This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and molecular subtypes.
Testing the association between copy number variation 67 arm-level events and 6 molecular subtypes across 159 patients, 21 significant findings detected with P value < 0.05 and Q value < 0.25.
-
1q gain cnv correlated to 'CN_CNMF'.
-
12p gain cnv correlated to 'MIRSEQ_MATURE_CNMF'.
-
13q gain cnv correlated to 'CN_CNMF'.
-
1p loss cnv correlated to 'CN_CNMF'.
-
3p loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
3q loss cnv correlated to 'CN_CNMF'.
-
6q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
11p loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
11q loss cnv correlated to 'CN_CNMF' and 'MIRSEQ_MATURE_CNMF'.
-
16q loss cnv correlated to 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 67 arm-level events and 6 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 21 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 6q loss | 19 (12%) | 140 |
0.000412 (0.152) |
0.000536 (0.196) |
0.00044 (0.162) |
9.39e-06 (0.00355) |
0.000367 (0.136) |
4.75e-06 (0.0018) |
| 11p loss | 52 (33%) | 107 |
5.84e-12 (2.25e-09) |
0.00106 (0.381) |
0.00206 (0.732) |
7.08e-06 (0.00268) |
0.000203 (0.0757) |
8.26e-05 (0.0311) |
| 3p loss | 61 (38%) | 98 |
2.4e-08 (9.18e-06) |
0.0305 (1.00) |
6.9e-05 (0.026) |
0.00053 (0.194) |
0.000918 (0.332) |
0.0112 (1.00) |
| 11q loss | 39 (25%) | 120 |
4.13e-11 (1.59e-08) |
0.049 (1.00) |
0.0301 (1.00) |
0.00644 (1.00) |
0.000182 (0.0681) |
0.000879 (0.32) |
| 1q gain | 16 (10%) | 143 |
1.33e-12 (5.15e-10) |
0.698 (1.00) |
0.525 (1.00) |
0.447 (1.00) |
0.638 (1.00) |
0.801 (1.00) |
| 12p gain | 11 (7%) | 148 |
0.00724 (1.00) |
0.00844 (1.00) |
0.182 (1.00) |
0.0743 (1.00) |
0.000209 (0.0779) |
0.0325 (1.00) |
| 13q gain | 9 (6%) | 150 |
0.000279 (0.103) |
0.0213 (1.00) |
0.142 (1.00) |
0.096 (1.00) |
0.00779 (1.00) |
0.0694 (1.00) |
| 1p loss | 96 (60%) | 63 |
1.46e-09 (5.58e-07) |
0.0482 (1.00) |
0.0103 (1.00) |
0.0137 (1.00) |
0.000883 (0.321) |
0.0248 (1.00) |
| 3q loss | 89 (56%) | 70 |
3.15e-07 (0.00012) |
0.522 (1.00) |
0.0522 (1.00) |
