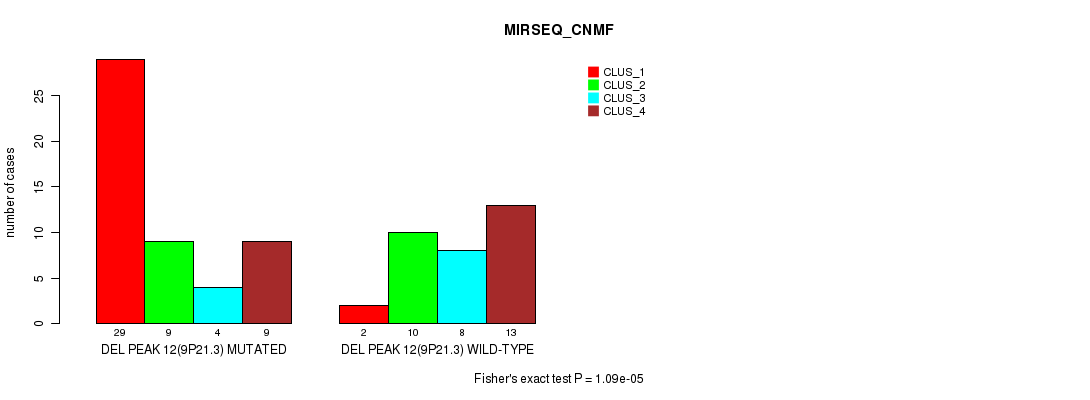
P value = 0.000183 (Fisher's exact test), Q value = 0.058
Table S32. Gene #28: 'del_9p21.3' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 37 | 17 | 30 |
| DEL PEAK 12(9P21.3) MUTATED | 31 | 5 | 15 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 6 | 12 | 15 |
Figure S32. Get High-res Image Gene #28: 'del_9p21.3' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
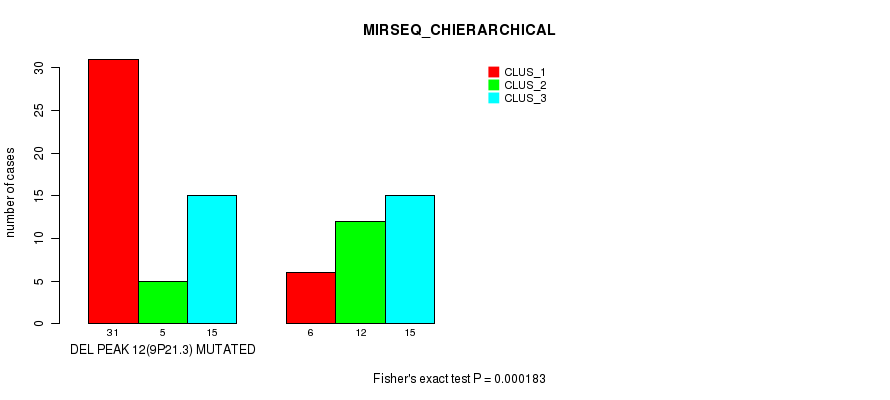
P value = 2.56e-05 (Fisher's exact test), Q value = 0.0084
Table S33. Gene #28: 'del_9p21.3' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 38 | 19 | 27 |
| DEL PEAK 12(9P21.3) MUTATED | 33 | 8 | 10 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 5 | 11 | 17 |
Figure S33. Get High-res Image Gene #28: 'del_9p21.3' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
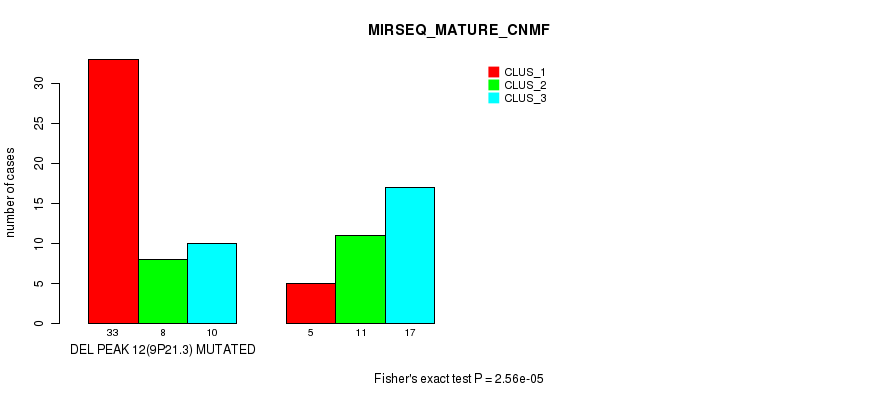
P value = 2.54e-05 (Fisher's exact test), Q value = 0.0083
