This pipeline computes the correlation between significant copy number variation (cnv focal) genes and molecular subtypes.
Testing the association between copy number variation 47 focal events and 8 molecular subtypes across 102 patients, 46 significant findings detected with P value < 0.05 and Q value < 0.25.
-
amp_7q22.1 cnv correlated to 'CN_CNMF'.
-
amp_8q24.21 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
amp_12p13.33 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
amp_12p12.1 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
amp_12p11.21 cnv correlated to 'CN_CNMF'.
-
amp_18q11.2 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_MATURE_CNMF'.
-
amp_19q13.2 cnv correlated to 'CN_CNMF'.
-
amp_20q13.2 cnv correlated to 'CN_CNMF'.
-
del_1p36.11 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
del_1p21.2 cnv correlated to 'CN_CNMF'.
-
del_5q14.2 cnv correlated to 'CN_CNMF'.
-
del_6p25.2 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
del_7q36.1 cnv correlated to 'CN_CNMF'.
-
del_8p21.3 cnv correlated to 'CN_CNMF'.
-
del_9p21.3 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
del_10q23.2 cnv correlated to 'CN_CNMF'.
-
del_12p12.3 cnv correlated to 'CN_CNMF'.
-
del_15q15.1 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
del_17q22 cnv correlated to 'CN_CNMF'.
-
del_18q21.2 cnv correlated to 'CN_CNMF'.
-
del_19p13.3 cnv correlated to 'CN_CNMF' and 'MIRSEQ_CHIERARCHICAL'.
-
del_21q21.1 cnv correlated to 'CN_CNMF'.
-
del_22q13.31 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 47 focal events and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 46 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| del 9p21 3 | 67 (66%) | 35 |
1e-05 (0.00376) |
2e-05 (0.00726) |
1e-05 (0.00376) |
1e-05 (0.00376) |
2e-05 (0.00726) |
3e-05 (0.0107) |
2e-05 (0.00726) |
3e-05 (0.0107) |
| amp 18q11 2 | 34 (33%) | 68 |
1e-05 (0.00376) |
0.00014 (0.0482) |
0.00032 (0.108) |
0.00456 (1.00) |
0.0001 (0.0347) |
0.00385 (1.00) |
3e-05 (0.0107) |
0.00082 (0.27) |
| amp 12p13 33 | 16 (16%) | 86 |
1e-05 (0.00376) |
0.00018 (0.0616) |
0.188 (1.00) |
0.307 (1.00) |
0.647 (1.00) |
0.00051 (0.17) |
0.0521 (1.00) |
0.0828 (1.00) |
| amp 12p12 1 | 22 (22%) | 80 |
1e-05 (0.00376) |
2e-05 (0.00726) |
0.0115 (1.00) |
0.0341 (1.00) |
0.145 (1.00) |
5e-05 (0.0175) |
0.00204 (0.647) |
0.00658 (1.00) |
| del 1p36 11 | 35 (34%) | 67 |
0.00024 (0.0818) |
0.00052 (0.173) |
6e-05 (0.021) |
0.0302 (1.00) |
0.0142 (1.00) |
0.00415 (1.00) |
0.00503 (1.00) |
0.0114 (1.00) |
| del 22q13 31 | 35 (34%) | 67 |
6e-05 (0.021) |
0.0001 (0.0347) |
0.00729 (1.00) |
0.0232 (1.00) |
0.188 (1.00) |
0.00063 (0.209) |
0.0148 (1.00) |
0.0291 (1.00) |
| amp 8q24 21 | 42 (41%) | 60 |
1e-05 (0.00376) |
0.00028 (0.0949) |
0.079 (1.00) |
0.144 (1.00) |
0.264 (1.00) |
0.00462 (1.00) |
0.0161 (1.00) |
0.0133 (1.00) |
| del 6p25 2 | 42 (41%) | 60 |
2e-05 (0.00726) |
8e-05 (0.0278) |
0.0395 (1.00) |
0.123 (1.00) |
0.1 (1.00) |
0.00592 (1.00) |
0.233 (1.00) |
0.0214 (1.00) |
| del 15q15 1 | 23 (23%) | 79 |
1e-05 (0.00376) |
0.00026 (0.0884) |
0.00461 (1.00) |
0.00898 (1.00) |
0.17 (1.00) |
0.0168 (1.00) |
0.0469 (1.00) |
