This pipeline computes the correlation between significant copy number variation (cnv focal) genes and molecular subtypes.
Testing the association between copy number variation 27 focal events and 6 molecular subtypes across 159 patients, 29 significant findings detected with P value < 0.05 and Q value < 0.25.
-
amp_1q21.3 cnv correlated to 'CN_CNMF'.
-
amp_14q24.3 cnv correlated to 'CN_CNMF'.
-
del_1p36.32 cnv correlated to 'CN_CNMF' and 'MIRSEQ_MATURE_CNMF'.
-
del_1p12 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_MATURE_CNMF'.
-
del_3p24.1 cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
del_3q26.1 cnv correlated to 'CN_CNMF' and 'MIRSEQ_CHIERARCHICAL'.
-
del_11p15.4 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
del_11q22.1 cnv correlated to 'CN_CNMF' and 'MIRSEQ_MATURE_CNMF'.
-
del_12q21.32 cnv correlated to 'CN_CNMF'.
-
del_16q21 cnv correlated to 'CN_CNMF'.
-
del_17p13.2 cnv correlated to 'METHLYATION_CNMF' and 'MIRSEQ_CNMF'.
-
del_17q11.2 cnv correlated to 'MIRSEQ_MATURE_CNMF'.
-
del_22q12.3 cnv correlated to 'MIRSEQ_MATURE_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 27 focal events and 6 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 29 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Chi-square test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| del 11p15 4 | 56 (35%) | 103 |
3.96e-15 (6.18e-13) |
3.07e-05 (0.00449) |
0.000981 (0.131) |
1.3e-06 (0.000195) |
2.43e-06 (0.000363) |
5.62e-05 (0.00809) |
| del 3p24 1 | 60 (38%) | 99 |
2.93e-09 (4.48e-07) |
0.00353 (0.434) |
7.42e-07 (0.000112) |
9.23e-05 (0.0132) |
1.19e-05 (0.00175) |
0.0006 (0.0823) |
| del 1p12 | 111 (70%) | 48 |
6.7e-13 (1.04e-10) |
9.17e-06 (0.00136) |
0.000344 (0.0485) |
0.00454 (0.536) |
0.000848 (0.115) |
0.00429 (0.51) |
| del 1p36 32 | 101 (64%) | 58 |
2.46e-17 (3.86e-15) |
0.00951 (0.989) |
0.102 (1.00) |
0.00358 (0.436) |
0.00156 (0.203) |
0.0109 (1.00) |
| del 3q26 1 | 93 (58%) | 66 |
2.49e-08 (3.79e-06) |
0.647 (1.00) |
0.14 (1.00) |
0.000844 (0.115) |
0.251 (1.00) |
0.0147 (1.00) |
| del 11q22 1 | 44 (28%) | 115 |