Table S34. Gene #28: 'del_9p21.3' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 26 | 20 | 38 |
| DEL PEAK 12(9P21.3) MUTATED | 11 | 7 | 33 |
| DEL PEAK 12(9P21.3) WILD-TYPE | 15 | 13 | 5 |
Figure S34. Get High-res Image Gene #28: 'del_9p21.3' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
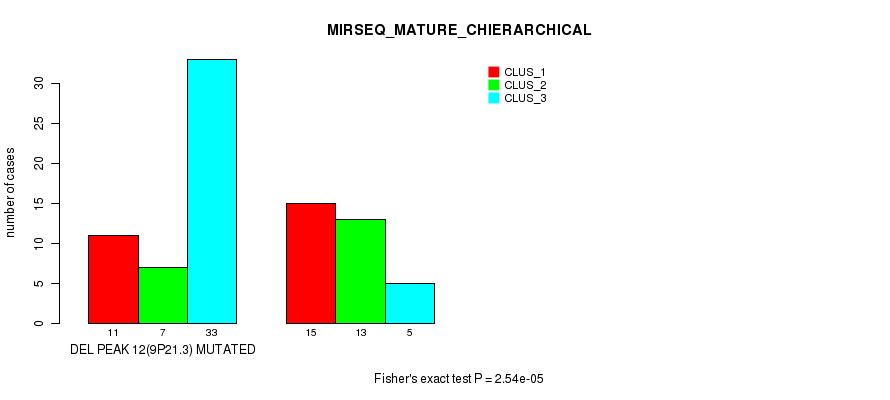
P value = 7.66e-05 (Fisher's exact test), Q value = 0.025
Table S35. Gene #31: 'del_10q23.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 15(10Q23.2) MUTATED | 7 | 1 | 8 |
| DEL PEAK 15(10Q23.2) WILD-TYPE | 17 | 45 | 13 |
Figure S35. Get High-res Image Gene #31: 'del_10q23.2' versus Molecular Subtype #1: 'CN_CNMF'
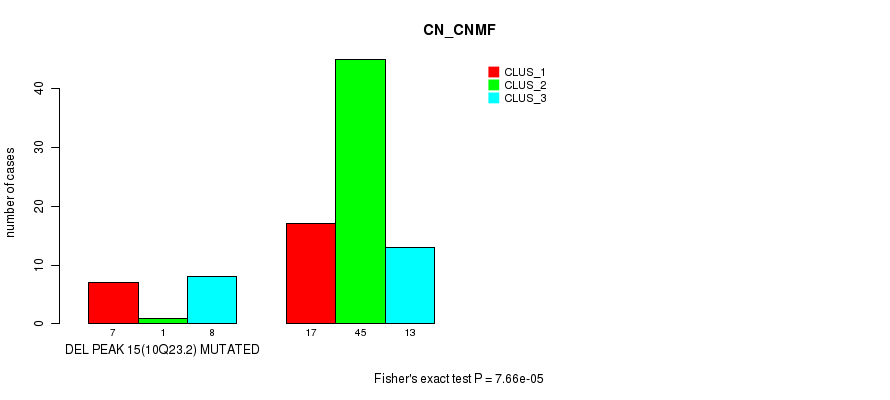
P value = 0.00034 (Fisher's exact test), Q value = 0.11
Table S36. Gene #33: 'del_12p12.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 17(12P12.1) MUTATED | 6 | 0 | 5 |
| DEL PEAK 17(12P12.1) WILD-TYPE | 18 | 46 | 16 |