0.000727 (0.265) |
0.244 (1.00) |
0.0118 (1.00) |
| 16q loss | 4 (3%) | 155 |
0.000193 (0.0723) |
0.274 (1.00) |
1 (1.00) |
0.441 (1.00) |
0.348 (1.00) |
0.471 (1.00) |
| 1p gain | 5 (3%) | 154 |
0.00188 (0.671) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.292 (1.00) |
0.681 (1.00) |
| 2p gain | 6 (4%) | 153 |
0.134 (1.00) |
0.00595 (1.00) |
0.184 (1.00) |
0.285 (1.00) |
0.00537 (1.00) |
0.138 (1.00) |
| 4p gain | 4 (3%) | 155 |
0.806 (1.00) |
0.471 (1.00) |
0.218 (1.00) |
0.376 (1.00) |
0.348 (1.00) |
0.181 (1.00) |
| 4q gain | 3 (2%) | 156 |
0.361 (1.00) |
0.803 (1.00) |
||||
| 5p gain | 11 (7%) | 148 |
0.08 (1.00) |
0.367 (1.00) |
0.567 (1.00) |
0.569 (1.00) |
1 (1.00) |
0.902 (1.00) |
| 5q gain | 7 (4%) | 152 |
0.218 (1.00) |
0.00526 (1.00) |
0.157 (1.00) |
0.227 (1.00) |
0.61 (1.00) |
0.166 (1.00) |
| 6p gain | 13 (8%) | 146 |
0.49 (1.00) |
0.293 (1.00) |
0.911 (1.00) |
0.609 (1.00) |
0.547 (1.00) |
0.645 (1.00) |
| 6q gain | 7 (4%) | 152 |
0.0835 (1.00) |
0.000935 (0.337) |
0.157 (1.00) |
0.191 (1.00) |
0.0607 (1.00) |
0.166 (1.00) |
| 7p gain | 26 (16%) | 133 |
0.0376 (1.00) |
0.118 (1.00) |
0.41 (1.00) |
0.288 (1.00) |
0.201 (1.00) |
0.179 (1.00) |
| 7q gain | 21 (13%) | 138 |
0.00702 (1.00) |
0.0113 (1.00) |
0.375 (1.00) |
0.487 (1.00) |
0.0345 (1.00) |
0.246 (1.00) |
| 8p gain | 10 (6%) | 149 |
0.0256 (1.00) |
0.334 (1.00) |
0.361 (1.00) |
0.286 (1.00) |
0.109 (1.00) |
0.217 (1.00) |
| 8q gain | 13 (8%) | 146 |
0.152 (1.00) |
0.753 (1.00) |
0.309 (1.00) |
0.253 (1.00) |
0.144 (1.00) |
0.268 (1.00) |
| 9p gain | 3 (2%) | 156 |
0.185 (1.00) |
0.135 (1.00) |
0.402 (1.00) |
0.441 (1.00) |
0.112 (1.00) |
0.471 (1.00) |
| 9q gain | 3 (2%) | 156 |
0.185 (1.00) |
0.135 (1.00) |
0.402 (1.00) |
0.441 (1.00) |
0.112 (1.00) |
0.471 (1.00) |
| 10p gain | 12 (8%) | 147 |
0.816 (1.00) |
0.0487 (1.00) |
0.819 (1.00) |
1 (1.00) |
0.907 (1.00) |
0.905 (1.00) |
| 10q gain | 10 (6%) | 149 |
0.542 (1.00) |
0.691 (1.00) |
0.507 (1.00) |
0.718 (1.00) |
0.423 (1.00) |
0.807 (1.00) |
| 11p gain | 4 (3%) | 155 |
0.534 (1.00) |
0.808 (1.00) |
0.15 (1.00) |
0.139 (1.00) |
0.184 (1.00) |