5.56e-11 (8.56e-09) |
0.201 (1.00) |
0.108 (1.00) |
0.0111 (1.00) |
0.000574 (0.0792) |
0.00995 (1.00) |
| del 17p13 2 | 64 (40%) | 95 |
0.0205 (1.00) |
0.000461 (0.064) |
0.00131 (0.173) |
0.00754 (0.822) |
0.00589 (0.677) |
0.0133 (1.00) |
| amp 1q21 3 | 30 (19%) | 129 |
1.82e-18 (2.88e-16) |
0.621 (1.00) |
1 (1.00) |
0.28 (1.00) |
0.286 (1.00) |
0.495 (1.00) |
| amp 14q24 3 | 10 (6%) | 149 |
0.00155 (0.203) |
0.198 (1.00) |
0.759 (1.00) |
0.722 (1.00) |
0.0249 (1.00) |
0.593 (1.00) |
| del 12q21 32 | 3 (2%) | 156 |
5.33e-05 (0.00773) |
0.438 (1.00) |
||||
| del 16q21 | 5 (3%) | 154 |
0.00039 (0.0545) |
0.115 (1.00) |
0.524 (1.00) |
0.289 (1.00) |
0.292 (1.00) |
0.359 (1.00) |
| del 17q11 2 | 42 (26%) | 117 |
0.117 (1.00) |
0.00349 (0.433) |
0.00214 (0.274) |
0.00328 (0.41) |
0.00017 (0.0241) |
0.00215 (0.274) |
| del 22q12 3 | 59 (37%) | 100 |
0.0376 (1.00) |
0.00888 (0.932) |
0.00773 (0.835) |
0.0195 (1.00) |
0.00815 (0.864) |
0.000931 (0.125) |
| amp 4q31 1 | 12 (8%) | 147 |
0.0901 (1.00) |
0.0838 (1.00) |
0.819 (1.00) |
0.00369 (0.446) |
0.192 (1.00) |
0.689 (1.00) |
| amp 12q13 3 | 16 (10%) | 143 |
0.0378 (1.00) |
0.132 (1.00) |
0.125 (1.00) |
0.149 (1.00) |
0.00802 (0.858) |
0.12 (1.00) |
| amp 17q21 31 | 15 (9%) | 144 |
0.103 (1.00) |
0.828 (1.00) |
0.675 (1.00) |
0.00632 (0.714) |
0.376 (1.00) |
0.583 (1.00) |
| del 1q44 | 21 (13%) | 138 |
0.0773 (1.00) |
0.332 (1.00) |
1 (1.00) |
0.906 (1.00) |
0.0777 (1.00) |
0.253 (1.00) |
| del 4q22 1 | 14 (9%) | 145 |
0.0306 (1.00) |
0.356 (1.00) |
0.23 (1.00) |
0.507 (1.00) |
0.0262 (1.00) |
0.47 (1.00) |
| del 4q32 3 | 14 (9%) | 145 |
0.0506 (1.00) |
0.165 (1.00) |
0.44 (1.00) |
0.697 (1.00) |
0.156 (1.00) |
0.161 (1.00) |
| del 5q23 1 | 11 (7%) | 148 |
0.00621 (0.708) |
0.922 (1.00) |
0.45 (1.00) |
0.185 (1.00) |
0.373 (1.00) |
0.723 (1.00) |
| del 6p12 3 | 8 (5%) | 151 |
0.0724 (1.00) |
0.288 (1.00) |
0.2 (1.00) |
0.0392 (1.00) |
0.705 (1.00) |
0.161 (1.00) |
| del 6q16 1 | 23 (14%) | 136 |
0.0176 (1.00) |
0.061 (1.00) |
0.0157 (1.00) |
0.00209 (0.27) |
0.0469 (1.00) |
0.00498 (0.577) |
| del 8p22 | 24 (15%) | 135 |