Figure S36. Get High-res Image Gene #33: 'del_12p12.1' versus Molecular Subtype #1: 'CN_CNMF'
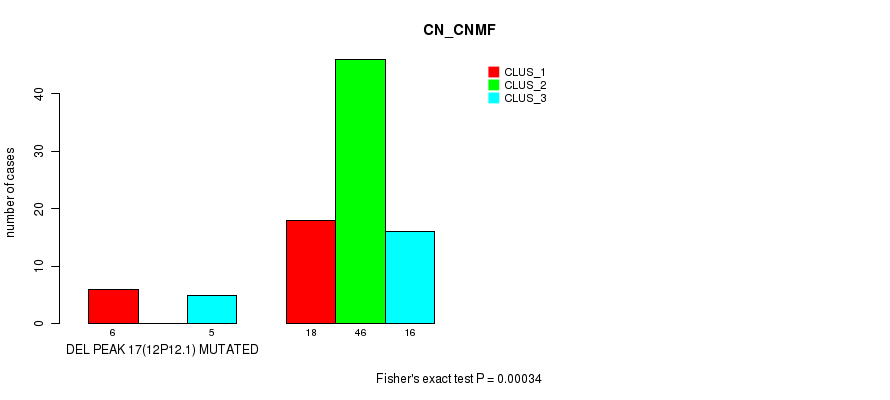
P value = 0.000179 (Fisher's exact test), Q value = 0.057
Table S37. Gene #34: 'del_12q12' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 18(12Q12) MUTATED | 7 | 0 | 4 |
| DEL PEAK 18(12Q12) WILD-TYPE | 17 | 46 | 17 |
Figure S37. Get High-res Image Gene #34: 'del_12q12' versus Molecular Subtype #1: 'CN_CNMF'
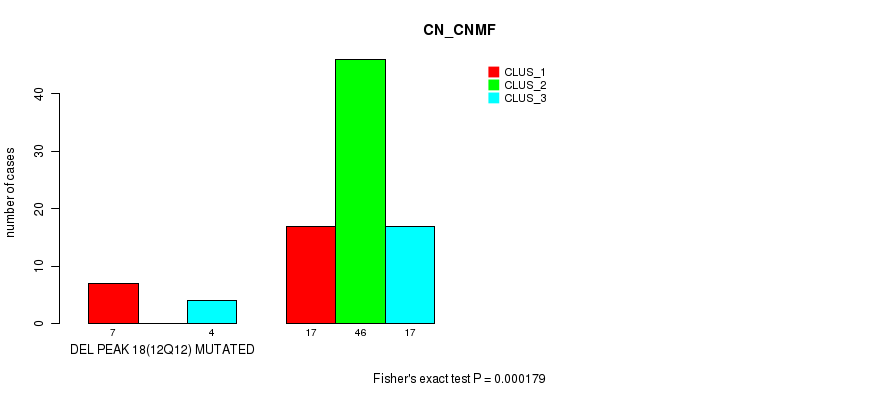
P value = 0.000131 (Fisher's exact test), Q value = 0.042
Table S38. Gene #35: 'del_12q24.33' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 19(12Q24.33) MUTATED | 5 | 3 | 11 |
| DEL PEAK 19(12Q24.33) WILD-TYPE | 19 | 43 | 10 |
Figure S38. Get High-res Image Gene #35: 'del_12q24.33' versus Molecular Subtype #1: 'CN_CNMF'
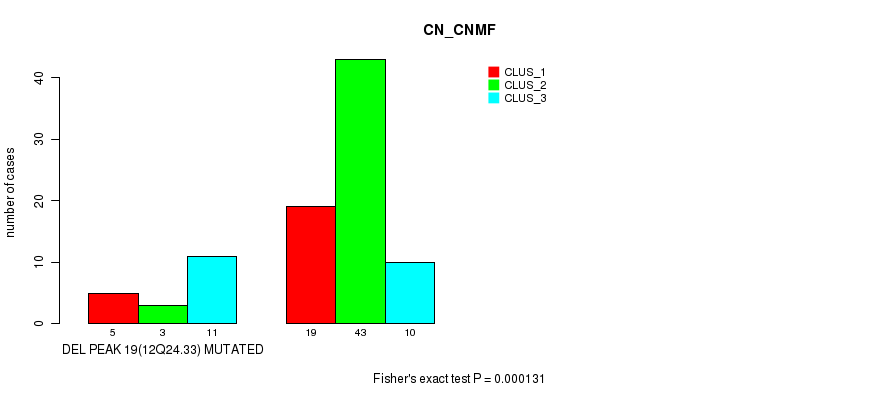
P value = 1.17e-07 (Fisher's exact test), Q value = 3.9e-05