0.0473 (1.00) |
| 12q gain | 13 (8%) | 146 |
0.0617 (1.00) |
0.102 (1.00) |
0.263 (1.00) |
0.171 (1.00) |
0.00933 (1.00) |
0.0912 (1.00) |
| 14q gain | 4 (3%) | 155 |
0.468 (1.00) |
0.471 (1.00) |
0.832 (1.00) |
0.731 (1.00) |
0.0631 (1.00) |
0.46 (1.00) |
| 15q gain | 14 (9%) | 145 |
0.214 (1.00) |
0.0266 (1.00) |
1 (1.00) |
1 (1.00) |
0.595 (1.00) |
1 (1.00) |
| 16p gain | 6 (4%) | 153 |
0.152 (1.00) |
0.551 (1.00) |
0.412 (1.00) |
1 (1.00) |
0.167 (1.00) |
0.681 (1.00) |
| 16q gain | 6 (4%) | 153 |
0.191 (1.00) |
1 (1.00) |
0.507 (1.00) |
0.567 (1.00) |
0.61 (1.00) |
0.369 (1.00) |
| 17q gain | 4 (3%) | 155 |
0.369 (1.00) |
0.185 (1.00) |
0.15 (1.00) |
0.619 (1.00) |
0.184 (1.00) |
0.336 (1.00) |
| 18p gain | 8 (5%) | 151 |
0.1 (1.00) |
0.257 (1.00) |
1 (1.00) |
0.569 (1.00) |
0.0751 (1.00) |
0.651 (1.00) |
| 18q gain | 10 (6%) | 149 |
0.0892 (1.00) |
0.585 (1.00) |
1 (1.00) |
0.672 (1.00) |
0.146 (1.00) |
0.619 (1.00) |
| 19p gain | 20 (13%) | 139 |
0.0221 (1.00) |
0.0746 (1.00) |
0.274 (1.00) |
0.63 (1.00) |
0.254 (1.00) |
0.654 (1.00) |
| 19q gain | 14 (9%) | 145 |
0.0715 (1.00) |
0.00761 (1.00) |
0.151 (1.00) |
0.263 (1.00) |
0.0849 (1.00) |
0.232 (1.00) |
| 20p gain | 11 (7%) | 148 |
0.166 (1.00) |
0.19 (1.00) |
0.735 (1.00) |
0.392 (1.00) |
0.023 (1.00) |
0.454 (1.00) |
| 20q gain | 9 (6%) | 150 |
0.218 (1.00) |
0.0358 (1.00) |
0.236 (1.00) |
0.0625 (1.00) |
0.00702 (1.00) |
0.0487 (1.00) |
| 21q gain | 3 (2%) | 156 |
0.551 (1.00) |
0.135 (1.00) |
||||
| 22q gain | 3 (2%) | 156 |
0.185 (1.00) |
0.0823 (1.00) |
0.218 (1.00) |
0.376 (1.00) |
0.0163 (1.00) |
0.181 (1.00) |
| xq gain | 5 (3%) | 154 |
0.541 (1.00) |
0.054 (1.00) |
0.832 (1.00) |
0.289 (1.00) |
0.167 (1.00) |
0.359 (1.00) |
| 1q loss | 28 (18%) | 131 |
0.217 (1.00) |
0.0741 (1.00) |
0.918 (1.00) |
0.387 (1.00) |
0.351 (1.00) |
0.125 (1.00) |
| 2p loss | 10 (6%) | 149 |
0.463 (1.00) |
0.0815 (1.00) |
0.419 (1.00) |
0.00286 (1.00) |
0.454 (1.00) |
0.00121 (0.433) |
| 2q loss | 13 (8%) | 146 |
0.559 (1.00) |
0.448 (1.00) |
0.645 (1.00) |
0.0974 (1.00) |
0.933 (1.00) |
0.327 (1.00) |
| 4p loss | 10 (6%) | 149 |
