This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 17 genes and 12 molecular subtypes across 178 patients, 7 significant findings detected with P value < 0.05 and Q value < 0.25.
-
TP53 mutation correlated to 'CN_CNMF'.
-
NFE2L2 mutation correlated to 'MRNA_CNMF', 'MRNA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
KEAP1 mutation correlated to 'MRNA_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between mutation status of 17 genes and 12 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 7 significant findings detected.
|
Clinical Features |
MRNA CNMF |
MRNA CHIERARCHICAL |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| NFE2L2 | 27 (15%) | 151 |
0.00027 (0.0486) |
0.00013 (0.0235) |
0.0138 (1.00) |
0.0361 (1.00) |
0.253 (1.00) |
0.738 (1.00) |
1e-05 (0.00184) |
1e-05 (0.00184) |
0.00598 (1.00) |
5e-05 (0.0091) |
0.628 (1.00) |
0.866 (1.00) |
| TP53 | 141 (79%) | 37 |
0.044 (1.00) |
0.0263 (1.00) |
0.0006 (0.107) |
0.00147 (0.259) |
1 (1.00) |
0.7 (1.00) |
0.00501 (0.872) |
0.0233 (1.00) |
0.407 (1.00) |
0.441 (1.00) |
0.0895 (1.00) |
0.0639 (1.00) |
| KEAP1 | 22 (12%) | 156 |
0.00145 (0.257) |
0.00062 (0.11) |
0.119 (1.00) |
0.0754 (1.00) |
0.202 (1.00) |
0.148 (1.00) |
0.187 (1.00) |
0.0297 (1.00) |
0.181 (1.00) |
0.447 (1.00) |
0.481 (1.00) |
0.669 (1.00) |
| CDKN2A | 26 (15%) | 152 |
0.00162 (0.283) |
0.00555 (0.96) |
0.0245 (1.00) |
0.667 (1.00) |
0.339 (1.00) |
0.169 (1.00) |