0.242 (1.00) |
0.131 (1.00) |
0.236 (1.00) |
0.165 (1.00) |
0.119 (1.00) |
0.0448 (1.00) |
| del 9p24 2 | 15 (9%) | 144 |
0.0256 (1.00) |
0.363 (1.00) |
1 (1.00) |
0.0107 (1.00) |
0.585 (1.00) |
0.744 (1.00) |
| del 9q21 12 | 14 (9%) | 145 |
0.0546 (1.00) |
0.628 (1.00) |
0.296 (1.00) |
0.167 (1.00) |
0.616 (1.00) |
0.567 (1.00) |
| del 22q13 31 | 64 (40%) | 95 |
0.0249 (1.00) |
0.0109 (1.00) |
0.0164 (1.00) |
0.0355 (1.00) |
0.0134 (1.00) |
0.003 (0.378) |
| del xp21 1 | 46 (29%) | 113 |
0.00677 (0.759) |
0.00706 (0.783) |
0.00368 (0.446) |
0.0456 (1.00) |
0.0046 (0.538) |
0.0073 (0.803) |
P value = 1.82e-18 (Chi-square test), Q value = 2.9e-16
Table S1. Gene #1: 'amp_1q21.3' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| AMP PEAK 1(1Q21.3) MUTATED | 21 | 1 | 5 | 3 | 0 | 0 |
| AMP PEAK 1(1Q21.3) WILD-TYPE | 2 | 23 | 52 | 43 | 7 | 2 |
Figure S1. Get High-res Image Gene #1: 'amp_1q21.3' versus Molecular Subtype #1: 'CN_CNMF'
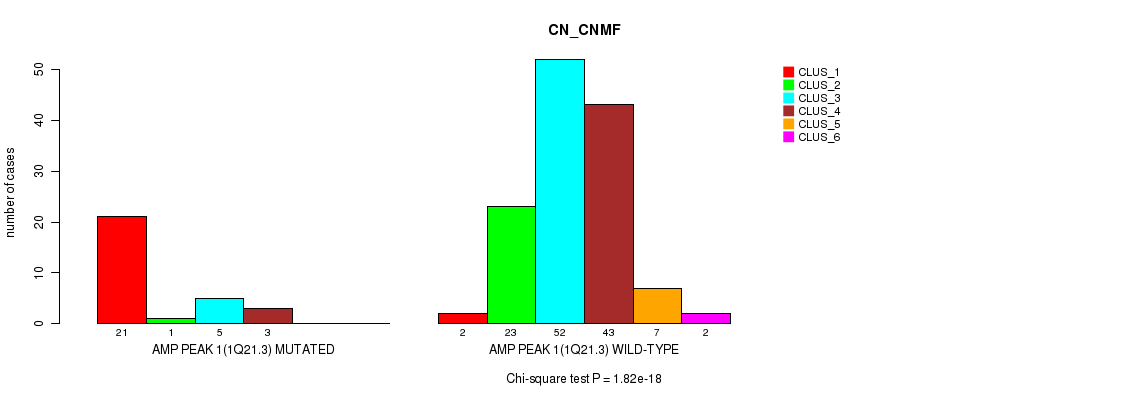
P value = 0.00155 (Chi-square test), Q value = 0.2
Table S2. Gene #4: 'amp_14q24.3' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| AMP PEAK 4(14Q24.3) MUTATED | 0 | 0 | 1 | 9 | 0 | 0 |
| AMP PEAK 4(14Q24.3) WILD-TYPE | 23 | 24 | 56 | 37 | 7 | 2 |
Figure S2. Get High-res Image Gene #4: 'amp_14q24.3' versus Molecular Subtype #1: 'CN_CNMF'
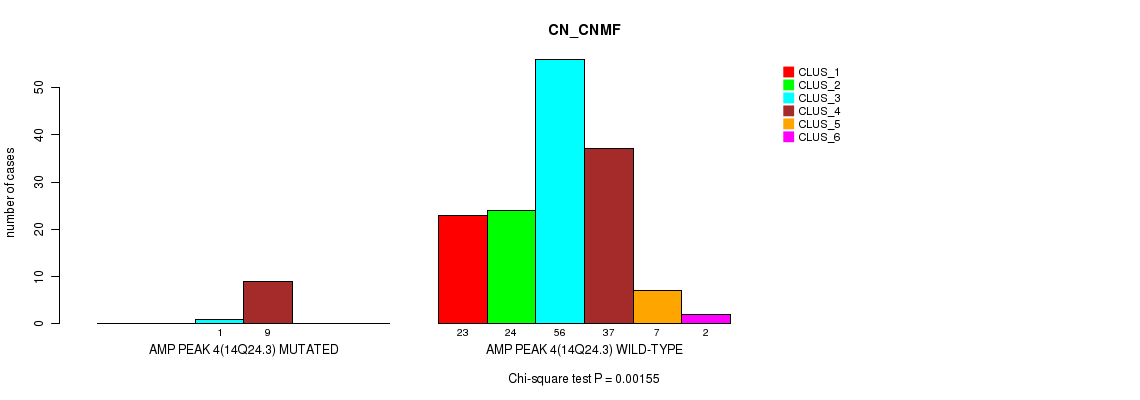
P value = 2.46e-17 (Chi-square test), Q value = 3.9e-15
Table S3. Gene #6: 'del_1p36.32' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| DEL PEAK 1(1P36.32) MUTATED | 21 | 2 | 54 | 15 | 7 | 2 |
| DEL PEAK 1(1P36.32) WILD-TYPE | 2 | 22 | 3 | 31 | 0 | 0 |
Figure S3. Get High-res Image Gene #6: 'del_1p36.32' versus Molecular Subtype #1: 'CN_CNMF'
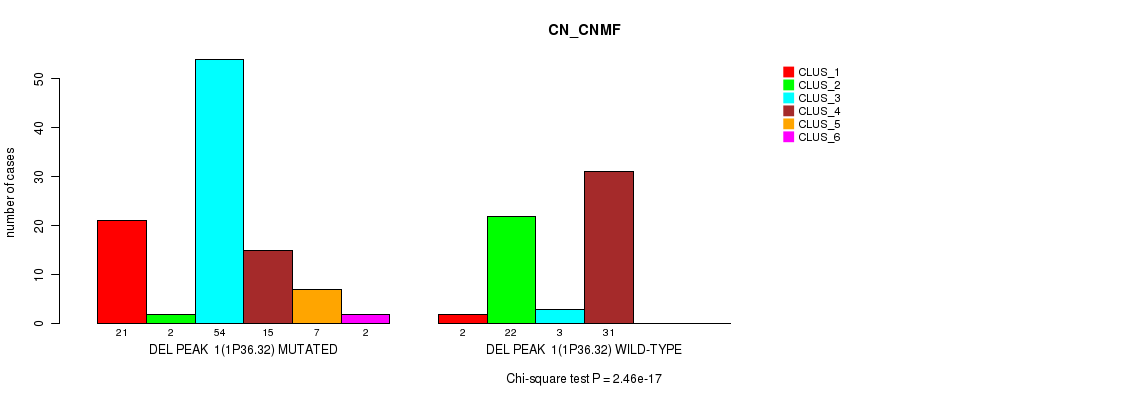
P value = 0.00156 (Fisher's exact test), Q value = 0.2
Table S4. Gene #6: 'del_1p36.32' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 55 | 42 |
| DEL PEAK 1(1P36.32) MUTATED | 12 | 40 | 28 |
| DEL PEAK 1(1P36.32) WILD-TYPE | 22 | 15 | 14 |
Figure S4. Get High-res Image Gene #6: 'del_1p36.32' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