0.0134 (1.00) |
0.239 (1.00) |
0.396 (1.00) |
0.569 (1.00) |
0.0238 (1.00) |
0.651 (1.00) |
| 4q loss | 10 (6%) | 149 |
0.103 (1.00) |
0.239 (1.00) |
0.396 (1.00) |
0.569 (1.00) |
0.0238 (1.00) |
0.651 (1.00) |
| 5p loss | 5 (3%) | 154 |
0.0266 (1.00) |
0.307 (1.00) |
1 (1.00) |
0.441 (1.00) |
0.348 (1.00) |
0.471 (1.00) |
| 5q loss | 7 (4%) | 152 |
0.0747 (1.00) |
0.785 (1.00) |
0.457 (1.00) |
0.557 (1.00) |
0.108 (1.00) |
1 (1.00) |
| 7q loss | 6 (4%) | 153 |
0.221 (1.00) |
0.426 (1.00) |
0.131 (1.00) |
0.0588 (1.00) |
0.0306 (1.00) |
0.042 (1.00) |
| 8p loss | 19 (12%) | 140 |
0.218 (1.00) |
0.575 (1.00) |
0.753 (1.00) |
0.645 (1.00) |
0.34 (1.00) |
0.414 (1.00) |
| 8q loss | 12 (8%) | 147 |
0.564 (1.00) |
0.803 (1.00) |
0.396 (1.00) |
0.26 (1.00) |
0.902 (1.00) |
0.266 (1.00) |
| 9p loss | 12 (8%) | 147 |
0.0468 (1.00) |
0.686 (1.00) |
1 (1.00) |
0.186 (1.00) |
0.249 (1.00) |
0.306 (1.00) |
| 9q loss | 11 (7%) | 148 |
0.0439 (1.00) |
0.663 (1.00) |
0.439 (1.00) |
0.476 (1.00) |
0.417 (1.00) |
0.833 (1.00) |
| 13q loss | 7 (4%) | 152 |
0.757 (1.00) |
0.611 (1.00) |
0.49 (1.00) |
0.203 (1.00) |
0.571 (1.00) |
0.489 (1.00) |
| 14q loss | 20 (13%) | 139 |
0.00984 (1.00) |
0.0474 (1.00) |
0.36 (1.00) |
0.125 (1.00) |
0.722 (1.00) |
0.279 (1.00) |
| 15q loss | 3 (2%) | 156 |
0.159 (1.00) |
0.327 (1.00) |
||||
| 16p loss | 8 (5%) | 151 |
0.0118 (1.00) |
0.0244 (1.00) |
0.312 (1.00) |
0.341 (1.00) |
0.479 (1.00) |
0.791 (1.00) |
| 17p loss | 61 (38%) | 98 |
0.0351 (1.00) |
0.00258 (0.913) |
0.00496 (1.00) |
0.0178 (1.00) |
0.00802 (1.00) |
0.0234 (1.00) |
| 17q loss | 20 (13%) | 139 |
0.0102 (1.00) |
0.919 (1.00) |
0.204 (1.00) |
0.0421 (1.00) |
0.0241 (1.00) |
0.0682 (1.00) |
| 18p loss | 16 (10%) | 143 |
0.783 (1.00) |
0.744 (1.00) |
0.302 (1.00) |
0.328 (1.00) |
0.496 (1.00) |
0.707 (1.00) |
| 18q loss | 6 (4%) | 153 |
0.413 (1.00) |
0.876 (1.00) |
0.157 (1.00) |
0.227 (1.00) |
0.218 (1.00) |
0.166 (1.00) |
| 19q loss | 6 (4%) | 153 |
0.0178 (1.00) |
1 (1.00) |
0.236 (1.00) |
0.814 (1.00) |
0.819 (1.00) |
1 (1.00) |
| 20q loss | 3 (2%) | 156 |
0.763 (1.00) |
1 (1.00) |
||||