0.38 (1.00) |
0.0891 (1.00) |
0.664 (1.00) |
0.819 (1.00) |
0.843 (1.00) |
0.0299 (1.00) |
| PTEN | 14 (8%) | 164 |
0.381 (1.00) |
0.323 (1.00) |
0.412 (1.00) |
0.135 (1.00) |
0.722 (1.00) |
0.961 (1.00) |
0.594 (1.00) |
0.511 (1.00) |
0.573 (1.00) |
0.513 (1.00) |
0.199 (1.00) |
1 (1.00) |
| PIK3CA | 27 (15%) | 151 |
0.69 (1.00) |
0.754 (1.00) |
0.291 (1.00) |
0.546 (1.00) |
1 (1.00) |
0.666 (1.00) |
0.339 (1.00) |
0.586 (1.00) |
0.364 (1.00) |
0.379 (1.00) |
0.724 (1.00) |
0.753 (1.00) |
| MLL2 | 35 (20%) | 143 |
0.168 (1.00) |
0.464 (1.00) |
0.646 (1.00) |
0.314 (1.00) |
0.697 (1.00) |
0.363 (1.00) |
0.324 (1.00) |
0.856 (1.00) |
0.93 (1.00) |
0.941 (1.00) |
0.743 (1.00) |
1 (1.00) |
| RB1 | 12 (7%) | 166 |
0.313 (1.00) |
0.646 (1.00) |
0.524 (1.00) |
0.107 (1.00) |
0.656 (1.00) |
0.48 (1.00) |
0.775 (1.00) |
0.241 (1.00) |
0.322 (1.00) |
0.754 (1.00) |
||
| IBTK | 5 (3%) | 173 |
0.489 (1.00) |
0.847 (1.00) |
0.739 (1.00) |
0.807 (1.00) |
0.842 (1.00) |
1 (1.00) |
0.608 (1.00) |
|||||
| CYP11B1 | 15 (8%) | 163 |
0.653 (1.00) |
0.677 (1.00) |
0.659 (1.00) |
0.125 (1.00) |
0.24 (1.00) |
0.0944 (1.00) |
0.57 (1.00) |
0.368 (1.00) |
0.562 (1.00) |
0.977 (1.00) |
||
| NOTCH1 | 14 (8%) | 164 |
0.816 (1.00) |
0.603 (1.00) |
0.468 (1.00) |
0.0345 (1.00) |
0.59 (1.00) |
0.628 (1.00) |
0.756 (1.00) |
0.392 (1.00) |
0.349 (1.00) |
0.32 (1.00) |
0.0103 (1.00) |
0.0296 (1.00) |
| SLC28A1 | 9 (5%) | 169 |
0.0136 (1.00) |