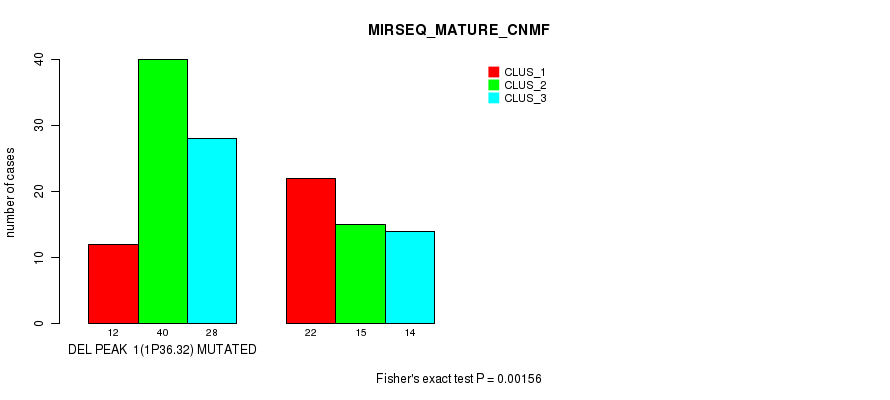
P value = 6.7e-13 (Chi-square test), Q value = 1e-10
Table S5. Gene #7: 'del_1p12' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| DEL PEAK 2(1P12) MUTATED | 21 | 3 | 53 | 25 | 7 | 2 |
| DEL PEAK 2(1P12) WILD-TYPE | 2 | 21 | 4 | 21 | 0 | 0 |
Figure S5. Get High-res Image Gene #7: 'del_1p12' versus Molecular Subtype #1: 'CN_CNMF'
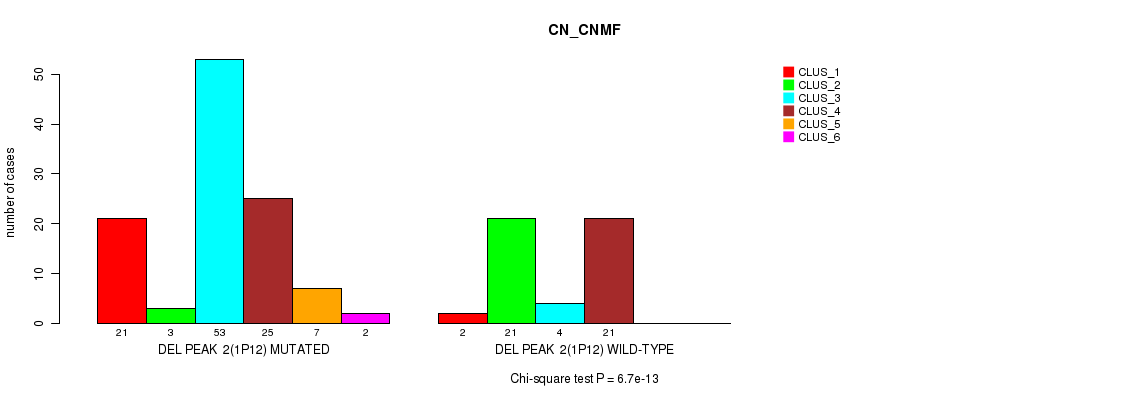
P value = 9.17e-06 (Fisher's exact test), Q value = 0.0014
Table S6. Gene #7: 'del_1p12' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 40 | 77 | 42 |
| DEL PEAK 2(1P12) MUTATED | 24 | 67 | 20 |
| DEL PEAK 2(1P12) WILD-TYPE | 16 | 10 | 22 |
Figure S6. Get High-res Image Gene #7: 'del_1p12' versus Molecular Subtype #2: 'METHLYATION_CNMF'
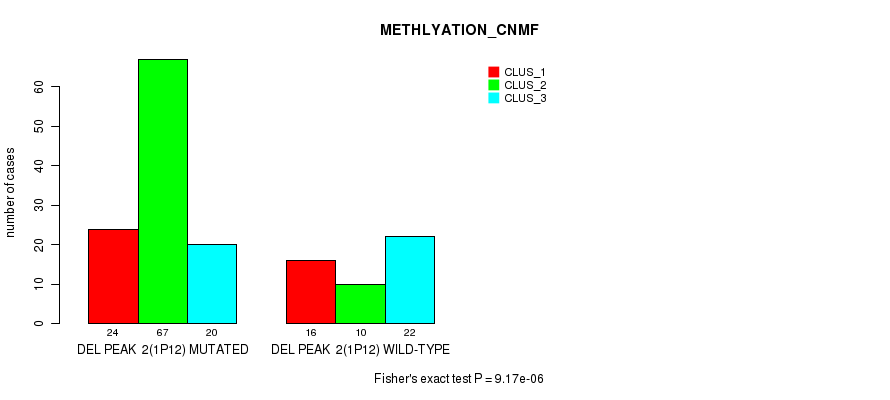
P value = 0.000344 (Fisher's exact test), Q value = 0.048
Table S7. Gene #7: 'del_1p12' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 53 | 24 |
| DEL PEAK 2(1P12) MUTATED | 31 | 46 | 12 |
| DEL PEAK 2(1P12) WILD-TYPE | 23 | 7 | 12 |
Figure S7. Get High-res Image Gene #7: 'del_1p12' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
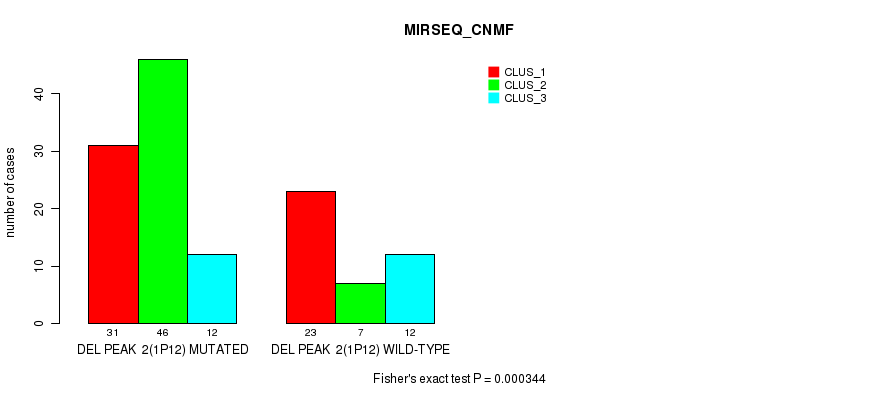
P value = 0.000848 (Fisher's exact test), Q value = 0.11
Table S8. Gene #7: 'del_1p12' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 55 | 42 |
| DEL PEAK 2(1P12) MUTATED | 18 | 47 | 24 |
| DEL PEAK 2(1P12) WILD-TYPE | 16 | 8 | 18 |
Figure S8. Get High-res Image Gene #7: 'del_1p12' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
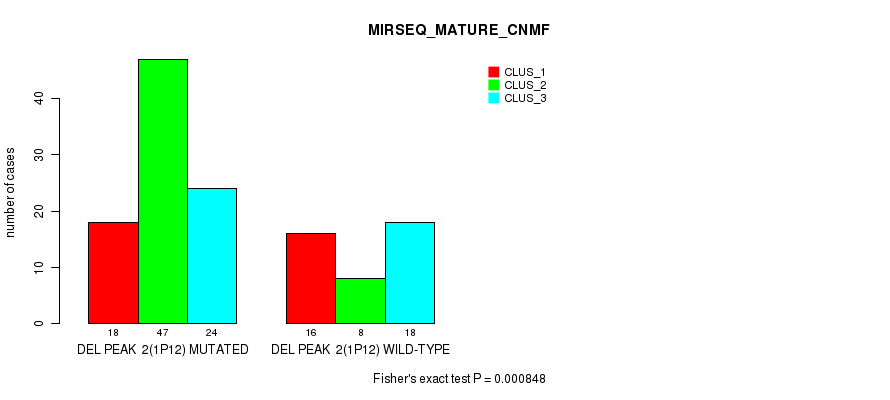
P value = 2.93e-09 (Chi-square test), Q value = 4.5e-07
Table S9. Gene #9: 'del_3p24.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| DEL PEAK 4(3P24.1) MUTATED | 12 | 1 | 10 | 33 | 4 | 0 |
| DEL PEAK 4(3P24.1) WILD-TYPE | 11 | 23 | 47 | 13 | 3 | 2 |
Figure S9. Get High-res Image Gene #9: 'del_3p24.1' versus Molecular Subtype #1: 'CN_CNMF'
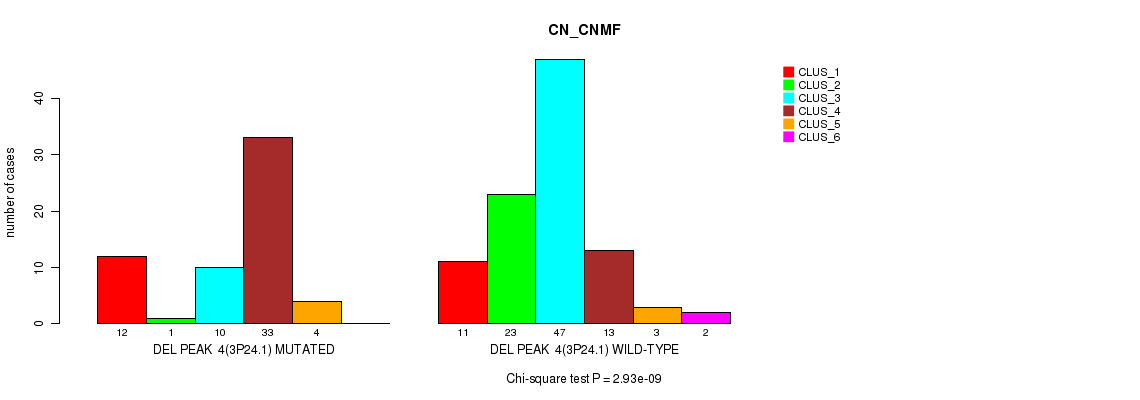
P value = 7.42e-07 (Fisher's exact test), Q value = 0.00011