| 21q loss | 34 (21%) | 125 |
0.0985 (1.00) |
0.0639 (1.00) |
0.763 (1.00) |
0.491 (1.00) |
0.631 (1.00) |
0.126 (1.00) |
| 22q loss | 56 (35%) | 103 |
0.027 (1.00) |
0.0358 (1.00) |
0.0404 (1.00) |
0.102 (1.00) |
0.0399 (1.00) |
0.0075 (1.00) |
| xq loss | 49 (31%) | 110 |
0.00477 (1.00) |
0.00139 (0.496) |
0.00869 (1.00) |
0.0176 (1.00) |
0.00184 (0.657) |
0.00276 (0.974) |
P value = 1.33e-12 (Chi-square test), Q value = 5.2e-10
Table S1. Gene #2: '1q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| 1Q GAIN MUTATED | 13 | 1 | 2 | 0 | 0 | 0 |
| 1Q GAIN WILD-TYPE | 10 | 23 | 55 | 46 | 7 | 2 |
Figure S1. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #1: 'CN_CNMF'
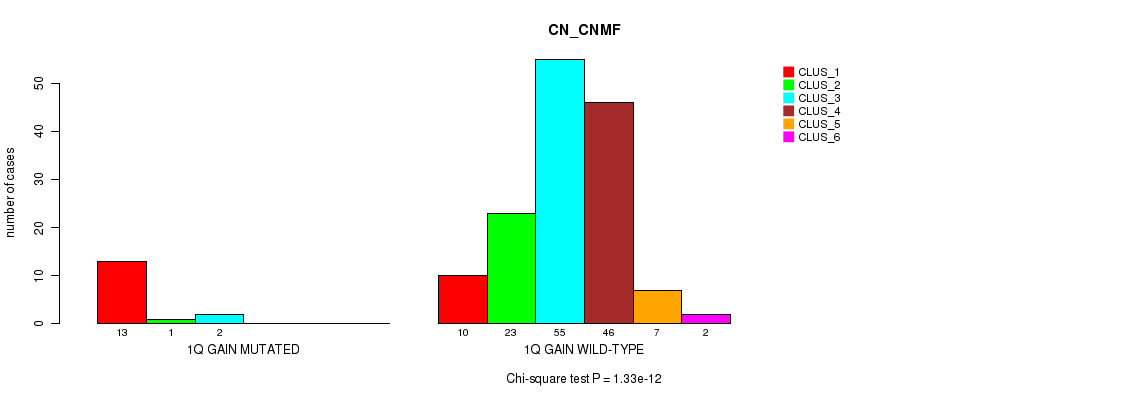
P value = 0.000209 (Fisher's exact test), Q value = 0.078
Table S2. Gene #19: '12p gain' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 55 | 42 |
| 12P GAIN MUTATED | 7 | 0 | 1 |
| 12P GAIN WILD-TYPE | 27 | 55 | 41 |
Figure S2. Get High-res Image Gene #19: '12p gain' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
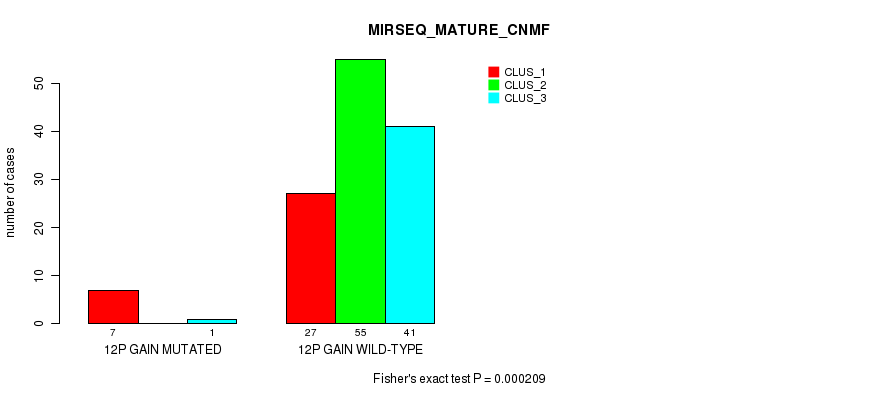
P value = 0.000279 (Chi-square test), Q value = 0.1
Table S3. Gene #21: '13q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| 13Q GAIN MUTATED | 0 | 0 | 0 | 9 | 0 | 0 |
| 13Q GAIN WILD-TYPE | 23 | 24 | 57 | 37 | 7 | 2 |
Figure S3. Get High-res Image Gene #21: '13q gain' versus Molecular Subtype #1: 'CN_CNMF'
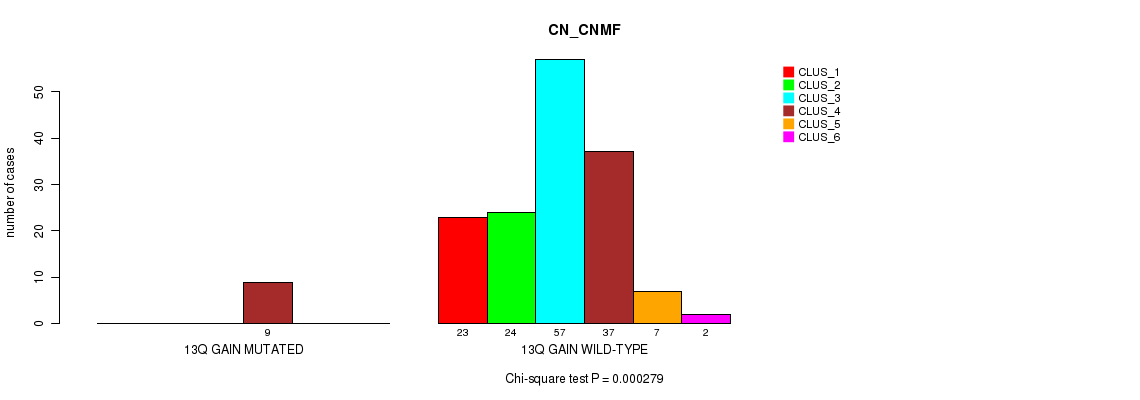
P value = 1.46e-09 (Chi-square test), Q value = 5.6e-07
Table S4. Gene #36: '1p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| 1P LOSS MUTATED | 17 | 4 | 49 | 18 | 7 | 1 |
| 1P LOSS WILD-TYPE | 6 | 20 | 8 | 28 | 0 | 1 |
Figure S4. Get High-res Image Gene #36: '1p loss' versus Molecular Subtype #1: 'CN_CNMF'
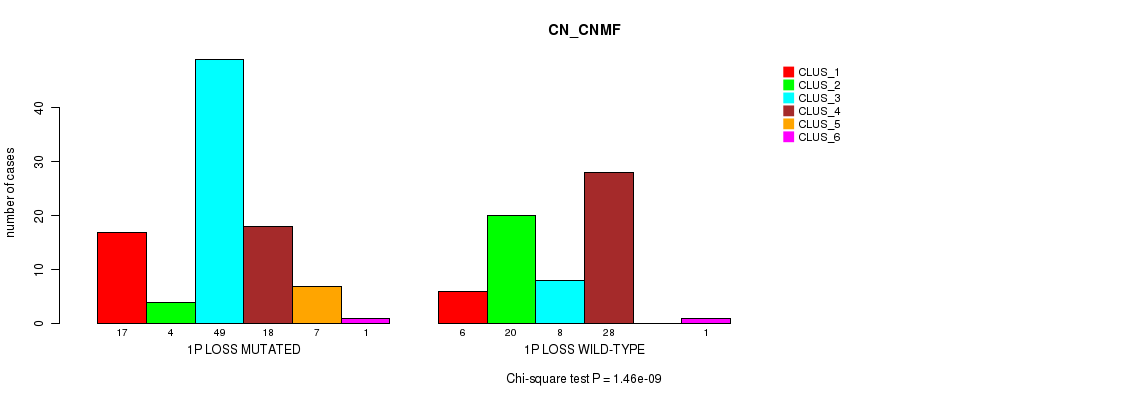
P value = 2.4e-08 (Chi-square test), Q value = 9.2e-06
Table S5. Gene #40: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| 3P LOSS MUTATED | 12 | 1 | 12 | 33 | 3 | 0 |
| 3P LOSS WILD-TYPE | 11 | 23 | 45 | 13 | 4 | 2 |
Figure S5. Get High-res Image Gene #40: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