Table S39. Gene #37: 'del_17q22' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 21(17Q22) MUTATED | 9 | 2 | 14 |
| DEL PEAK 21(17Q22) WILD-TYPE | 15 | 44 | 7 |
Figure S39. Get High-res Image Gene #37: 'del_17q22' versus Molecular Subtype #1: 'CN_CNMF'
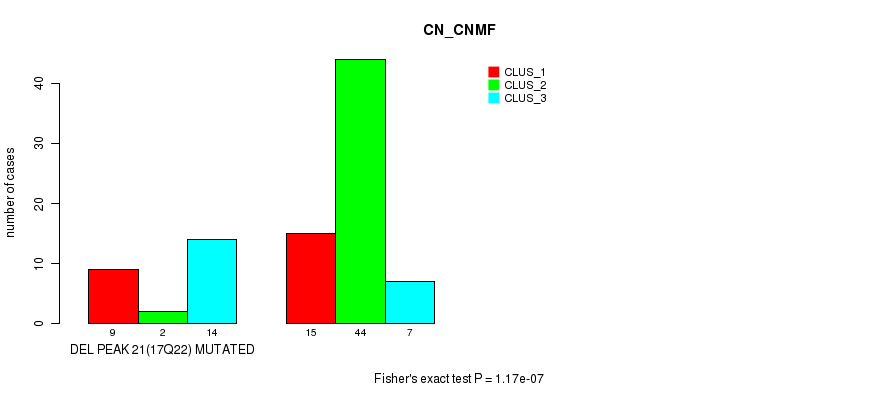
P value = 0.000359 (Fisher's exact test), Q value = 0.11
Table S40. Gene #37: 'del_17q22' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| DEL PEAK 21(17Q22) MUTATED | 14 | 2 | 9 |
| DEL PEAK 21(17Q22) WILD-TYPE | 17 | 33 | 15 |
Figure S40. Get High-res Image Gene #37: 'del_17q22' versus Molecular Subtype #2: 'METHLYATION_CNMF'
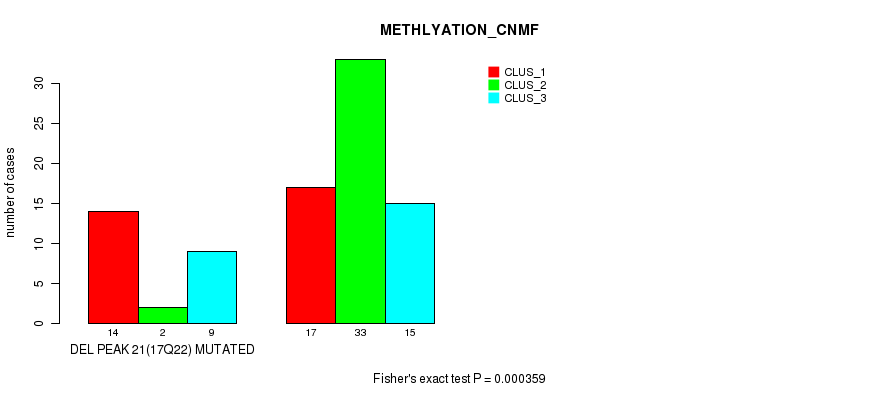
P value = 3.56e-06 (Fisher's exact test), Q value = 0.0012
Table S41. Gene #39: 'del_18q21.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 23(18Q21.2) MUTATED | 24 | 23 | 18 |
| DEL PEAK 23(18Q21.2) WILD-TYPE | 0 | 23 | 3 |
Figure S41. Get High-res Image Gene #39: 'del_18q21.2' versus Molecular Subtype #1: 'CN_CNMF'