0.0148 (1.00) |
| del 19p13 3 | 20 (20%) | 82 |
1e-05 (0.00376) |
0.00116 (0.377) |
0.0567 (1.00) |
0.0349 (1.00) |
0.716 (1.00) |
0.00044 (0.147) |
0.0585 (1.00) |
0.199 (1.00) |
| amp 7q22 1 | 29 (28%) | 73 |
1e-05 (0.00376) |
0.0224 (1.00) |
0.0449 (1.00) |
0.147 (1.00) |
0.333 (1.00) |
0.179 (1.00) |
0.00469 (1.00) |
0.0231 (1.00) |
| amp 12p11 21 | 17 (17%) | 85 |
1e-05 (0.00376) |
0.0011 (0.36) |
0.152 (1.00) |
0.261 (1.00) |
0.707 (1.00) |
0.00725 (1.00) |
0.0427 (1.00) |
0.16 (1.00) |
| amp 19q13 2 | 26 (25%) | 76 |
0.00048 (0.16) |
0.00281 (0.885) |
0.159 (1.00) |
0.162 (1.00) |
0.623 (1.00) |
0.182 (1.00) |
0.222 (1.00) |
0.288 (1.00) |
| amp 20q13 2 | 31 (30%) | 71 |
1e-05 (0.00376) |
0.0192 (1.00) |
0.217 (1.00) |
0.615 (1.00) |
0.657 (1.00) |
0.00494 (1.00) |
0.155 (1.00) |
0.0496 (1.00) |
| del 1p21 2 | 23 (23%) | 79 |
2e-05 (0.00726) |
0.00592 (1.00) |
0.0048 (1.00) |
0.084 (1.00) |
0.163 (1.00) |
0.00457 (1.00) |
0.0143 (1.00) |
0.125 (1.00) |
| del 5q14 2 | 24 (24%) | 78 |
3e-05 (0.0107) |
0.0529 (1.00) |
0.108 (1.00) |
0.138 (1.00) |
0.357 (1.00) |
0.0392 (1.00) |
0.0126 (1.00) |
0.187 (1.00) |
| del 7q36 1 | 11 (11%) | 91 |
0.00039 (0.131) |
0.268 (1.00) |
0.835 (1.00) |
0.23 (1.00) |
0.769 (1.00) |
0.0203 (1.00) |
1 (1.00) |
0.826 (1.00) |
| del 8p21 3 | 32 (31%) | 70 |
0.00016 (0.0549) |
0.00176 (0.563) |
0.00584 (1.00) |
0.0728 (1.00) |
0.0961 (1.00) |
0.0013 (0.419) |
0.00744 (1.00) |
0.0374 (1.00) |
| del 10q23 2 | 19 (19%) | 83 |
0.00031 (0.105) |
0.102 (1.00) |
0.0872 (1.00) |
0.0385 (1.00) |
0.677 (1.00) |
0.0278 (1.00) |
0.745 (1.00) |
0.0213 (1.00) |
| del 12p12 3 | 15 (15%) | 87 |
0.0001 (0.0347) |
0.0024 (0.758) |
0.106 (1.00) |
0.11 (1.00) |
0.0418 (1.00) |
0.068 (1.00) |
0.146 (1.00) |
0.0377 (1.00) |
| del 17q22 | 28 (27%) | 74 |
1e-05 (0.00376) |
0.00085 (0.279) |
0.0381 (1.00) |
0.0391 (1.00) |
0.138 (1.00) |
0.00926 (1.00) |
0.00994 (1.00) |
0.0632 (1.00) |
| del 18q21 2 | 73 (72%) | 29 |
4e-05 (0.0141) |
0.0031 (0.973) |
0.00639 (1.00) |
0.0186 (1.00) |
0.0369 (1.00) |
0.0903 (1.00) |
0.0123 (1.00) |
0.0432 (1.00) |
| del 21q21 1 | 40 (39%) | 62 |
3e-05 (0.0107) |
0.0012 (0.389) |
0.0131 (1.00) |
0.0108 (1.00) |
0.0796 (1.00) |
0.00403 (1.00) |
0.0213 (1.00) |
0.0131 (1.00) |
| amp 1p34 2 | 7 (7%) | 95 |
0.656 (1.00) |
0.886 (1.00) |
0.755 (1.00) |
0.417 (1.00) |
0.939 (1.00) |
0.212 (1.00) |
0.562 (1.00) |
0.858 (1.00) |
| amp 1p12 | 16 (16%) | 86 |
0.782 (1.00) |
1 (1.00) |
0.59 (1.00) |
0.979 (1.00) |
0.974 (1.00) |
0.374 (1.00) |
0.748 (1.00) |
0.728 (1.00) |
| amp 6p21 31 | 7 (7%) | 95 |
0.0709 (1.00) |
0.342 (1.00) |
0.303 (1.00) |
0.623 (1.00) |
0.704 (1.00) |
0.0168 (1.00) |
0.133 (1.00) |
0.451 (1.00) |
| amp 9p13 3 | 12 (12%) | 90 |
0.0451 (1.00) |
0.175 (1.00) |
0.11 (1.00) |
0.56 (1.00) |
0.442 (1.00) |
0.152 (1.00) |
0.32 (1.00) |
0.0706 (1.00) |
| amp 11p11 2 | 12 (12%) | 90 |
0.0215 (1.00) |
0.174 (1.00) |
0.454 (1.00) |
0.786 (1.00) |
0.694 (1.00) |
0.196 (1.00) |
0.359 (1.00) |
0.396 (1.00) |
| amp 12q15 | 11 (11%) | 91 |
0.0008 (0.264) |
0.00135 (0.433) |
0.0205 (1.00) |
0.0477 (1.00) |
0.197 (1.00) |
0.00613 (1.00) |
0.00828 (1.00) |