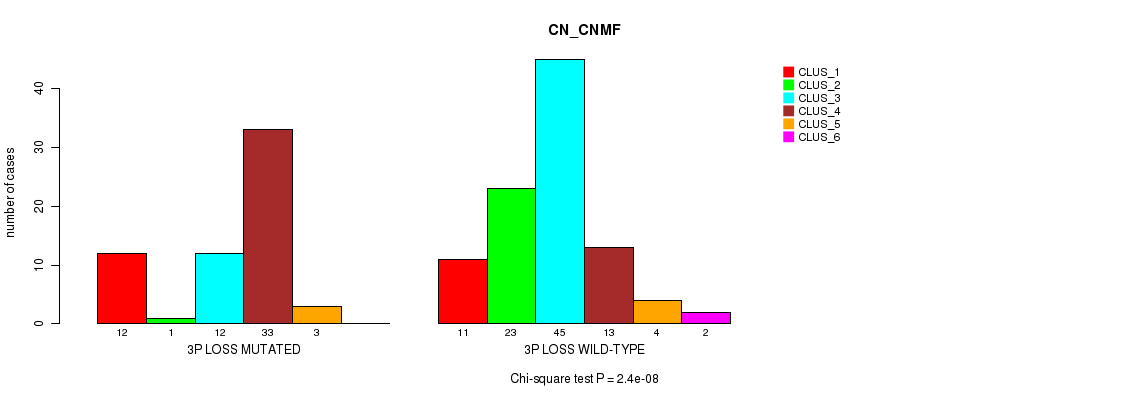
P value = 6.9e-05 (Fisher's exact test), Q value = 0.026
Table S6. Gene #40: '3p loss' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 53 | 24 |
| 3P LOSS MUTATED | 32 | 13 | 4 |
| 3P LOSS WILD-TYPE | 22 | 40 | 20 |
Figure S6. Get High-res Image Gene #40: '3p loss' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
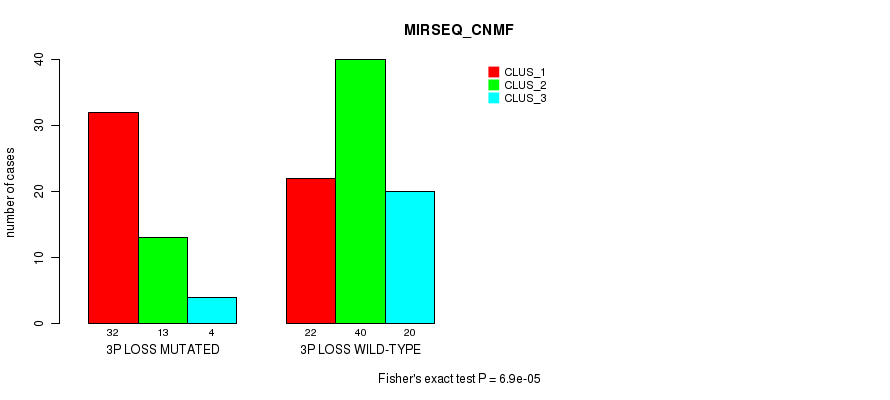
P value = 0.00053 (Fisher's exact test), Q value = 0.19
Table S7. Gene #40: '3p loss' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 41 | 18 | 52 |
| 3P LOSS MUTATED | 2 | 12 | 5 | 30 |
| 3P LOSS WILD-TYPE | 18 | 29 | 13 | 22 |
Figure S7. Get High-res Image Gene #40: '3p loss' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
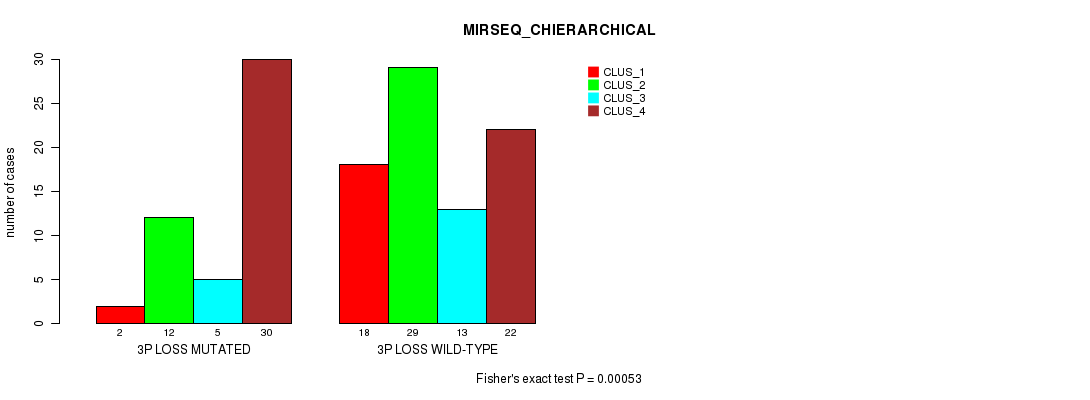
P value = 3.15e-07 (Chi-square test), Q value = 0.00012
Table S8. Gene #41: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| 3Q LOSS MUTATED | 12 | 0 | 39 | 32 | 5 | 1 |
| 3Q LOSS WILD-TYPE | 11 | 24 | 18 | 14 | 2 | 1 |
Figure S8. Get High-res Image Gene #41: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
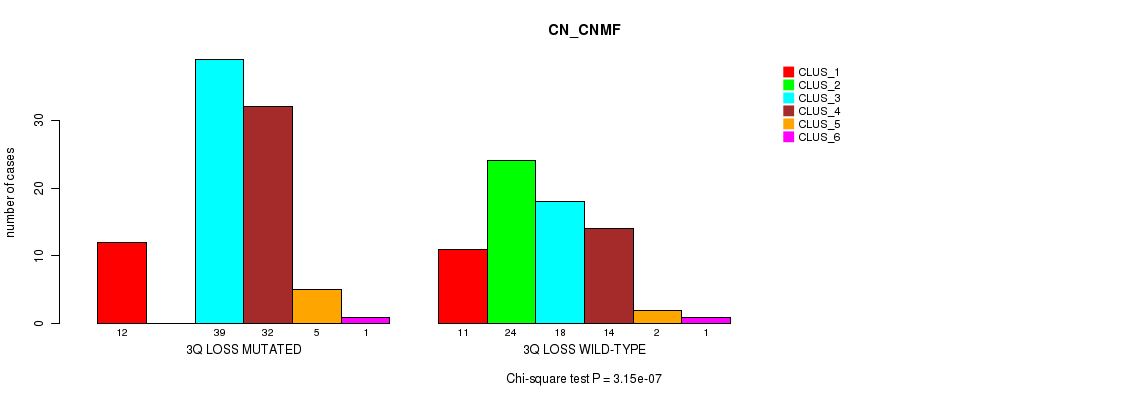
P value = 0.000412 (Chi-square test), Q value = 0.15
Table S9. Gene #46: '6q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| 6Q LOSS MUTATED | 0 | 0 | 15 | 3 | 0 | 1 |
| 6Q LOSS WILD-TYPE | 23 | 24 | 42 | 43 | 7 | 1 |
Figure S9. Get High-res Image Gene #46: '6q loss' versus Molecular Subtype #1: 'CN_CNMF'
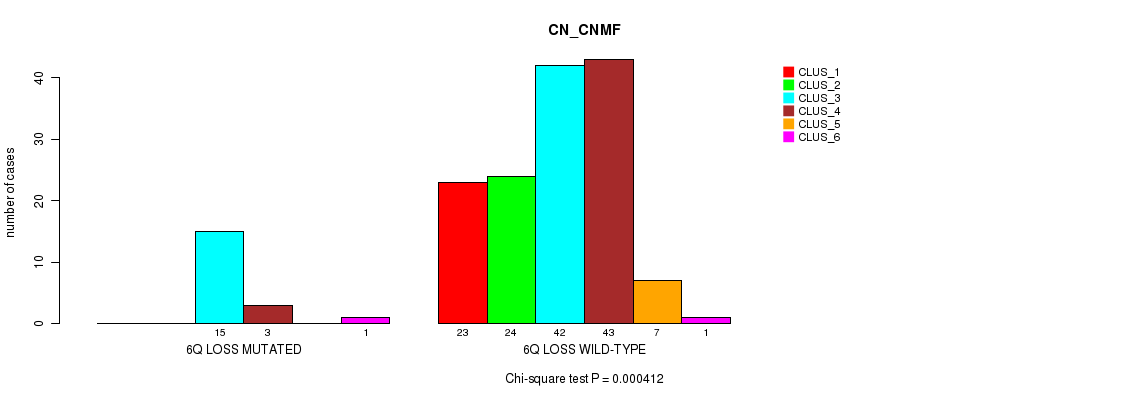
P value = 0.000536 (Fisher's exact test), Q value = 0.2
Table S10. Gene #46: '6q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 40 | 77 | 42 |
| 6Q LOSS MUTATED | 1 | 17 | 1 |
| 6Q LOSS WILD-TYPE | 39 | 60 | 41 |
Figure S10. Get High-res Image Gene #46: '6q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
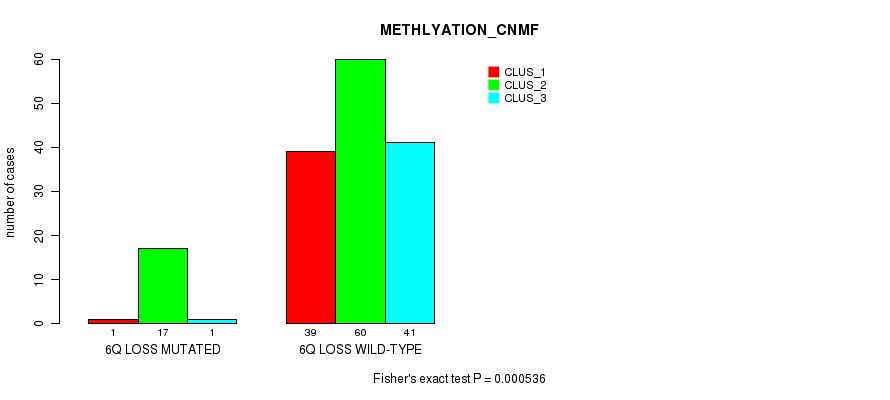
P value = 0.00044 (Fisher's exact test), Q value = 0.16
Table S11. Gene #46: '6q loss' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 53 | 24 |
| 6Q LOSS MUTATED | 0 | 11 | 2 |
| 6Q LOSS WILD-TYPE | 54 | 42 | 22 |