0.0501 (1.00) |
0.0793 (1.00) |
0.134 (1.00) |
0.32 (1.00) |
0.523 (1.00) |
0.0367 (1.00) |
0.0281 (1.00) |
0.185 (1.00) |
0.0169 (1.00) |
0.819 (1.00) |
0.183 (1.00) |
| ASB5 | 9 (5%) | 169 |
0.496 (1.00) |
0.55 (1.00) |
0.263 (1.00) |
0.472 (1.00) |
1 (1.00) |
0.48 (1.00) |
0.497 (1.00) |
0.252 (1.00) |
0.337 (1.00) |
|||
| CPS1 | 25 (14%) | 153 |
0.768 (1.00) |
0.235 (1.00) |
0.267 (1.00) |
0.892 (1.00) |
0.731 (1.00) |
0.498 (1.00) |
0.99 (1.00) |
0.726 (1.00) |
0.0379 (1.00) |
0.279 (1.00) |
1 (1.00) |
1 (1.00) |
| EP300 | 8 (4%) | 170 |
0.15 (1.00) |
0.118 (1.00) |
0.437 (1.00) |
0.357 (1.00) |
1 (1.00) |
0.0811 (1.00) |
0.083 (1.00) |
0.57 (1.00) |
0.822 (1.00) |
|||
| ARID1A | 12 (7%) | 166 |
0.313 (1.00) |
0.552 (1.00) |
0.452 (1.00) |
0.354 (1.00) |
0.495 (1.00) |
0.601 (1.00) |
0.111 (1.00) |
0.862 (1.00) |
0.53 (1.00) |
0.692 (1.00) |
||
| FBXW7 | 10 (6%) | 168 |
0.493 (1.00) |
0.83 (1.00) |
0.07 (1.00) |
0.623 (1.00) |
0.788 (1.00) |
0.833 (1.00) |
0.526 (1.00) |
0.962 (1.00) |
0.0672 (1.00) |
P value = 0.044 (Fisher's exact test), Q value = 1
Table S1. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| TP53 MUTATED | 29 | 43 | 10 | 14 |
| TP53 WILD-TYPE | 8 | 5 | 7 | 5 |
Figure S1. Get High-res Image Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
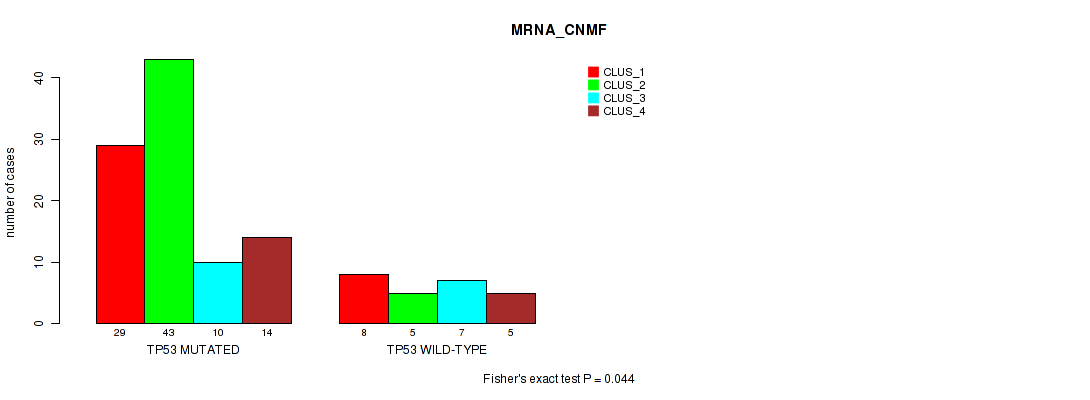
P value = 0.0263 (Fisher's exact test), Q value = 1
Table S2. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| TP53 MUTATED | 30 | 47 | 19 |