Table S10. Gene #9: 'del_3p24.1' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 53 | 24 |
| DEL PEAK 4(3P24.1) MUTATED | 34 | 11 | 3 |
| DEL PEAK 4(3P24.1) WILD-TYPE | 20 | 42 | 21 |
Figure S10. Get High-res Image Gene #9: 'del_3p24.1' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
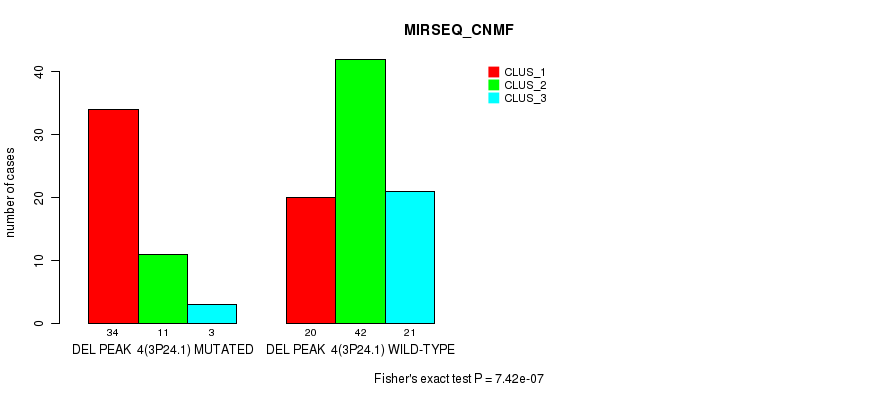
P value = 9.23e-05 (Fisher's exact test), Q value = 0.013
Table S11. Gene #9: 'del_3p24.1' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 41 | 18 | 52 |
| DEL PEAK 4(3P24.1) MUTATED | 2 | 11 | 4 | 31 |
| DEL PEAK 4(3P24.1) WILD-TYPE | 18 | 30 | 14 | 21 |
Figure S11. Get High-res Image Gene #9: 'del_3p24.1' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
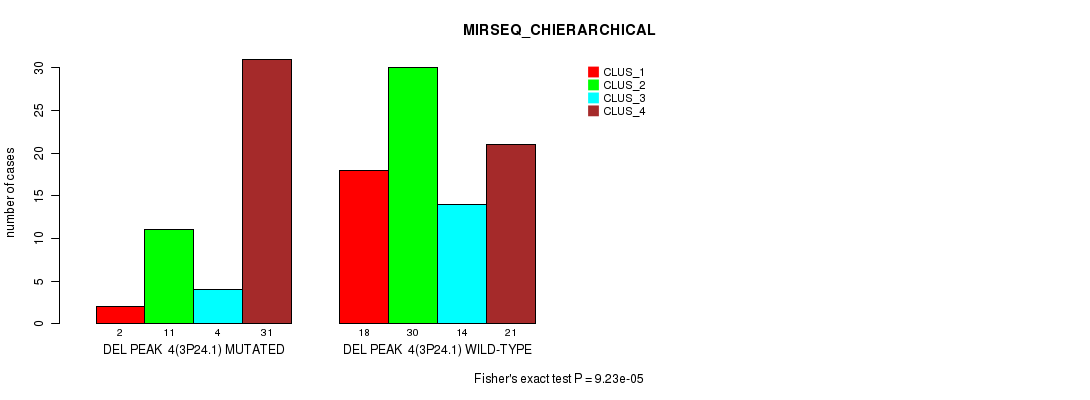
P value = 1.19e-05 (Fisher's exact test), Q value = 0.0017
Table S12. Gene #9: 'del_3p24.1' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 55 | 42 |
| DEL PEAK 4(3P24.1) MUTATED | 24 | 12 | 12 |
| DEL PEAK 4(3P24.1) WILD-TYPE | 10 | 43 | 30 |
Figure S12. Get High-res Image Gene #9: 'del_3p24.1' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
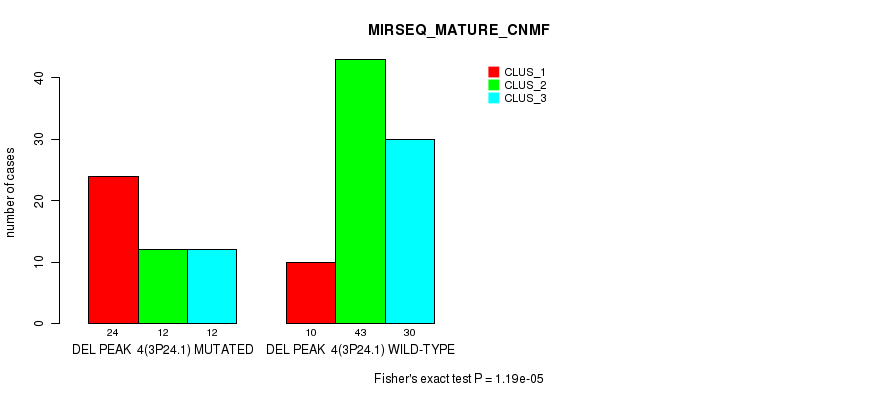
P value = 6e-04 (Fisher's exact test), Q value = 0.082
Table S13. Gene #9: 'del_3p24.1' versus Molecular Subtype #6: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 43 | 56 |
| DEL PEAK 4(3P24.1) MUTATED | 6 | 11 | 31 |
| DEL PEAK 4(3P24.1) WILD-TYPE | 26 | 32 | 25 |
Figure S13. Get High-res Image Gene #9: 'del_3p24.1' versus Molecular Subtype #6: 'MIRSEQ_MATURE_CHIERARCHICAL'
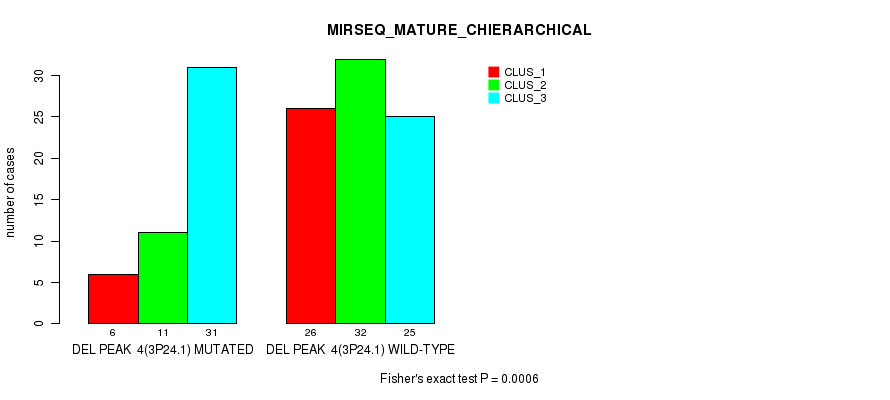
P value = 2.49e-08 (Chi-square test), Q value = 3.8e-06
Table S14. Gene #10: 'del_3q26.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| DEL PEAK 5(3Q26.1) MUTATED | 12 | 0 | 42 | 32 | 5 | 2 |
| DEL PEAK 5(3Q26.1) WILD-TYPE | 11 | 24 | 15 | 14 | 2 | 0 |
Figure S14. Get High-res Image Gene #10: 'del_3q26.1' versus Molecular Subtype #1: 'CN_CNMF'
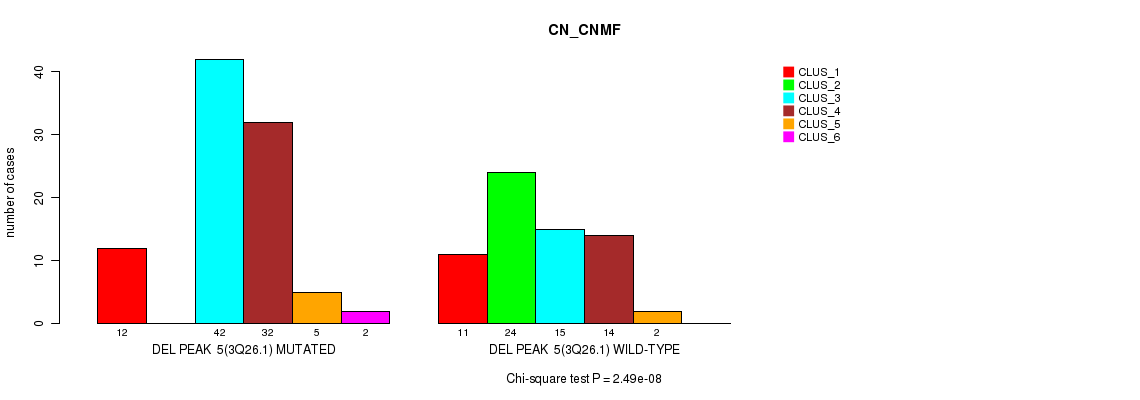
P value = 0.000844 (Fisher's exact test), Q value = 0.11
Table S15. Gene #10: 'del_3q26.1' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 41 | 18 | 52 |