0.0302 (1.00) |
| amp 17p11 2 | 5 (5%) | 97 |
0.906 (1.00) |
0.438 (1.00) |
0.594 (1.00) |
0.872 (1.00) |
0.596 (1.00) |
0.313 (1.00) |
0.122 (1.00) |
0.703 (1.00) |
| amp 17q21 32 | 11 (11%) | 91 |
0.0412 (1.00) |
0.494 (1.00) |
0.267 (1.00) |
0.562 (1.00) |
1 (1.00) |
0.208 (1.00) |
1 (1.00) |
0.643 (1.00) |
| amp xp22 11 | 5 (5%) | 97 |
0.57 (1.00) |
0.843 (1.00) |
1 (1.00) |
0.507 (1.00) |
1 (1.00) |
0.723 (1.00) |
0.848 (1.00) |
0.701 (1.00) |
| amp xq27 1 | 9 (9%) | 93 |
0.106 (1.00) |
1 (1.00) |
0.436 (1.00) |
0.572 (1.00) |
0.92 (1.00) |
0.562 (1.00) |
0.901 (1.00) |
0.521 (1.00) |
| del 1p36 32 | 26 (25%) | 76 |
0.0019 (0.604) |
0.0336 (1.00) |
0.00183 (0.584) |
0.0442 (1.00) |
0.0662 (1.00) |
0.036 (1.00) |
0.0469 (1.00) |
0.102 (1.00) |
| del 1q42 2 | 4 (4%) | 98 |
0.0089 (1.00) |
0.182 (1.00) |
0.807 (1.00) |
0.614 (1.00) |
1 (1.00) |
0.43 (1.00) |
0.357 (1.00) |
0.242 (1.00) |
| del 2p23 3 | 11 (11%) | 91 |
0.032 (1.00) |
0.27 (1.00) |
0.196 (1.00) |
0.131 (1.00) |
0.227 (1.00) |
0.354 (1.00) |
0.322 (1.00) |
0.11 (1.00) |
| del 2q37 3 | 4 (4%) | 98 |
0.0397 (1.00) |
0.18 (1.00) |
0.182 (1.00) |
0.401 (1.00) |
0.446 (1.00) |
0.3 (1.00) |
0.0833 (1.00) |
0.242 (1.00) |
| del 3p24 1 | 22 (22%) | 80 |
0.00115 (0.375) |
0.545 (1.00) |
0.531 (1.00) |
0.686 (1.00) |
0.78 (1.00) |
0.283 (1.00) |
0.337 (1.00) |
0.372 (1.00) |
| del 4q35 2 | 17 (17%) | 85 |
0.392 (1.00) |
0.397 (1.00) |
0.521 (1.00) |
0.78 (1.00) |
1 (1.00) |
0.717 (1.00) |
0.783 (1.00) |
0.585 (1.00) |
| del 8q12 1 | 9 (9%) | 93 |
0.594 (1.00) |
0.732 (1.00) |
0.272 (1.00) |
0.533 (1.00) |
0.728 (1.00) |
0.615 (1.00) |
0.483 (1.00) |
0.47 (1.00) |
| del 9q22 33 | 27 (26%) | 75 |
0.0712 (1.00) |
0.833 (1.00) |
0.438 (1.00) |
0.818 (1.00) |
0.647 (1.00) |
0.324 (1.00) |
0.244 (1.00) |
0.808 (1.00) |
| del 11p15 4 | 18 (18%) | 84 |
0.0319 (1.00) |
1 (1.00) |
0.429 (1.00) |
0.423 (1.00) |
0.632 (1.00) |
0.899 (1.00) |
0.78 (1.00) |
0.82 (1.00) |
| del 12q12 | 11 (11%) | 91 |
0.00126 (0.407) |
0.0111 (1.00) |
0.183 (1.00) |
0.0223 (1.00) |
0.256 (1.00) |
0.0653 (1.00) |
0.268 (1.00) |
0.0297 (1.00) |
| del 12q24 33 | 22 (22%) | 80 |
0.00444 (1.00) |
0.0401 (1.00) |
0.00567 (1.00) |
0.022 (1.00) |
0.382 (1.00) |
0.0273 (1.00) |
0.276 (1.00) |
0.0902 (1.00) |
| del 16q23 1 | 7 (7%) | 95 |
0.13 (1.00) |
0.888 (1.00) |
0.476 (1.00) |
0.526 (1.00) |
0.882 (1.00) |
0.443 (1.00) |
0.661 (1.00) |
1 (1.00) |
| del 18p11 32 | 38 (37%) | 64 |
0.0586 (1.00) |
0.386 (1.00) |
0.893 (1.00) |
0.784 (1.00) |
0.843 (1.00) |
0.404 (1.00) |
0.963 (1.00) |
0.673 (1.00) |
| del 19q13 41 | 11 (11%) | 91 |
0.0414 (1.00) |
0.496 (1.00) |
0.387 (1.00) |
0.265 (1.00) |
0.553 (1.00) |
0.388 (1.00) |
0.915 (1.00) |
0.905 (1.00) |
P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S1. Gene #4: 'amp_7q22.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| AMP PEAK 4(7Q22.1) MUTATED | 14 | 3 | 4 | 8 |
| AMP PEAK 4(7Q22.1) WILD-TYPE | 15 | 43 | 14 | 1 |
Figure S1. Get High-res Image Gene #4: 'amp_7q22.1' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S2. Gene #5: 'amp_8q24.21' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| AMP PEAK 5(8Q24.21) MUTATED | 25 | 8 | 6 | 3 |