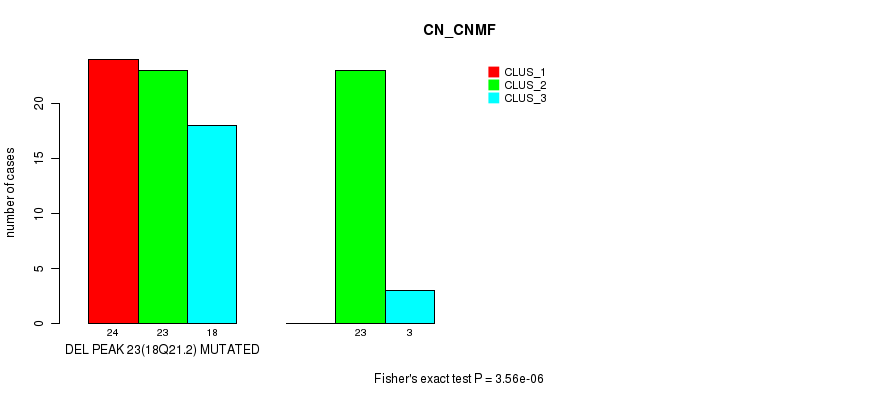
P value = 0.000175 (Fisher's exact test), Q value = 0.056
Table S42. Gene #39: 'del_18q21.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 31 | 35 | 24 |
| DEL PEAK 23(18Q21.2) MUTATED | 27 | 16 | 21 |
| DEL PEAK 23(18Q21.2) WILD-TYPE | 4 | 19 | 3 |
Figure S42. Get High-res Image Gene #39: 'del_18q21.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
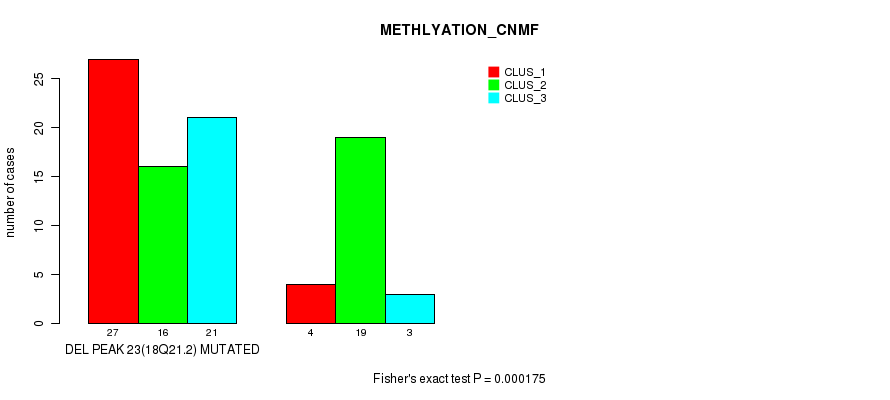
P value = 4.19e-06 (Fisher's exact test), Q value = 0.0014
Table S43. Gene #40: 'del_19p13.3' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 24(19P13.3) MUTATED | 5 | 1 | 11 |
| DEL PEAK 24(19P13.3) WILD-TYPE | 19 | 45 | 10 |
Figure S43. Get High-res Image Gene #40: 'del_19p13.3' versus Molecular Subtype #1: 'CN_CNMF'
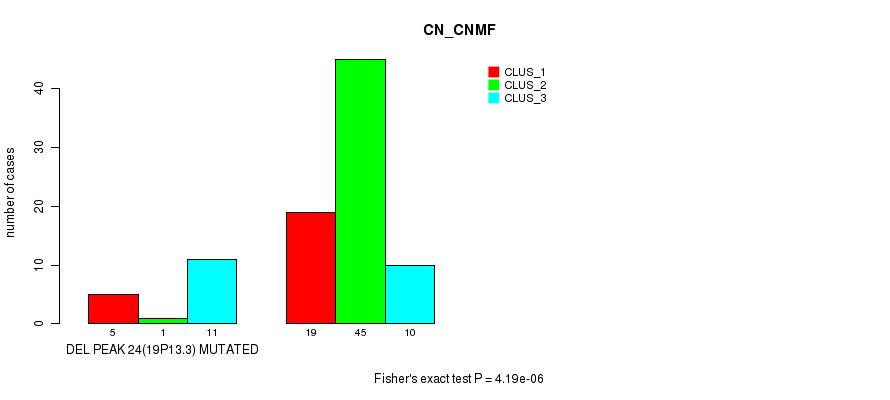
P value = 1.42e-06 (Fisher's exact test), Q value = 0.00048
Table S44. Gene #42: 'del_21q21.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 26(21Q21.1) MUTATED | 15 | 6 | 14 |