| DEL PEAK 5(3Q26.1) MUTATED | 7 | 29 | 4 | 33 |
| DEL PEAK 5(3Q26.1) WILD-TYPE | 13 | 12 | 14 | 19 |
Figure S15. Get High-res Image Gene #10: 'del_3q26.1' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
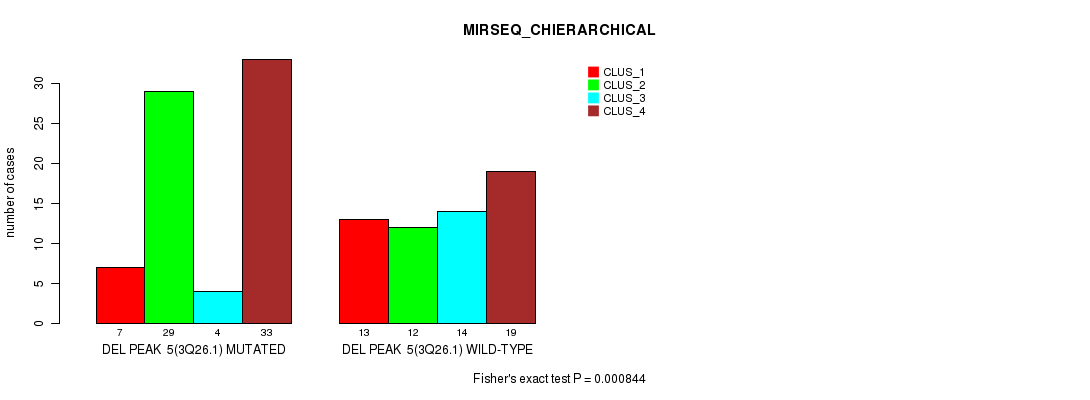
P value = 3.96e-15 (Chi-square test), Q value = 6.2e-13
Table S16. Gene #19: 'del_11p15.4' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| DEL PEAK 14(11P15.4) MUTATED | 8 | 4 | 1 | 35 | 6 | 2 |
| DEL PEAK 14(11P15.4) WILD-TYPE | 15 | 20 | 56 | 11 | 1 | 0 |
Figure S16. Get High-res Image Gene #19: 'del_11p15.4' versus Molecular Subtype #1: 'CN_CNMF'
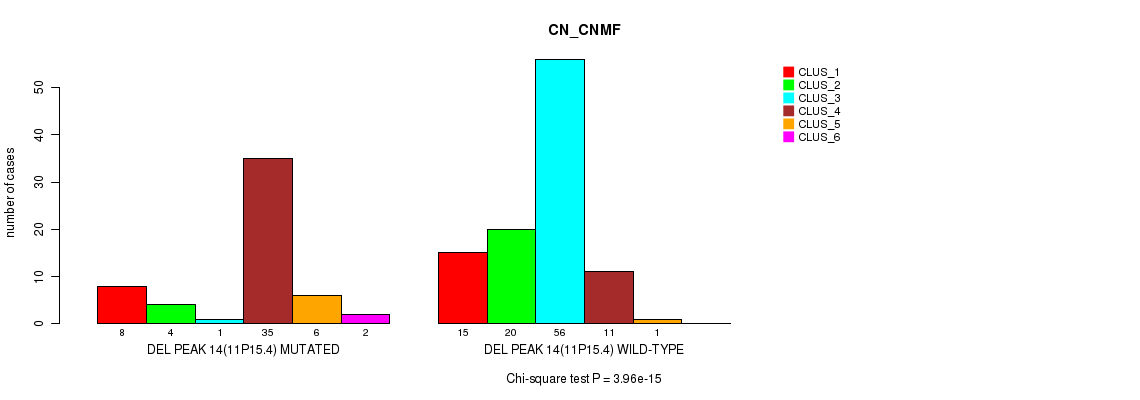
P value = 3.07e-05 (Fisher's exact test), Q value = 0.0045
Table S17. Gene #19: 'del_11p15.4' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 40 | 77 | 42 |
| DEL PEAK 14(11P15.4) MUTATED | 23 | 14 | 19 |
| DEL PEAK 14(11P15.4) WILD-TYPE | 17 | 63 | 23 |
Figure S17. Get High-res Image Gene #19: 'del_11p15.4' versus Molecular Subtype #2: 'METHLYATION_CNMF'
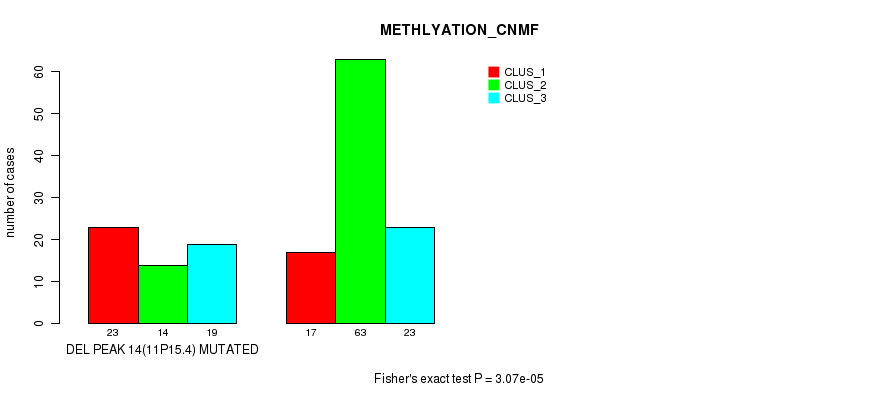
P value = 0.000981 (Fisher's exact test), Q value = 0.13
Table S18. Gene #19: 'del_11p15.4' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 53 | 24 |
| DEL PEAK 14(11P15.4) MUTATED | 29 | 11 | 6 |
| DEL PEAK 14(11P15.4) WILD-TYPE | 25 | 42 | 18 |
Figure S18. Get High-res Image Gene #19: 'del_11p15.4' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
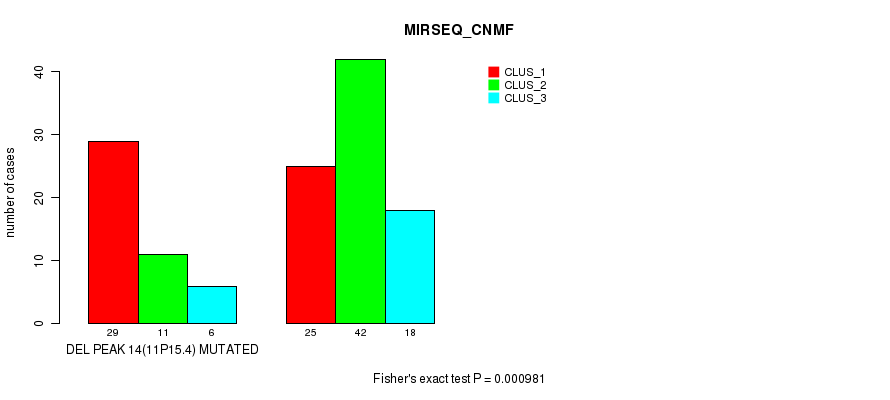
P value = 1.3e-06 (Fisher's exact test), Q value = 0.00019
Table S19. Gene #19: 'del_11p15.4' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 20 | 41 | 18 | 52 |
| DEL PEAK 14(11P15.4) MUTATED | 6 | 4 | 4 | 32 |
| DEL PEAK 14(11P15.4) WILD-TYPE | 14 | 37 | 14 | 20 |
Figure S19. Get High-res Image Gene #19: 'del_11p15.4' versus Molecular Subtype #4: 'MIRSEQ_CHIERARCHICAL'
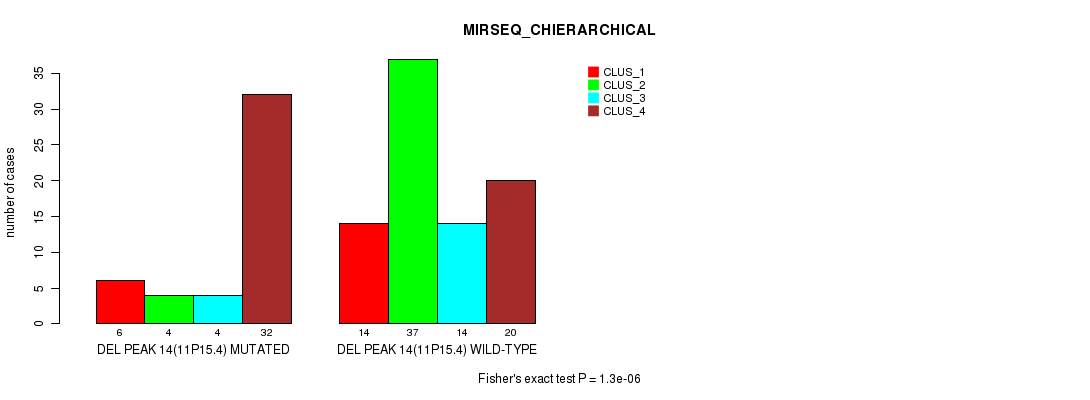
P value = 2.43e-06 (Fisher's exact test), Q value = 0.00036
Table S20. Gene #19: 'del_11p15.4' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 55 | 42 |
| DEL PEAK 14(11P15.4) MUTATED | 24 | 10 | 12 |
| DEL PEAK 14(11P15.4) WILD-TYPE | 10 | 45 | 30 |