| AMP PEAK 5(8Q24.21) WILD-TYPE | 4 | 38 | 12 | 6 |
Figure S2. Get High-res Image Gene #5: 'amp_8q24.21' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00028 (Fisher's exact test), Q value = 0.095
Table S3. Gene #5: 'amp_8q24.21' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 49 | 26 | 26 |
| AMP PEAK 5(8Q24.21) MUTATED | 30 | 6 | 5 |
| AMP PEAK 5(8Q24.21) WILD-TYPE | 19 | 20 | 21 |
Figure S3. Get High-res Image Gene #5: 'amp_8q24.21' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S4. Gene #8: 'amp_12p13.33' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| AMP PEAK 8(12P13.33) MUTATED | 15 | 0 | 0 | 1 |
| AMP PEAK 8(12P13.33) WILD-TYPE | 14 | 46 | 18 | 8 |
Figure S4. Get High-res Image Gene #8: 'amp_12p13.33' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00018 (Fisher's exact test), Q value = 0.062
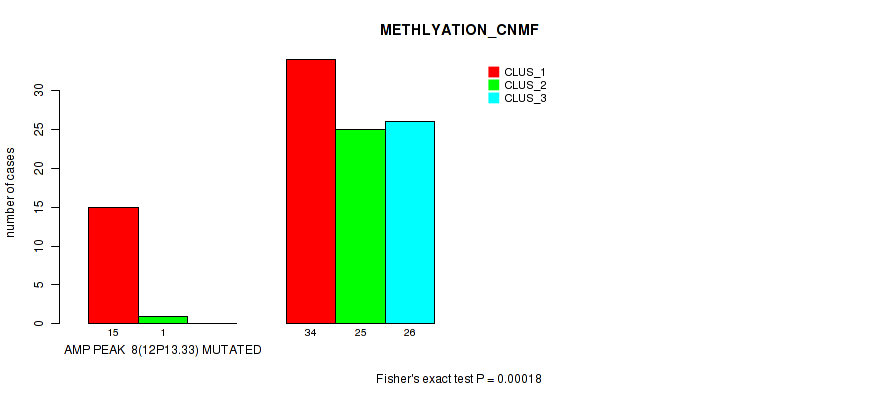
Table S5. Gene #8: 'amp_12p13.33' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 49 | 26 | 26 |
| AMP PEAK 8(12P13.33) MUTATED | 15 | 1 | 0 |
| AMP PEAK 8(12P13.33) WILD-TYPE | 34 | 25 | 26 |
Figure S5. Get High-res Image Gene #8: 'amp_12p13.33' versus Molecular Subtype #2: 'METHLYATION_CNMF'
P value = 0.00051 (Fisher's exact test), Q value = 0.17
Table S6. Gene #8: 'amp_12p13.33' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 17 | 20 | 18 | 8 | 20 | 12 |
| AMP PEAK 8(12P13.33) MUTATED | 7 | 1 | 0 | 4 | 2 | 1 |
| AMP PEAK 8(12P13.33) WILD-TYPE | 10 | 19 | 18 | 4 | 18 | 11 |
Figure S6. Get High-res Image Gene #8: 'amp_12p13.33' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S7. Gene #9: 'amp_12p12.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| AMP PEAK 9(12P12.1) MUTATED | 16 | 0 | 5 | 1 |
| AMP PEAK 9(12P12.1) WILD-TYPE | 13 | 46 | 13 | 8 |
Figure S7. Get High-res Image Gene #9: 'amp_12p12.1' versus Molecular Subtype #1: 'CN_CNMF'

P value = 2e-05 (Fisher's exact test), Q value = 0.0073
Table S8. Gene #9: 'amp_12p12.1' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 49 | 26 | 26 |
| AMP PEAK 9(12P12.1) MUTATED | 20 | 1 | 1 |
| AMP PEAK 9(12P12.1) WILD-TYPE | 29 | 25 | 25 |
Figure S8. Get High-res Image Gene #9: 'amp_12p12.1' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 5e-05 (Fisher's exact test), Q value = 0.018
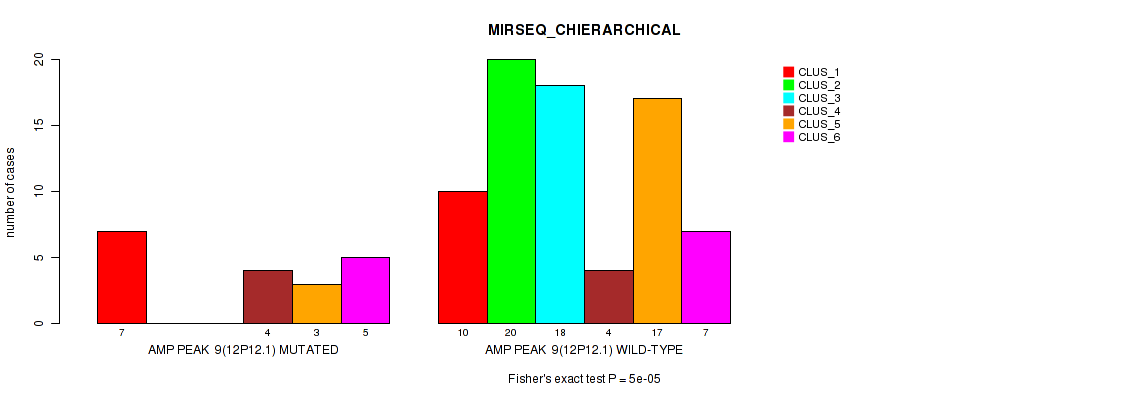
Table S9. Gene #9: 'amp_12p12.1' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 17 | 20 | 18 | 8 | 20 | 12 |
| AMP PEAK 9(12P12.1) MUTATED | 7 | 0 | 0 | 4 | 3 | 5 |
| AMP PEAK 9(12P12.1) WILD-TYPE | 10 | 20 | 18 | 4 | 17 | 7 |
Figure S9. Get High-res Image Gene #9: 'amp_12p12.1' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
P value = 1e-05 (Fisher's exact test), Q value = 0.0038
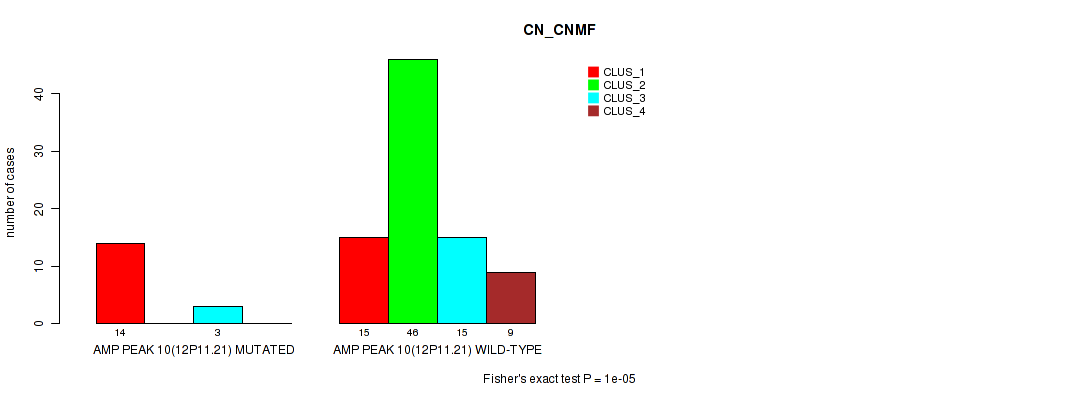
Table S10. Gene #10: 'amp_12p11.21' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| AMP PEAK 10(12P11.21) MUTATED | 14 | 0 | 3 | 0 |
| AMP PEAK 10(12P11.21) WILD-TYPE | 15 | 46 | 15 | 9 |
Figure S10. Get High-res Image Gene #10: 'amp_12p11.21' versus Molecular Subtype #1: 'CN_CNMF'
P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S11. Gene #14: 'amp_18q11.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| AMP PEAK 14(18Q11.2) MUTATED | 11 | 4 | 16 | 3 |