Figure S11. Get High-res Image Gene #46: '6q loss' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
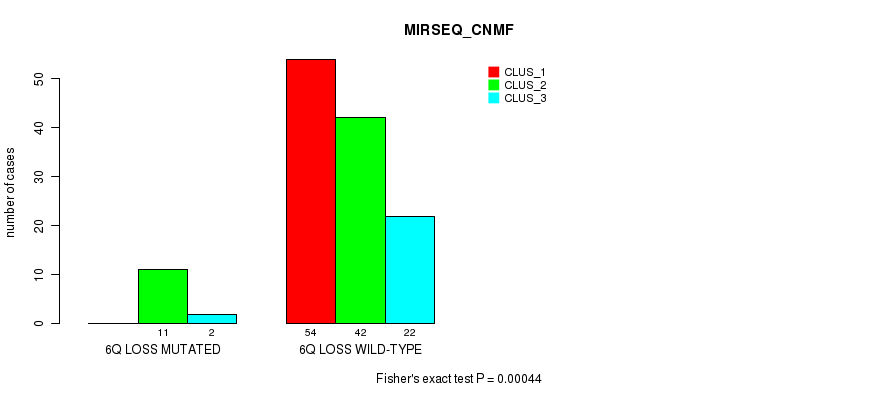
P value = 9.39e-06 (Fisher's exact test), Q value = 0.0035
Table S12. Gene #46: '6q loss' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 41 | 18 | 52 |
| 6Q LOSS MUTATED | 1 | 12 | 0 | 0 |
| 6Q LOSS WILD-TYPE | 19 | 29 | 18 | 52 |
Figure S12. Get High-res Image Gene #46: '6q loss' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
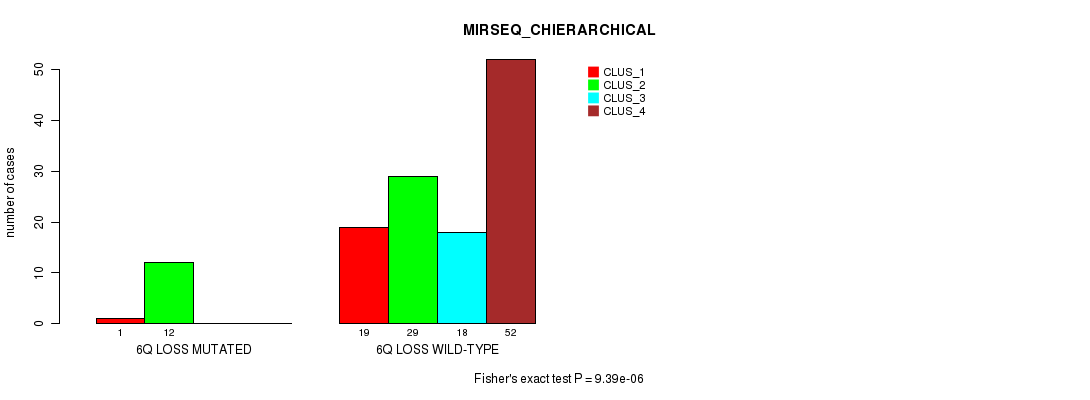
P value = 0.000367 (Fisher's exact test), Q value = 0.14
Table S13. Gene #46: '6q loss' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 55 | 42 |
| 6Q LOSS MUTATED | 0 | 12 | 1 |
| 6Q LOSS WILD-TYPE | 34 | 43 | 41 |
Figure S13. Get High-res Image Gene #46: '6q loss' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
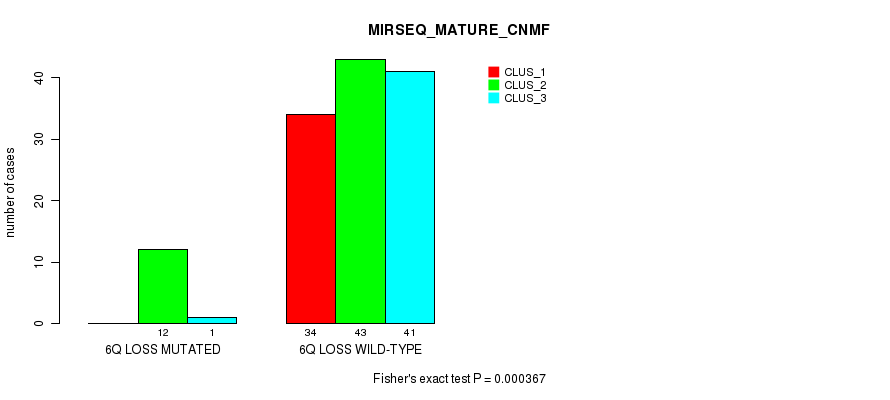
P value = 4.75e-06 (Fisher's exact test), Q value = 0.0018
Table S14. Gene #46: '6q loss' versus Molecular Subtype #6: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 43 | 56 |
| 6Q LOSS MUTATED | 1 | 12 | 0 |
| 6Q LOSS WILD-TYPE | 31 | 31 | 56 |
Figure S14. Get High-res Image Gene #46: '6q loss' versus Molecular Subtype #6: 'MIRSEQ_MATURE_CHIERARCHICAL'
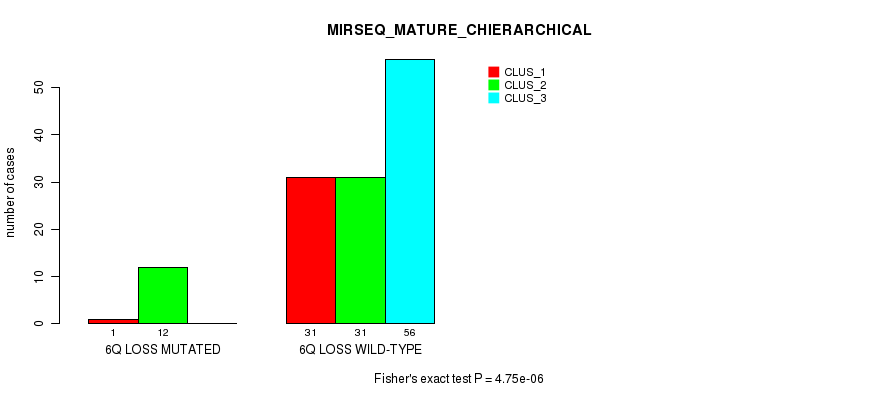
P value = 5.84e-12 (Chi-square test), Q value = 2.3e-09
Table S15. Gene #52: '11p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| 11P LOSS MUTATED | 8 | 5 | 1 | 30 | 6 | 2 |
| 11P LOSS WILD-TYPE | 15 | 19 | 56 | 16 | 1 | 0 |
Figure S15. Get High-res Image Gene #52: '11p loss' versus Molecular Subtype #1: 'CN_CNMF'
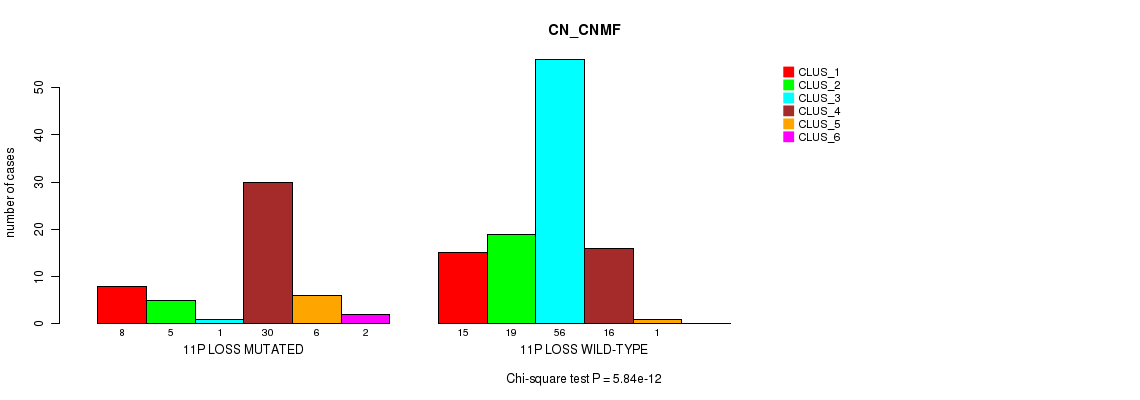
P value = 7.08e-06 (Fisher's exact test), Q value = 0.0027
Table S16. Gene #52: '11p loss' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 41 | 18 | 52 |
| 11P LOSS MUTATED | 6 | 3 | 5 | 29 |
| 11P LOSS WILD-TYPE | 14 | 38 | 13 | 23 |
Figure S16. Get High-res Image Gene #52: '11p loss' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
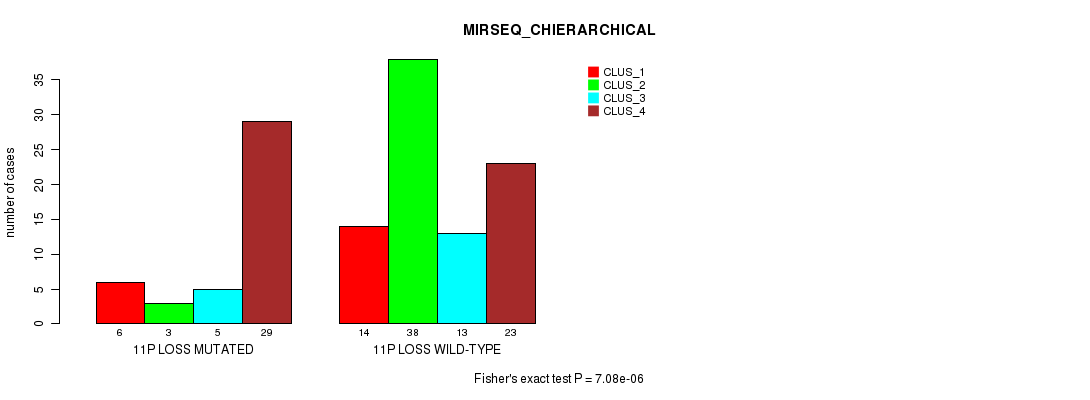
P value = 0.000203 (Fisher's exact test), Q value = 0.076