Figure S20. Get High-res Image Gene #19: 'del_11p15.4' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
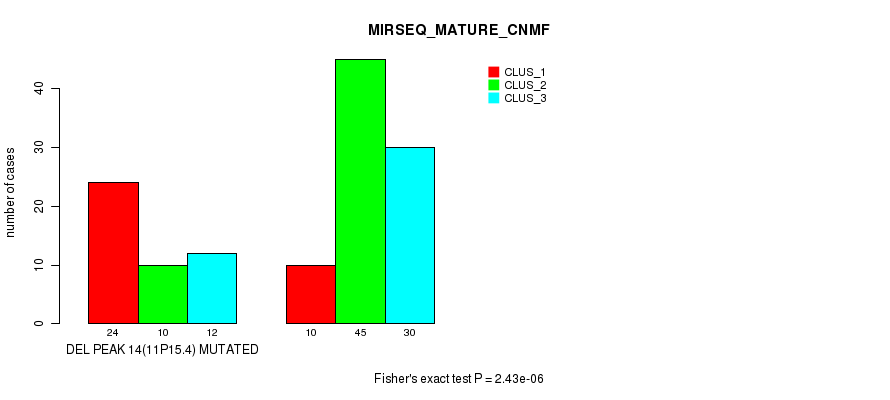
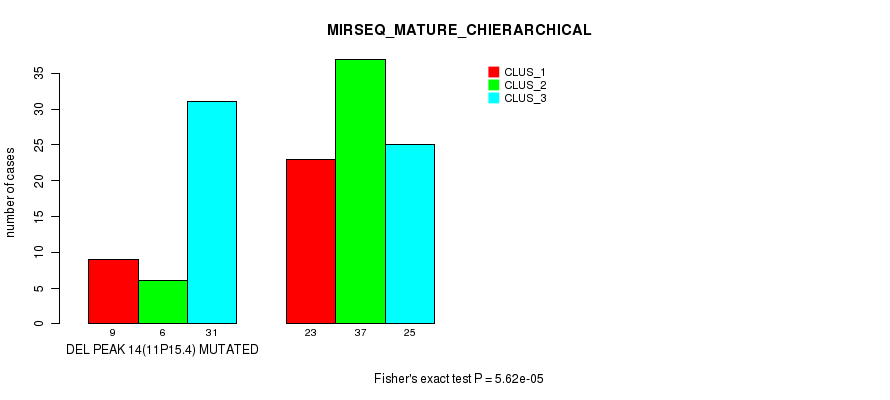
P value = 5.62e-05 (Fisher's exact test), Q value = 0.0081
Table S21. Gene #19: 'del_11p15.4' versus Molecular Subtype #6: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 43 | 56 |
| DEL PEAK 14(11P15.4) MUTATED | 9 | 6 | 31 |
| DEL PEAK 14(11P15.4) WILD-TYPE | 23 | 37 | 25 |
Figure S21. Get High-res Image Gene #19: 'del_11p15.4' versus Molecular Subtype #6: 'MIRSEQ_MATURE_CHIERARCHICAL'
P value = 5.56e-11 (Chi-square test), Q value = 8.6e-09
Table S22. Gene #20: 'del_11q22.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| DEL PEAK 15(11Q22.1) MUTATED | 6 | 3 | 2 | 31 | 1 | 1 |
| DEL PEAK 15(11Q22.1) WILD-TYPE | 17 | 21 | 55 | 15 | 6 | 1 |
Figure S22. Get High-res Image Gene #20: 'del_11q22.1' versus Molecular Subtype #1: 'CN_CNMF'
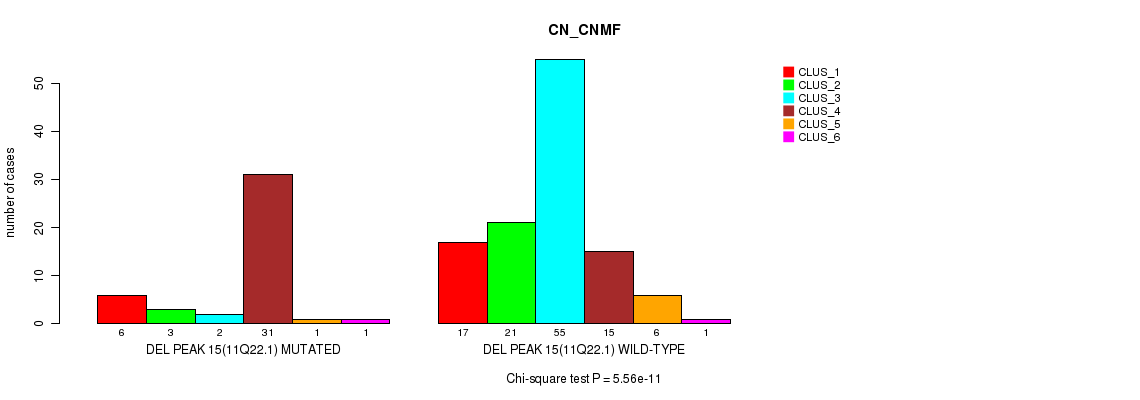
P value = 0.000574 (Fisher's exact test), Q value = 0.079
Table S23. Gene #20: 'del_11q22.1' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 55 | 42 |
| DEL PEAK 15(11Q22.1) MUTATED | 19 | 11 | 8 |
| DEL PEAK 15(11Q22.1) WILD-TYPE | 15 | 44 | 34 |
Figure S23. Get High-res Image Gene #20: 'del_11q22.1' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
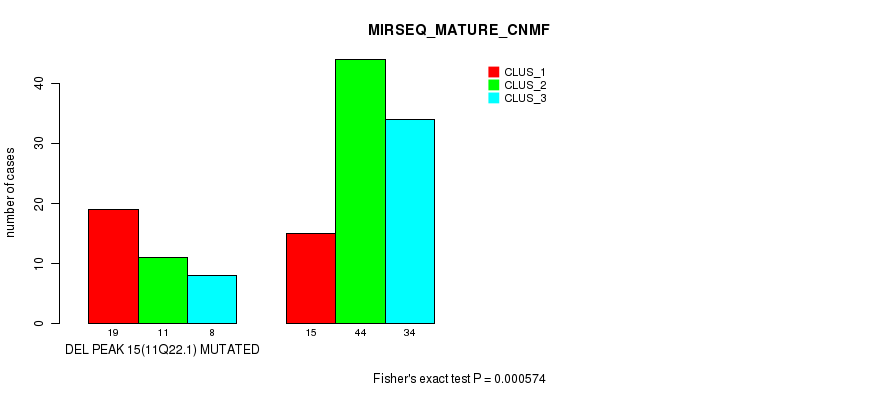
P value = 5.33e-05 (Chi-square test), Q value = 0.0077
Table S24. Gene #21: 'del_12q21.32' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| DEL PEAK 16(12Q21.32) MUTATED | 0 | 1 | 1 | 0 | 0 | 1 |
| DEL PEAK 16(12Q21.32) WILD-TYPE | 23 | 23 | 56 | 46 | 7 | 1 |
Figure S24. Get High-res Image Gene #21: 'del_12q21.32' versus Molecular Subtype #1: 'CN_CNMF'
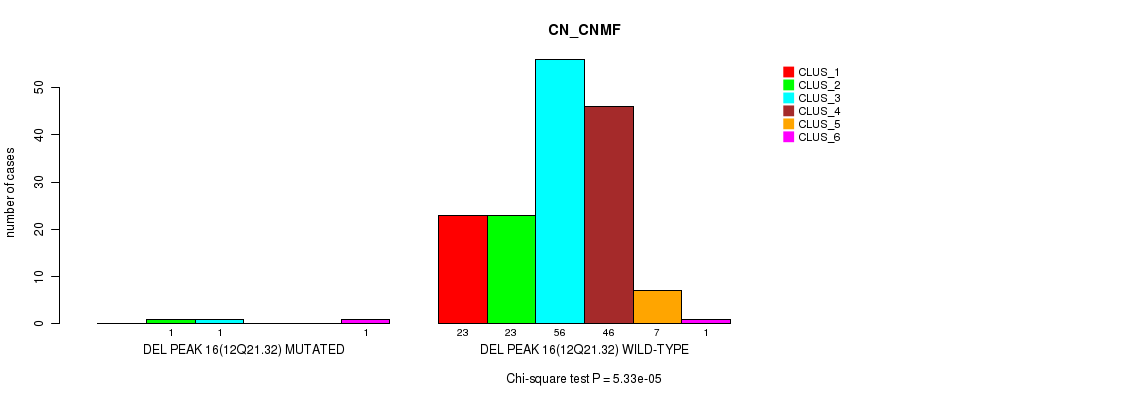
P value = 0.00039 (Chi-square test), Q value = 0.055
Table S25. Gene #22: 'del_16q21' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 23 | 24 | 57 | 46 | 7 | 2 |
| DEL PEAK 17(16Q21) MUTATED | 0 | 0 | 0 | 4 | 0 | 1 |
| DEL PEAK 17(16Q21) WILD-TYPE | 23 | 24 | 57 | 42 | 7 | 1 |