| AMP PEAK 14(18Q11.2) WILD-TYPE | 18 | 42 | 2 | 6 |
Figure S11. Get High-res Image Gene #14: 'amp_18q11.2' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00014 (Fisher's exact test), Q value = 0.048
Table S12. Gene #14: 'amp_18q11.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 49 | 26 | 26 |
| AMP PEAK 14(18Q11.2) MUTATED | 26 | 4 | 3 |
| AMP PEAK 14(18Q11.2) WILD-TYPE | 23 | 22 | 23 |
Figure S12. Get High-res Image Gene #14: 'amp_18q11.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00032 (Fisher's exact test), Q value = 0.11
Table S13. Gene #14: 'amp_18q11.2' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 46 | 26 | 23 |
| AMP PEAK 14(18Q11.2) MUTATED | 23 | 8 | 1 |
| AMP PEAK 14(18Q11.2) WILD-TYPE | 23 | 18 | 22 |
Figure S13. Get High-res Image Gene #14: 'amp_18q11.2' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 1e-04 (Fisher's exact test), Q value = 0.035
Table S14. Gene #14: 'amp_18q11.2' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 25 | 13 | 20 |
| AMP PEAK 14(18Q11.2) MUTATED | 22 | 7 | 1 | 2 |
| AMP PEAK 14(18Q11.2) WILD-TYPE | 15 | 18 | 12 | 18 |
Figure S14. Get High-res Image Gene #14: 'amp_18q11.2' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 3e-05 (Fisher's exact test), Q value = 0.011
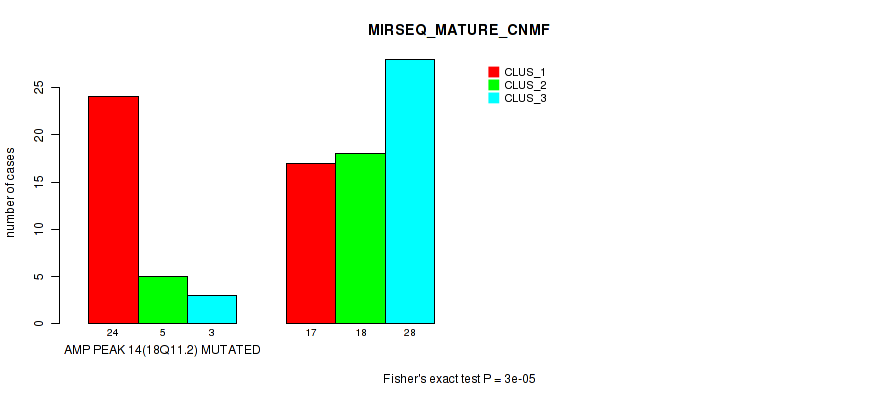
Table S15. Gene #14: 'amp_18q11.2' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 23 | 31 |
| AMP PEAK 14(18Q11.2) MUTATED | 24 | 5 | 3 |
| AMP PEAK 14(18Q11.2) WILD-TYPE | 17 | 18 | 28 |
Figure S15. Get High-res Image Gene #14: 'amp_18q11.2' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
P value = 0.00048 (Fisher's exact test), Q value = 0.16
Table S16. Gene #15: 'amp_19q13.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| AMP PEAK 15(19Q13.2) MUTATED | 14 | 4 | 4 | 4 |
| AMP PEAK 15(19Q13.2) WILD-TYPE | 15 | 42 | 14 | 5 |
Figure S16. Get High-res Image Gene #15: 'amp_19q13.2' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S17. Gene #16: 'amp_20q13.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| AMP PEAK 16(20Q13.2) MUTATED | 19 | 6 | 1 | 5 |
| AMP PEAK 16(20Q13.2) WILD-TYPE | 10 | 40 | 17 | 4 |
Figure S17. Get High-res Image Gene #16: 'amp_20q13.2' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00024 (Fisher's exact test), Q value = 0.082
Table S18. Gene #20: 'del_1p36.11' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 2(1P36.11) MUTATED | 18 | 7 | 8 | 2 |
| DEL PEAK 2(1P36.11) WILD-TYPE | 11 | 39 | 10 | 7 |
Figure S18. Get High-res Image Gene #20: 'del_1p36.11' versus Molecular Subtype #1: 'CN_CNMF'
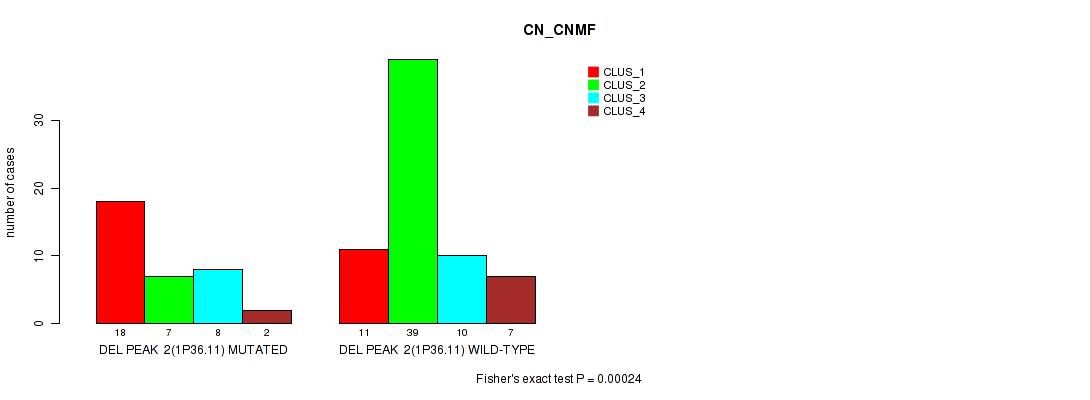
P value = 0.00052 (Fisher's exact test), Q value = 0.17
Table S19. Gene #20: 'del_1p36.11' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 49 | 26 | 26 |
| DEL PEAK 2(1P36.11) MUTATED | 25 | 8 | 2 |
| DEL PEAK 2(1P36.11) WILD-TYPE | 24 | 18 | 24 |
Figure S19. Get High-res Image Gene #20: 'del_1p36.11' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 6e-05 (Fisher's exact test), Q value = 0.021