| DEL PEAK 26(21Q21.1) WILD-TYPE | 9 | 40 | 7 |
Figure S44. Get High-res Image Gene #42: 'del_21q21.1' versus Molecular Subtype #1: 'CN_CNMF'
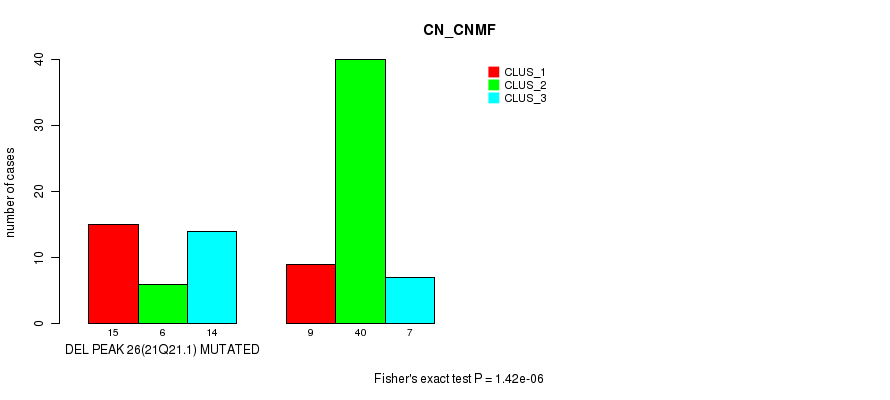
P value = 0.000211 (Fisher's exact test), Q value = 0.066
Table S45. Gene #43: 'del_22q13.31' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 24 | 46 | 21 |
| DEL PEAK 27(22Q13.31) MUTATED | 11 | 5 | 11 |
| DEL PEAK 27(22Q13.31) WILD-TYPE | 13 | 41 | 10 |
Figure S45. Get High-res Image Gene #43: 'del_22q13.31' versus Molecular Subtype #1: 'CN_CNMF'
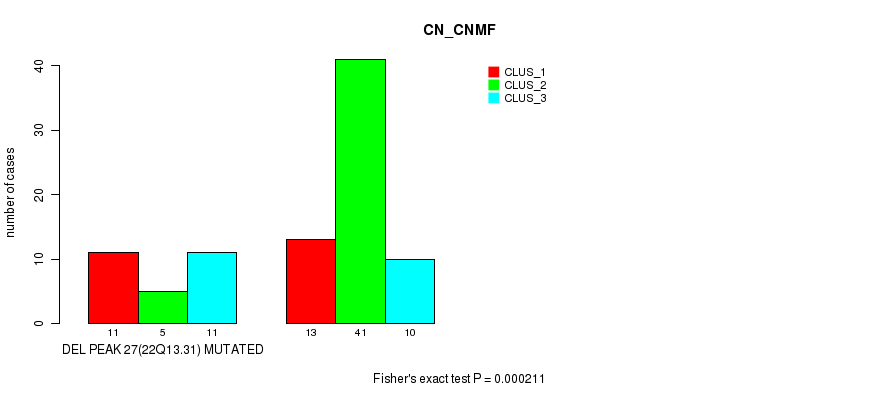
-
Copy number data file = transformed.cor.cli.txt
-
Molecular subtype file = PAAD-TP.transferedmergedcluster.txt
-
Number of patients = 91
-
Number of significantly focal cnvs = 43
-
Number of molecular subtypes = 8
-
Exclude genes that fewer than K tumors have alterations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.