Table S17. Gene #52: '11p loss' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 55 | 42 |
| 11P LOSS MUTATED | 20 | 9 | 14 |
| 11P LOSS WILD-TYPE | 14 | 46 | 28 |
Figure S17. Get High-res Image Gene #52: '11p loss' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
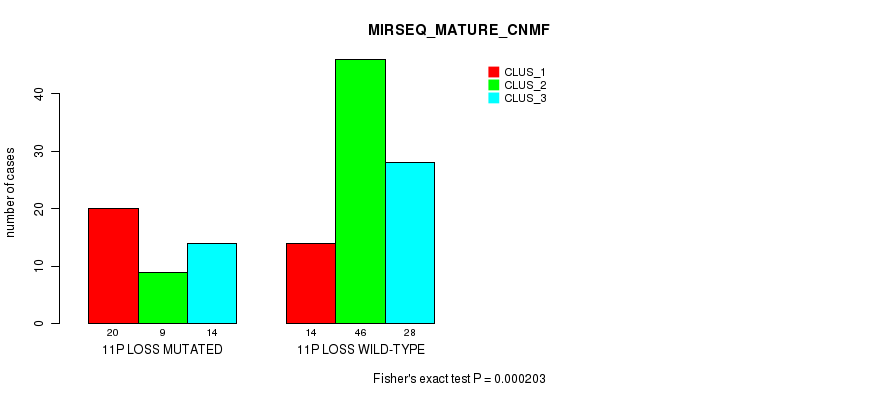
P value = 8.26e-05 (Fisher's exact test), Q value = 0.031
Table S18. Gene #52: '11p loss' versus Molecular Subtype #6: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 43 | 56 |
| 11P LOSS MUTATED | 9 | 5 | 29 |
| 11P LOSS WILD-TYPE | 23 | 38 | 27 |
Figure S18. Get High-res Image Gene #52: '11p loss' versus Molecular Subtype #6: 'MIRSEQ_MATURE_CHIERARCHICAL'
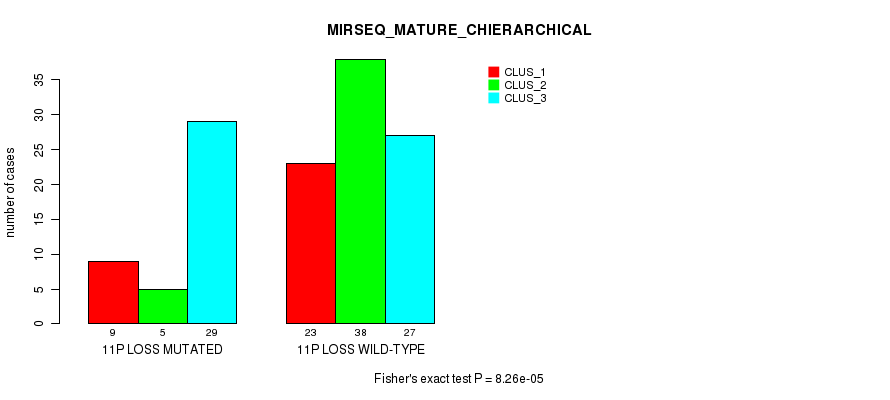
P value = 4.13e-11 (Chi-square test), Q value = 1.6e-08
Table S19. Gene #53: '11q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| 11Q LOSS MUTATED | 5 | 2 | 1 | 29 | 1 | 1 |
| 11Q LOSS WILD-TYPE | 18 | 22 | 56 | 17 | 6 | 1 |
Figure S19. Get High-res Image Gene #53: '11q loss' versus Molecular Subtype #1: 'CN_CNMF'
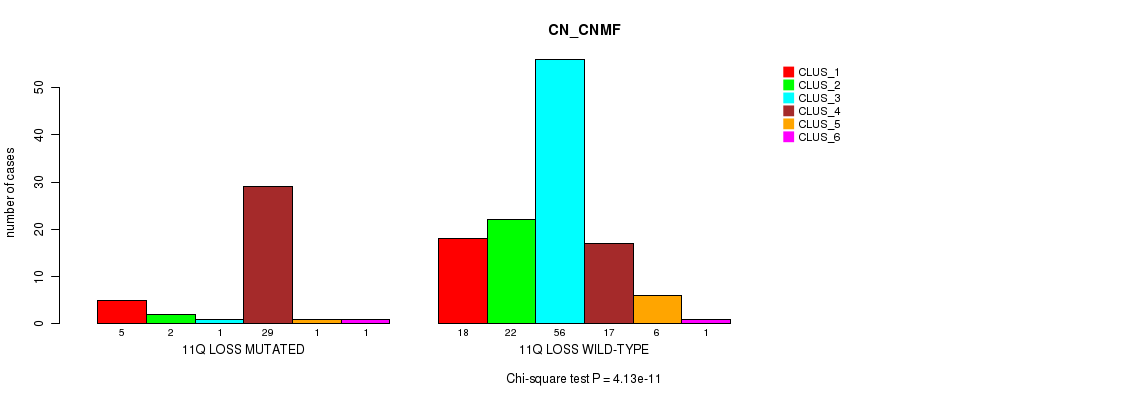
P value = 0.000182 (Fisher's exact test), Q value = 0.068
Table S20. Gene #53: '11q loss' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 55 | 42 |
| 11Q LOSS MUTATED | 18 | 8 | 7 |
| 11Q LOSS WILD-TYPE | 16 | 47 | 35 |
Figure S20. Get High-res Image Gene #53: '11q loss' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
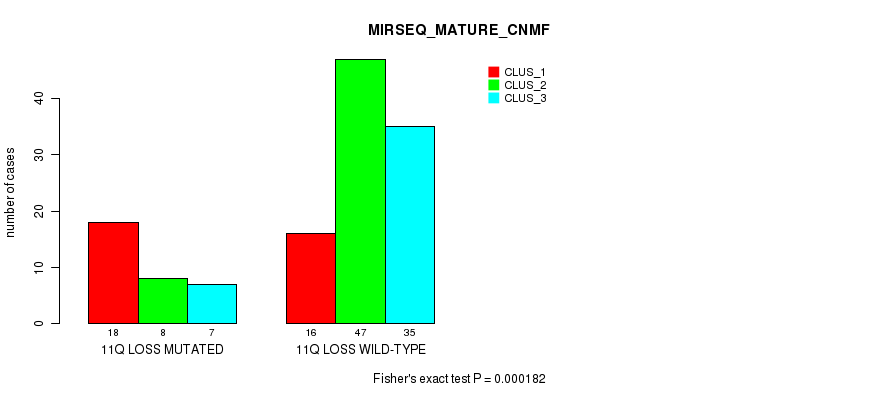
P value = 0.000193 (Chi-square test), Q value = 0.072
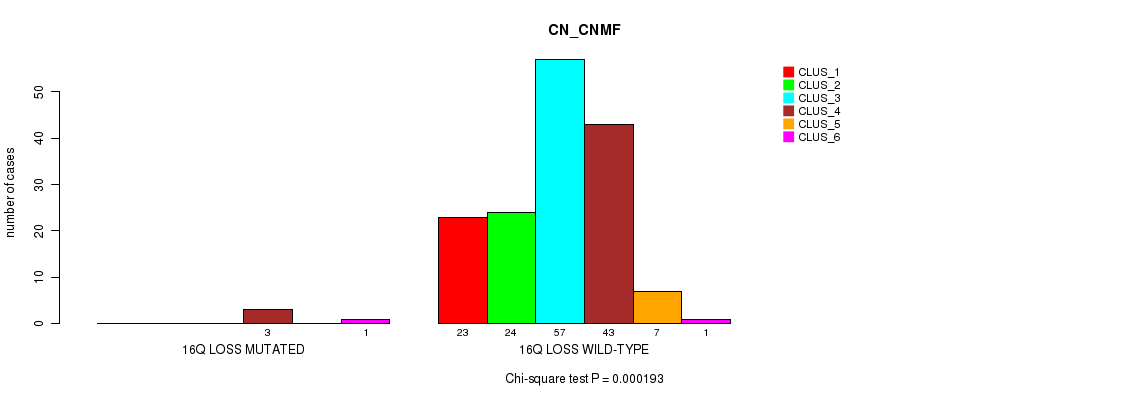
Table S21. Gene #58: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| 16Q LOSS MUTATED | 0 | 0 | 0 | 3 | 0 | 1 |
| 16Q LOSS WILD-TYPE | 23 | 24 | 57 | 43 | 7 | 1 |
Figure S21. Get High-res Image Gene #58: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
-
Copy number data file = transformed.cor.cli.txt
-
Molecular subtypes file = PCPG-TP.transferedmergedcluster.txt
-
Number of patients = 159
-
Number of significantly arm-level cnvs = 67
-
Number of molecular subtypes = 6
-
Exclude genes that fewer than K tumors have mutations, K = 3
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.