Table S20. Gene #20: 'del_1p36.11' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 46 | 26 | 23 |
| DEL PEAK 2(1P36.11) MUTATED | 18 | 13 | 0 |
| DEL PEAK 2(1P36.11) WILD-TYPE | 28 | 13 | 23 |
Figure S20. Get High-res Image Gene #20: 'del_1p36.11' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 2e-05 (Fisher's exact test), Q value = 0.0073
Table S21. Gene #21: 'del_1p21.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 3(1P21.2) MUTATED | 16 | 4 | 3 | 0 |
| DEL PEAK 3(1P21.2) WILD-TYPE | 13 | 42 | 15 | 9 |
Figure S21. Get High-res Image Gene #21: 'del_1p21.2' versus Molecular Subtype #1: 'CN_CNMF'

P value = 3e-05 (Fisher's exact test), Q value = 0.011
Table S22. Gene #27: 'del_5q14.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 9(5Q14.2) MUTATED | 16 | 3 | 4 | 1 |
| DEL PEAK 9(5Q14.2) WILD-TYPE | 13 | 43 | 14 | 8 |
Figure S22. Get High-res Image Gene #27: 'del_5q14.2' versus Molecular Subtype #1: 'CN_CNMF'
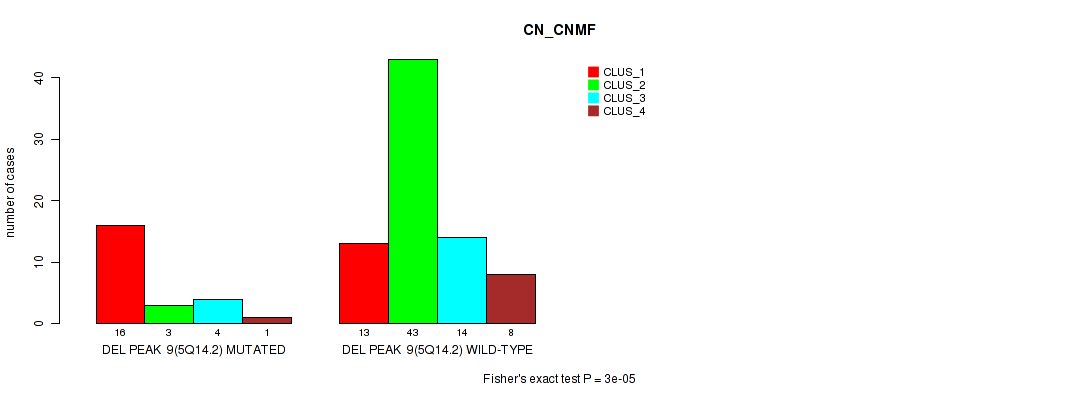
P value = 2e-05 (Fisher's exact test), Q value = 0.0073
Table S23. Gene #28: 'del_6p25.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 10(6P25.2) MUTATED | 21 | 7 | 7 | 7 |
| DEL PEAK 10(6P25.2) WILD-TYPE | 8 | 39 | 11 | 2 |
Figure S23. Get High-res Image Gene #28: 'del_6p25.2' versus Molecular Subtype #1: 'CN_CNMF'

P value = 8e-05 (Fisher's exact test), Q value = 0.028
Table S24. Gene #28: 'del_6p25.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 49 | 26 | 26 |
| DEL PEAK 10(6P25.2) MUTATED | 31 | 4 | 6 |
| DEL PEAK 10(6P25.2) WILD-TYPE | 18 | 22 | 20 |
Figure S24. Get High-res Image Gene #28: 'del_6p25.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00039 (Fisher's exact test), Q value = 0.13
Table S25. Gene #29: 'del_7q36.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 11(7Q36.1) MUTATED | 8 | 0 | 1 | 2 |
| DEL PEAK 11(7Q36.1) WILD-TYPE | 21 | 46 | 17 | 7 |
Figure S25. Get High-res Image Gene #29: 'del_7q36.1' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00016 (Fisher's exact test), Q value = 0.055
Table S26. Gene #30: 'del_8p21.3' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 12(8P21.3) MUTATED | 18 | 6 | 6 | 2 |
| DEL PEAK 12(8P21.3) WILD-TYPE | 11 | 40 | 12 | 7 |
Figure S26. Get High-res Image Gene #30: 'del_8p21.3' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S27. Gene #32: 'del_9p21.3' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 14(9P21.3) MUTATED | 27 | 15 | 17 | 8 |
| DEL PEAK 14(9P21.3) WILD-TYPE | 2 | 31 | 1 | 1 |
Figure S27. Get High-res Image Gene #32: 'del_9p21.3' versus Molecular Subtype #1: 'CN_CNMF'

P value = 2e-05 (Fisher's exact test), Q value = 0.0073
Table S28. Gene #32: 'del_9p21.3' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 49 | 26 | 26 |
| DEL PEAK 14(9P21.3) MUTATED | 43 | 10 | 13 |
| DEL PEAK 14(9P21.3) WILD-TYPE | 6 | 16 | 13 |
Figure S28. Get High-res Image Gene #32: 'del_9p21.3' versus Molecular Subtype #2: 'METHLYATION_CNMF'
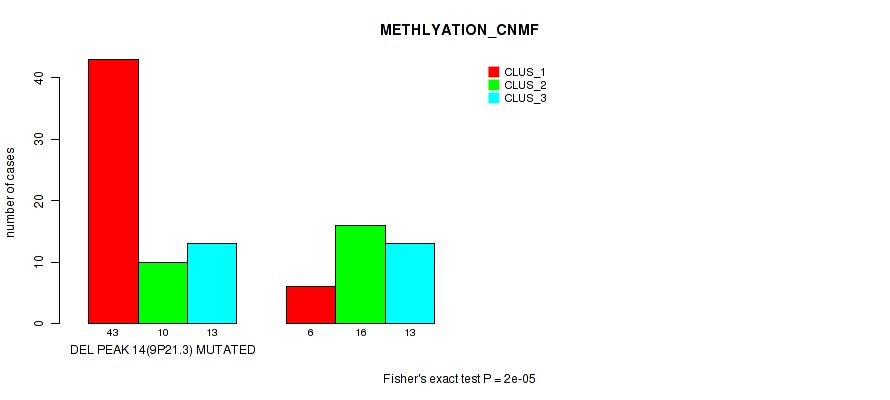
P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S29. Gene #32: 'del_9p21.3' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 46 | 26 | 23 |
| DEL PEAK 14(9P21.3) MUTATED | 39 | 15 | 6 |