Figure S25. Get High-res Image Gene #22: 'del_16q21' versus Molecular Subtype #1: 'CN_CNMF'
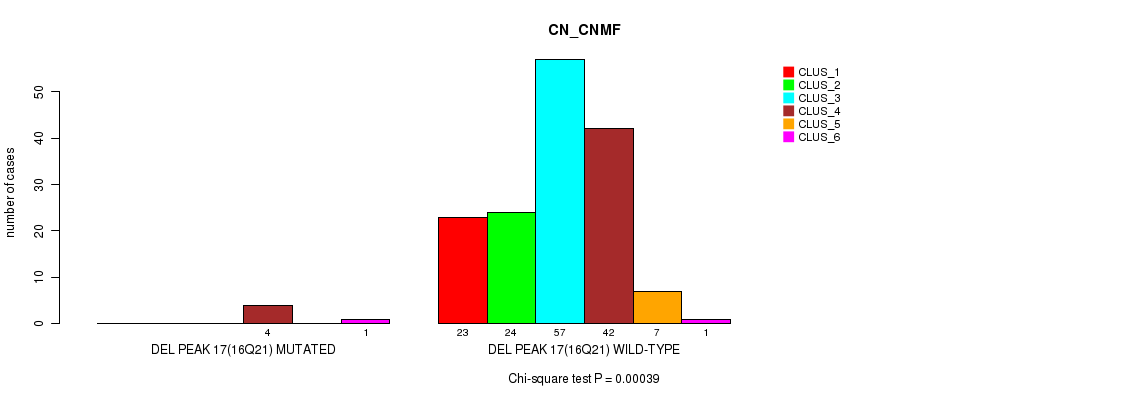
P value = 0.000461 (Fisher's exact test), Q value = 0.064
Table S26. Gene #23: 'del_17p13.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 40 | 77 | 42 |
| DEL PEAK 18(17P13.2) MUTATED | 9 | 43 | 12 |
| DEL PEAK 18(17P13.2) WILD-TYPE | 31 | 34 | 30 |
Figure S26. Get High-res Image Gene #23: 'del_17p13.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
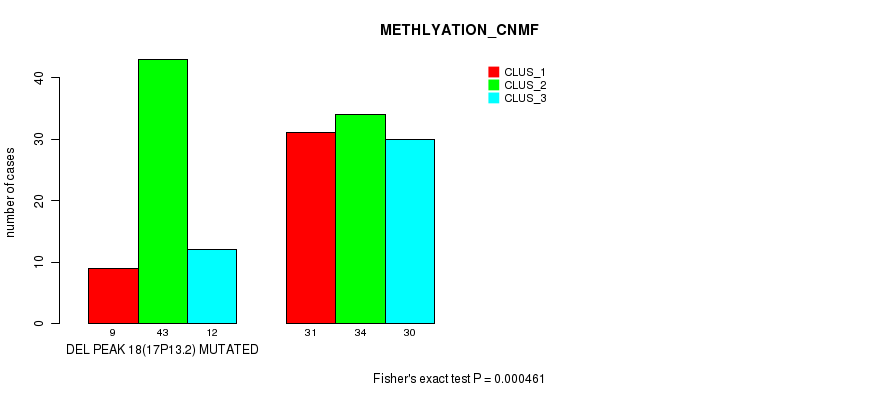
P value = 0.00131 (Fisher's exact test), Q value = 0.17
Table S27. Gene #23: 'del_17p13.2' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 54 | 53 | 24 |
| DEL PEAK 18(17P13.2) MUTATED | 17 | 30 | 4 |
| DEL PEAK 18(17P13.2) WILD-TYPE | 37 | 23 | 20 |
Figure S27. Get High-res Image Gene #23: 'del_17p13.2' versus Molecular Subtype #3: 'MIRSEQ_CNMF'
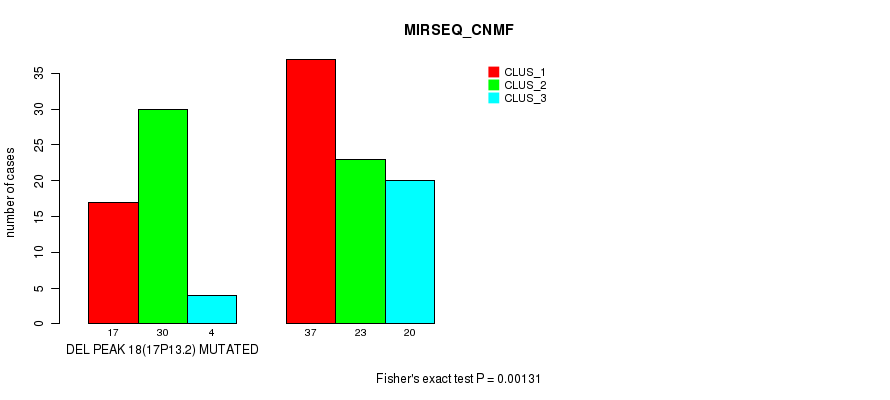
P value = 0.00017 (Fisher's exact test), Q value = 0.024
Table S28. Gene #24: 'del_17q11.2' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 55 | 42 |
| DEL PEAK 19(17Q11.2) MUTATED | 8 | 24 | 3 |
| DEL PEAK 19(17Q11.2) WILD-TYPE | 26 | 31 | 39 |
Figure S28. Get High-res Image Gene #24: 'del_17q11.2' versus Molecular Subtype #5: 'MIRSEQ_MATURE_CNMF'
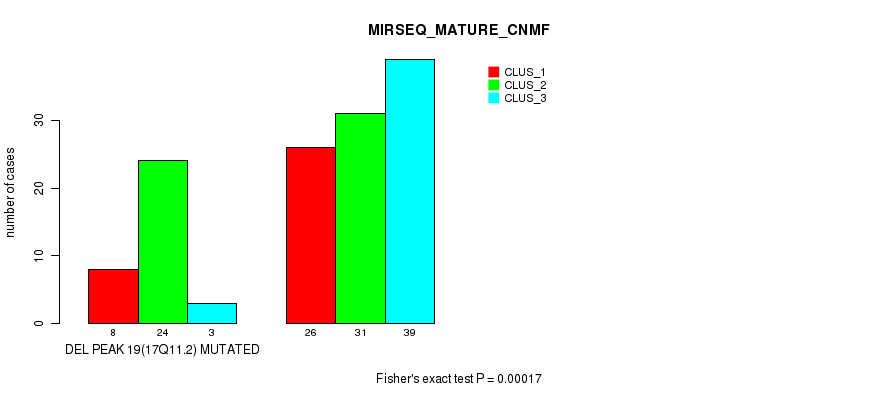
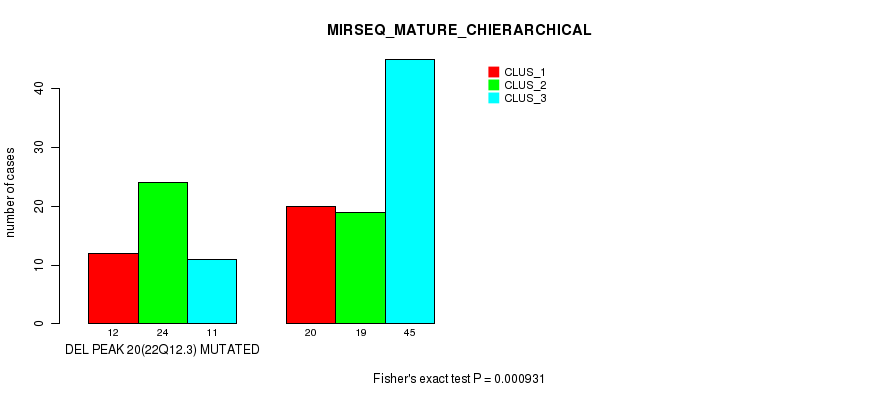
P value = 0.000931 (Fisher's exact test), Q value = 0.12
Table S29. Gene #25: 'del_22q12.3' versus Molecular Subtype #6: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 32 | 43 | 56 |
| DEL PEAK 20(22Q12.3) MUTATED | 12 | 24 | 11 |
| DEL PEAK 20(22Q12.3) WILD-TYPE | 20 | 19 | 45 |
Figure S29. Get High-res Image Gene #25: 'del_22q12.3' versus Molecular Subtype #6: 'MIRSEQ_MATURE_CHIERARCHICAL'
-
Copy number data file = transformed.cor.cli.txt
-
Molecular subtype file = PCPG-TP.transferedmergedcluster.txt
-
Number of patients = 159
-
Number of significantly focal cnvs = 27
-
Number of molecular subtypes = 6
-
Exclude genes that fewer than K tumors have alterations, K = 3
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.