| TP53 WILD-TYPE | 11 | 5 | 9 |
Figure S2. Get High-res Image Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
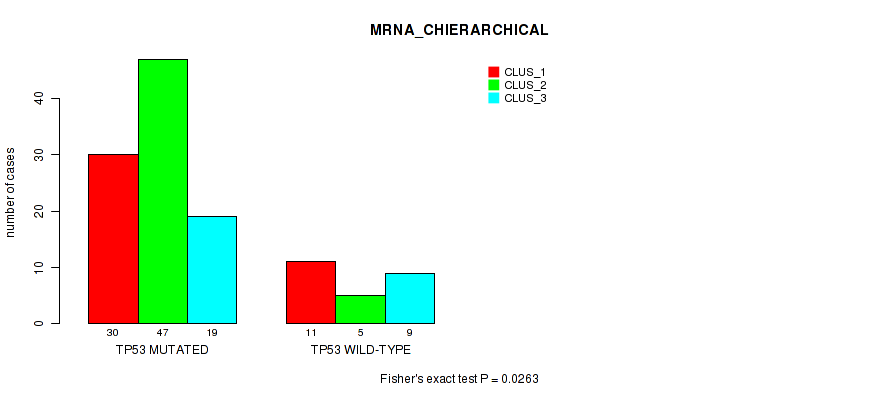
P value = 6e-04 (Fisher's exact test), Q value = 0.11
Table S3. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| TP53 MUTATED | 36 | 62 | 43 |
| TP53 WILD-TYPE | 22 | 7 | 8 |
Figure S3. Get High-res Image Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
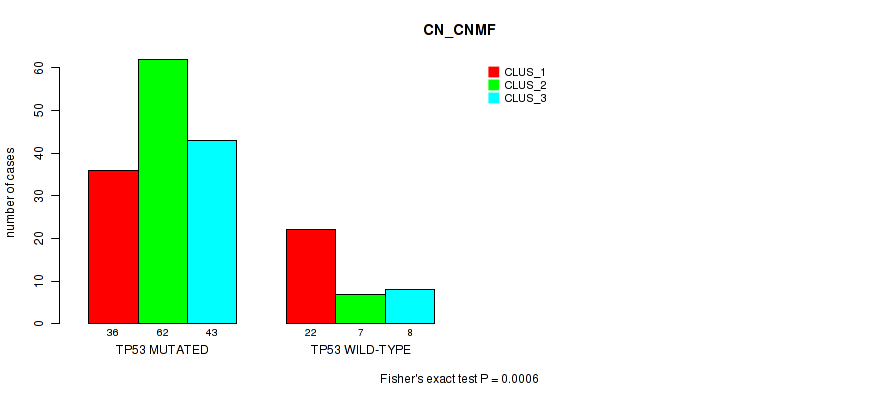
P value = 0.00147 (Fisher's exact test), Q value = 0.26
Table S4. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| TP53 MUTATED | 16 | 28 | 15 |
| TP53 WILD-TYPE | 11 | 1 | 3 |
Figure S4. Get High-res Image Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
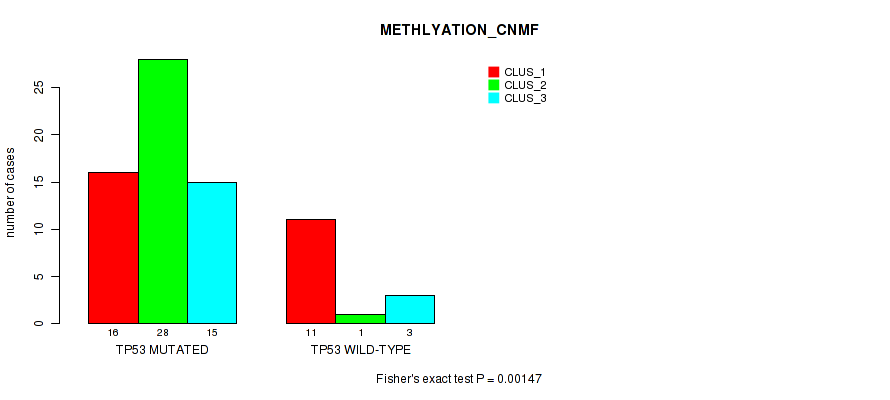
P value = 1 (Fisher's exact test), Q value = 1
Table S5. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| TP53 MUTATED | 26 | 37 | 24 |
| TP53 WILD-TYPE | 8 | 10 | 7 |
P value = 0.7 (Fisher's exact test), Q value = 1
Table S6. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| TP53 MUTATED | 21 | 16 | 28 | 22 |
| TP53 WILD-TYPE | 8 | 6 | 6 | 5 |
P value = 0.00501 (Fisher's exact test), Q value = 0.87
Table S7. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| TP53 MUTATED | 31 | 55 | 41 | 14 |
| TP53 WILD-TYPE | 9 | 7 | 9 | 12 |
Figure S5. Get High-res Image Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