| DEL PEAK 14(9P21.3) WILD-TYPE | 7 | 11 | 17 |
Figure S29. Get High-res Image Gene #32: 'del_9p21.3' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S30. Gene #32: 'del_9p21.3' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 27 | 13 | 14 | 12 | 22 | 7 |
| DEL PEAK 14(9P21.3) MUTATED | 26 | 10 | 4 | 7 | 10 | 3 |
| DEL PEAK 14(9P21.3) WILD-TYPE | 1 | 3 | 10 | 5 | 12 | 4 |
Figure S30. Get High-res Image Gene #32: 'del_9p21.3' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 2e-05 (Fisher's exact test), Q value = 0.0073
Table S31. Gene #32: 'del_9p21.3' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 25 | 13 | 20 |
| DEL PEAK 14(9P21.3) MUTATED | 34 | 12 | 4 | 10 |
| DEL PEAK 14(9P21.3) WILD-TYPE | 3 | 13 | 9 | 10 |
Figure S31. Get High-res Image Gene #32: 'del_9p21.3' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 3e-05 (Fisher's exact test), Q value = 0.011
Table S32. Gene #32: 'del_9p21.3' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 17 | 20 | 18 | 8 | 20 | 12 |
| DEL PEAK 14(9P21.3) MUTATED | 17 | 8 | 5 | 6 | 14 | 10 |
| DEL PEAK 14(9P21.3) WILD-TYPE | 0 | 12 | 13 | 2 | 6 | 2 |
Figure S32. Get High-res Image Gene #32: 'del_9p21.3' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 2e-05 (Fisher's exact test), Q value = 0.0073
Table S33. Gene #32: 'del_9p21.3' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 23 | 31 |
| DEL PEAK 14(9P21.3) MUTATED | 36 | 13 | 11 |
| DEL PEAK 14(9P21.3) WILD-TYPE | 5 | 10 | 20 |
Figure S33. Get High-res Image Gene #32: 'del_9p21.3' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'

P value = 3e-05 (Fisher's exact test), Q value = 0.011
Table S34. Gene #32: 'del_9p21.3' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 48 | 28 | 19 |
| DEL PEAK 14(9P21.3) MUTATED | 41 | 13 | 6 |
| DEL PEAK 14(9P21.3) WILD-TYPE | 7 | 15 | 13 |
Figure S34. Get High-res Image Gene #32: 'del_9p21.3' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 0.00031 (Fisher's exact test), Q value = 0.1
Table S35. Gene #34: 'del_10q23.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 16(10Q23.2) MUTATED | 12 | 2 | 2 | 3 |
| DEL PEAK 16(10Q23.2) WILD-TYPE | 17 | 44 | 16 | 6 |
Figure S35. Get High-res Image Gene #34: 'del_10q23.2' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-04 (Fisher's exact test), Q value = 0.035
Table S36. Gene #36: 'del_12p12.3' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 18(12P12.3) MUTATED | 6 | 0 | 6 | 3 |
| DEL PEAK 18(12P12.3) WILD-TYPE | 23 | 46 | 12 | 6 |
Figure S36. Get High-res Image Gene #36: 'del_12p12.3' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S37. Gene #39: 'del_15q15.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 21(15Q15.1) MUTATED | 15 | 1 | 4 | 3 |
| DEL PEAK 21(15Q15.1) WILD-TYPE | 14 | 45 | 14 | 6 |
Figure S37. Get High-res Image Gene #39: 'del_15q15.1' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00026 (Fisher's exact test), Q value = 0.088
Table S38. Gene #39: 'del_15q15.1' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 49 | 26 | 26 |
| DEL PEAK 21(15Q15.1) MUTATED | 19 | 1 | 2 |
| DEL PEAK 21(15Q15.1) WILD-TYPE | 30 | 25 | 24 |