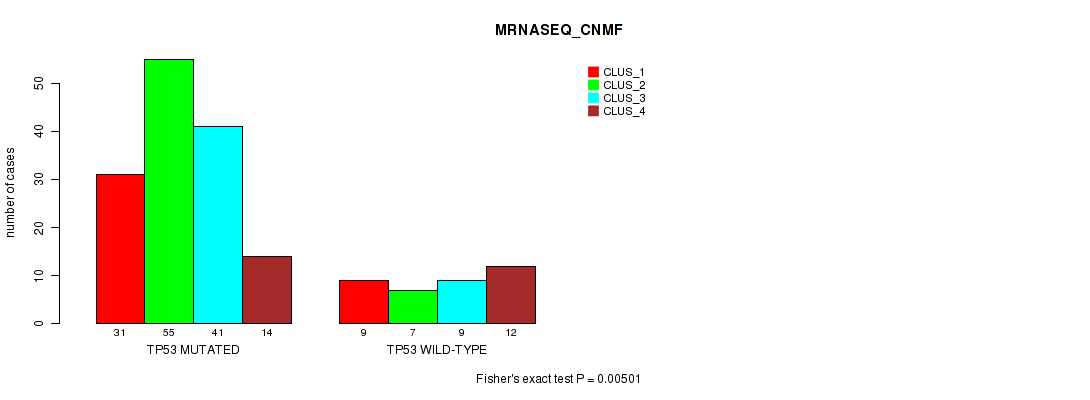
P value = 0.0233 (Fisher's exact test), Q value = 1
Table S8. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| TP53 MUTATED | 47 | 65 | 29 |
| TP53 WILD-TYPE | 21 | 9 | 7 |
Figure S6. Get High-res Image Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
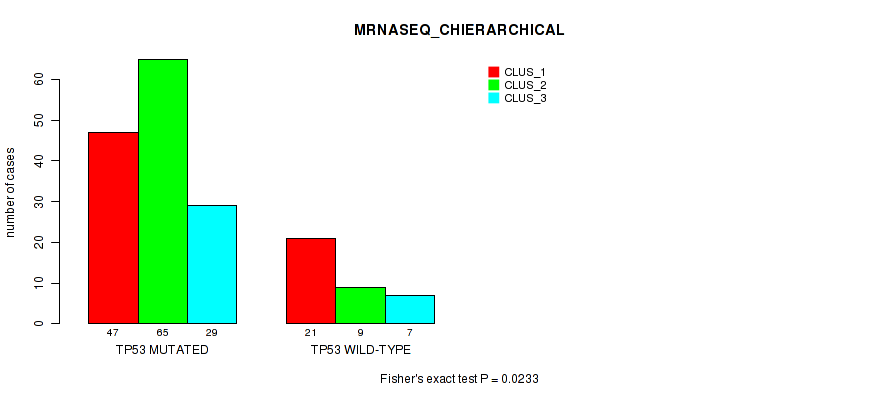
P value = 0.407 (Fisher's exact test), Q value = 1
Table S9. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| TP53 MUTATED | 79 | 12 | 14 | 20 | 8 |
| TP53 WILD-TYPE | 14 | 2 | 7 | 4 | 2 |
P value = 0.441 (Fisher's exact test), Q value = 1
Table S10. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| TP53 MUTATED | 31 | 39 | 46 | 17 |
| TP53 WILD-TYPE | 4 | 12 | 8 | 5 |
P value = 0.0895 (Fisher's exact test), Q value = 1
Table S11. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 16 | 19 | 13 |
| TP53 MUTATED | 10 | 17 | 12 |
| TP53 WILD-TYPE | 6 | 2 | 1 |
P value = 0.0639 (Fisher's exact test), Q value = 1
Table S12. Gene #1: 'TP53 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 4 | 19 | 25 |
| TP53 MUTATED | 4 | 18 | 17 |
| TP53 WILD-TYPE | 0 | 1 | 8 |
P value = 0.00027 (Fisher's exact test), Q value = 0.049
Table S13. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| NFE2L2 MUTATED | 0 | 14 | 1 | 1 |
| NFE2L2 WILD-TYPE | 37 | 34 | 16 | 18 |
Figure S7. Get High-res Image Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
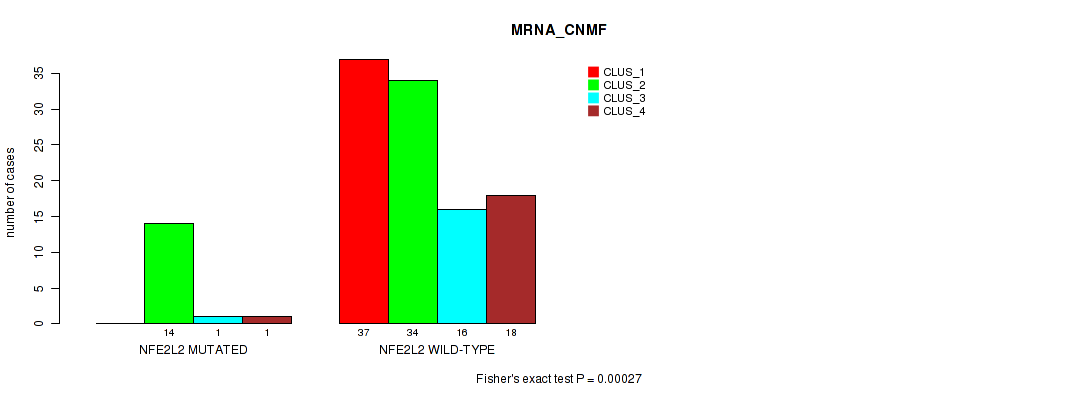
P value = 0.00013 (Fisher's exact test), Q value = 0.024
Table S14. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| NFE2L2 MUTATED | 0 | 14 | 2 |
| NFE2L2 WILD-TYPE | 41 | 38 | 26 |
Figure S8. Get High-res Image Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
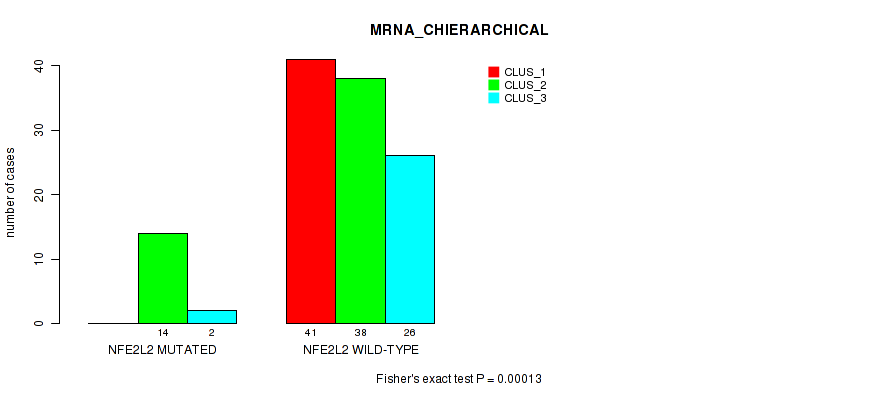
P value = 0.0138 (Fisher's exact test), Q value = 1
Table S15. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| NFE2L2 MUTATED | 7 | 17 | 3 |
| NFE2L2 WILD-TYPE | 51 | 52 | 48 |
Figure S9. Get High-res Image Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
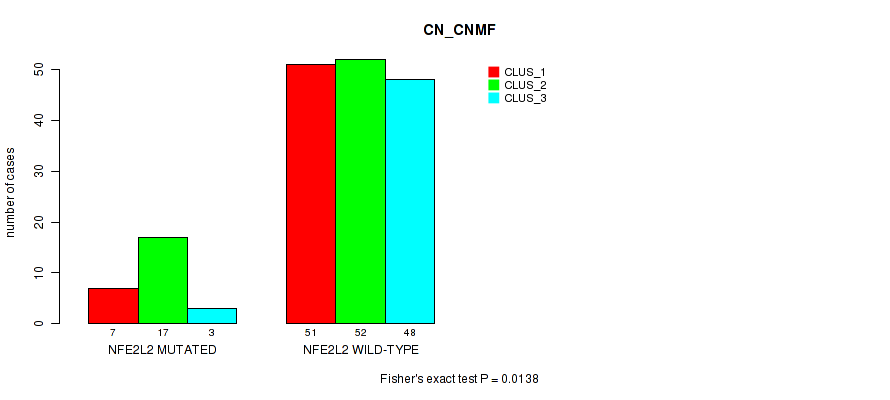
P value = 0.0361 (Fisher's exact test), Q value = 1
Table S16. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| NFE2L2 MUTATED | 4 | 8 | 0 |
| NFE2L2 WILD-TYPE | 23 | 21 | 18 |
Figure S10. Get High-res Image Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
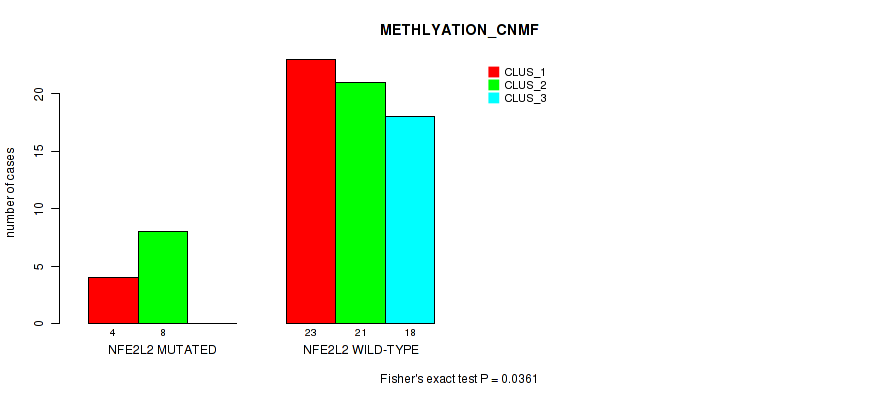
P value = 0.253 (Fisher's exact test), Q value = 1
Table S17. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| NFE2L2 MUTATED | 8 | 5 | 6 |
| NFE2L2 WILD-TYPE | 26 | 42 | 25 |
P value = 0.738 (Fisher's exact test), Q value = 1
Table S18. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| NFE2L2 MUTATED | 5 | 5 | 4 | 5 |
| NFE2L2 WILD-TYPE | 24 | 17 | 30 | 22 |
P value = 1e-05 (Fisher's exact test), Q value = 0.0018
Table S19. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| NFE2L2 MUTATED | 2 | 22 | 1 | 2 |