Figure S38. Get High-res Image Gene #39: 'del_15q15.1' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S39. Gene #41: 'del_17q22' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 23(17Q22) MUTATED | 18 | 1 | 5 | 4 |
| DEL PEAK 23(17Q22) WILD-TYPE | 11 | 45 | 13 | 5 |
Figure S39. Get High-res Image Gene #41: 'del_17q22' versus Molecular Subtype #1: 'CN_CNMF'

P value = 4e-05 (Fisher's exact test), Q value = 0.014
Table S40. Gene #43: 'del_18q21.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 25(18Q21.2) MUTATED | 25 | 23 | 18 | 7 |
| DEL PEAK 25(18Q21.2) WILD-TYPE | 4 | 23 | 0 | 2 |
Figure S40. Get High-res Image Gene #43: 'del_18q21.2' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.0038
Table S41. Gene #44: 'del_19p13.3' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 26(19P13.3) MUTATED | 15 | 1 | 2 | 2 |
| DEL PEAK 26(19P13.3) WILD-TYPE | 14 | 45 | 16 | 7 |
Figure S41. Get High-res Image Gene #44: 'del_19p13.3' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00044 (Fisher's exact test), Q value = 0.15
Table S42. Gene #44: 'del_19p13.3' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 17 | 20 | 18 | 8 | 20 | 12 |
| DEL PEAK 26(19P13.3) MUTATED | 8 | 1 | 1 | 5 | 3 | 1 |
| DEL PEAK 26(19P13.3) WILD-TYPE | 9 | 19 | 17 | 3 | 17 | 11 |
Figure S42. Get High-res Image Gene #44: 'del_19p13.3' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 3e-05 (Fisher's exact test), Q value = 0.011
Table S43. Gene #46: 'del_21q21.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 28(21Q21.1) MUTATED | 18 | 7 | 9 | 6 |
| DEL PEAK 28(21Q21.1) WILD-TYPE | 11 | 39 | 9 | 3 |
Figure S43. Get High-res Image Gene #46: 'del_21q21.1' versus Molecular Subtype #1: 'CN_CNMF'

P value = 6e-05 (Fisher's exact test), Q value = 0.021
Table S44. Gene #47: 'del_22q13.31' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 46 | 18 | 9 |
| DEL PEAK 29(22Q13.31) MUTATED | 18 | 5 | 9 | 3 |
| DEL PEAK 29(22Q13.31) WILD-TYPE | 11 | 41 | 9 | 6 |
Figure S44. Get High-res Image Gene #47: 'del_22q13.31' versus Molecular Subtype #1: 'CN_CNMF'
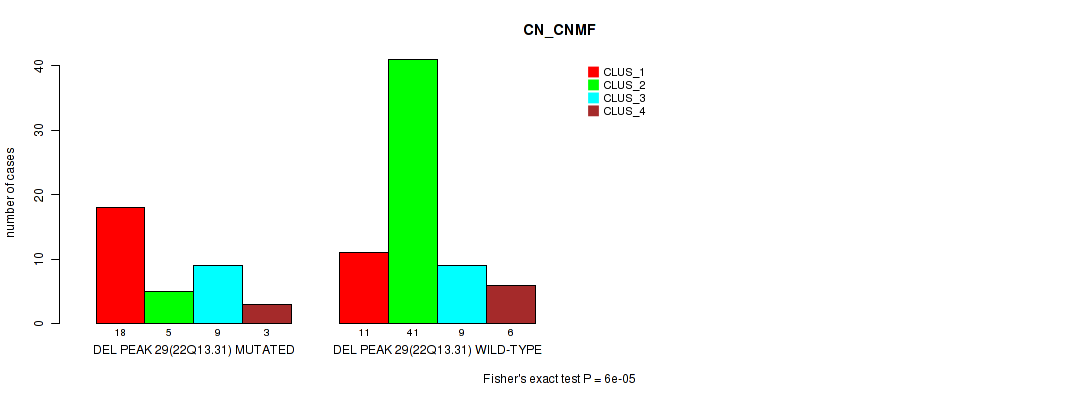
P value = 1e-04 (Fisher's exact test), Q value = 0.035
Table S45. Gene #47: 'del_22q13.31' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 49 | 26 | 26 |
| DEL PEAK 29(22Q13.31) MUTATED | 27 | 3 | 5 |
| DEL PEAK 29(22Q13.31) WILD-TYPE | 22 | 23 | 21 |
Figure S45. Get High-res Image Gene #47: 'del_22q13.31' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00063 (Fisher's exact test), Q value = 0.21
Table S46. Gene #47: 'del_22q13.31' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 17 | 20 | 18 | 8 | 20 | 12 |
| DEL PEAK 29(22Q13.31) MUTATED | 9 | 5 | 1 | 5 | 4 | 8 |
| DEL PEAK 29(22Q13.31) WILD-TYPE | 8 | 15 | 17 | 3 | 16 | 4 |
Figure S46. Get High-res Image Gene #47: 'del_22q13.31' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

-
Copy number data file = transformed.cor.cli.txt
-
Molecular subtype file = PAAD-TP.transferedmergedcluster.txt
-
Number of patients = 102
-
Number of significantly focal cnvs = 47
-
Number of molecular subtypes = 8
-
Exclude genes that fewer than K tumors have alterations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.