| NFE2L2 WILD-TYPE | 38 | 40 | 49 | 24 |
Figure S11. Get High-res Image Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
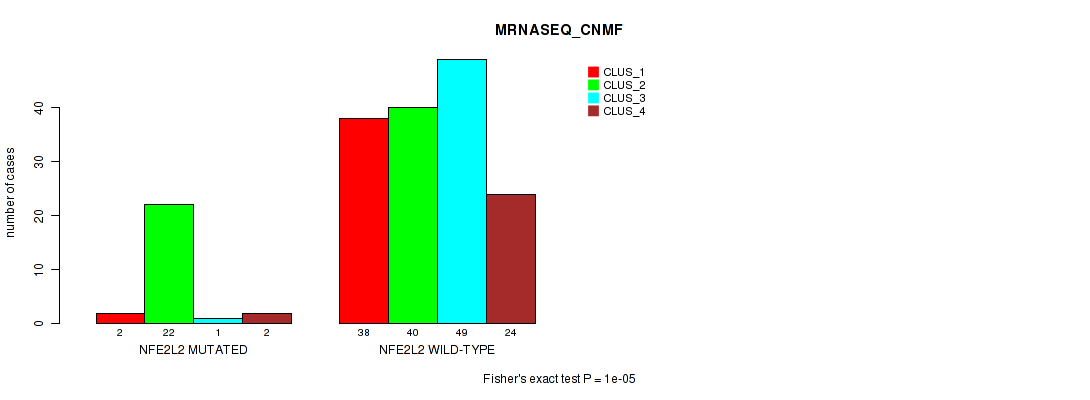
P value = 1e-05 (Fisher's exact test), Q value = 0.0018
Table S20. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| NFE2L2 MUTATED | 2 | 24 | 1 |
| NFE2L2 WILD-TYPE | 66 | 50 | 35 |
Figure S12. Get High-res Image Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
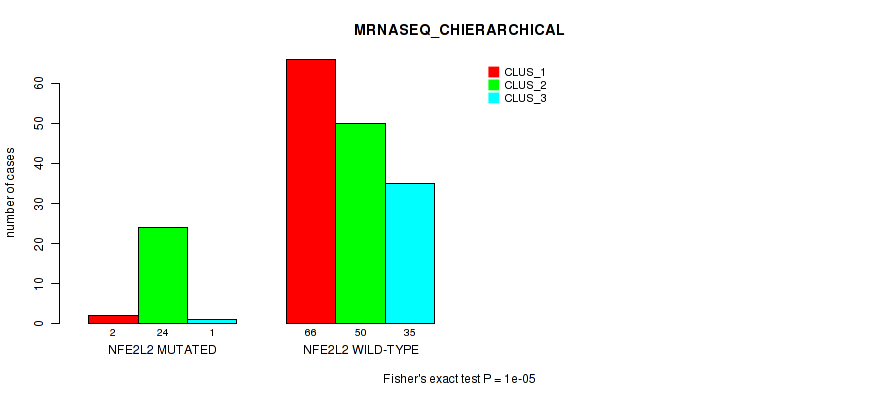
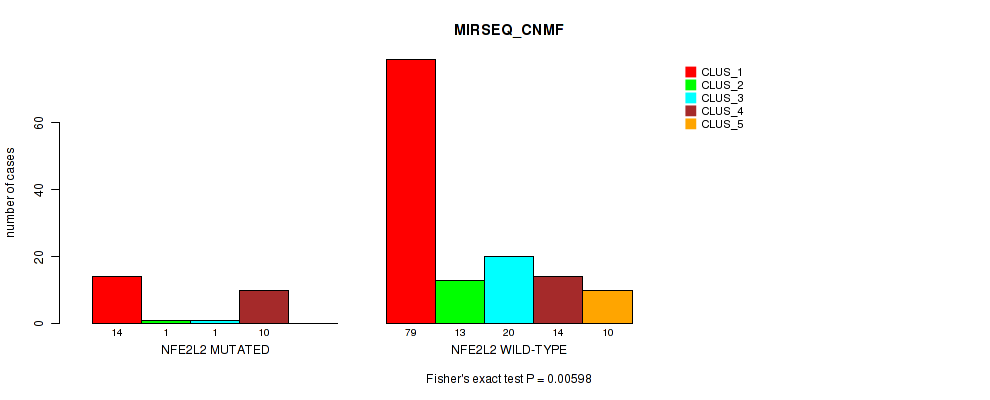
P value = 0.00598 (Fisher's exact test), Q value = 1
Table S21. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| NFE2L2 MUTATED | 14 | 1 | 1 | 10 | 0 |
| NFE2L2 WILD-TYPE | 79 | 13 | 20 | 14 | 10 |
Figure S13. Get High-res Image Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
P value = 5e-05 (Fisher's exact test), Q value = 0.0091
Table S22. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| NFE2L2 MUTATED | 15 | 6 | 2 | 3 |
| NFE2L2 WILD-TYPE | 20 | 45 | 52 | 19 |
Figure S14. Get High-res Image Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
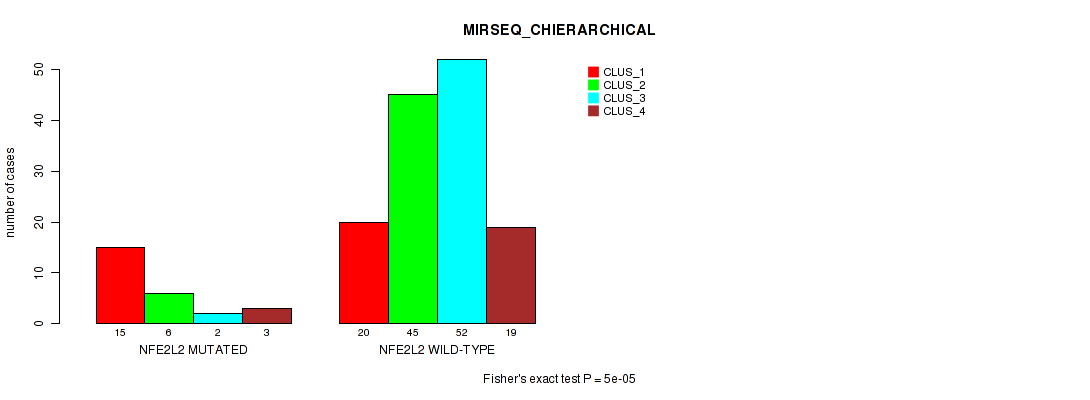
P value = 0.628 (Fisher's exact test), Q value = 1
Table S23. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 16 | 19 | 13 |
| NFE2L2 MUTATED | 3 | 2 | 3 |
| NFE2L2 WILD-TYPE | 13 | 17 | 10 |
P value = 0.866 (Fisher's exact test), Q value = 1
Table S24. Gene #2: 'NFE2L2 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 4 | 19 | 25 |
| NFE2L2 MUTATED | 0 | 4 | 4 |
| NFE2L2 WILD-TYPE | 4 | 15 | 21 |
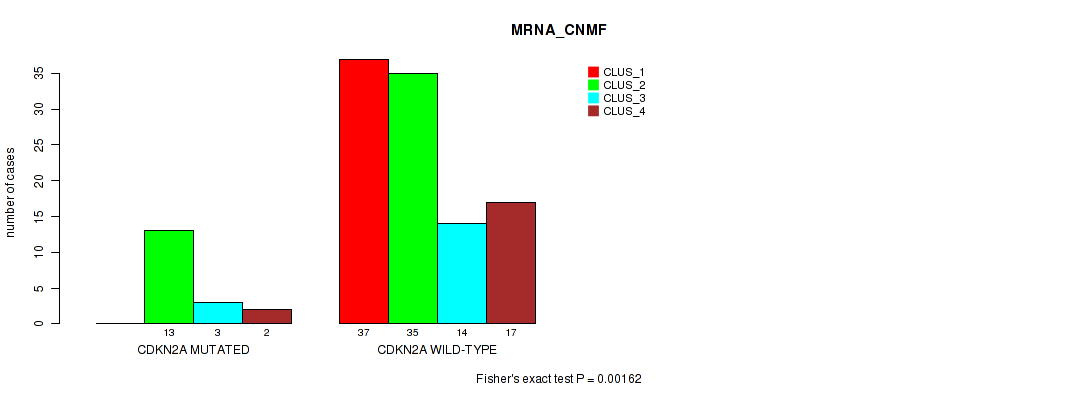
P value = 0.00162 (Fisher's exact test), Q value = 0.28
Table S25. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| CDKN2A MUTATED | 0 | 13 | 3 | 2 |
| CDKN2A WILD-TYPE | 37 | 35 | 14 | 17 |
Figure S15. Get High-res Image Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
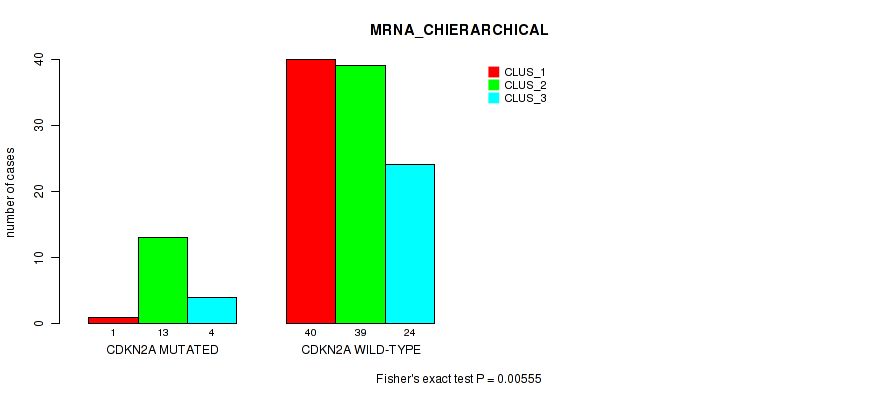
P value = 0.00555 (Fisher's exact test), Q value = 0.96
Table S26. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| CDKN2A MUTATED | 1 | 13 | 4 |
| CDKN2A WILD-TYPE | 40 | 39 | 24 |
Figure S16. Get High-res Image Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
P value = 0.0245 (Fisher's exact test), Q value = 1
Table S27. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| CDKN2A MUTATED | 3 | 15 | 8 |
| CDKN2A WILD-TYPE | 55 | 54 | 43 |
Figure S17. Get High-res Image Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
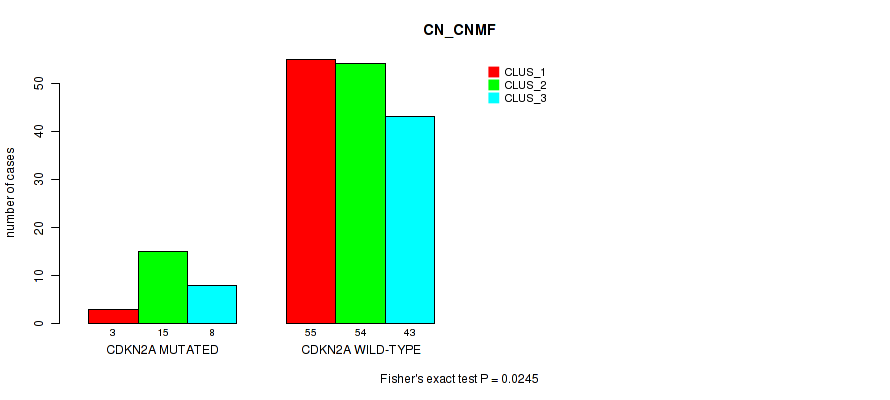
P value = 0.667 (Fisher's exact test), Q value = 1
Table S28. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| CDKN2A MUTATED | 2 | 4 | 3 |
| CDKN2A WILD-TYPE | 25 | 25 | 15 |
P value = 0.339 (Fisher's exact test), Q value = 1
Table S29. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| CDKN2A MUTATED | 2 | 8 | 3 |
| CDKN2A WILD-TYPE | 32 | 39 | 28 |
P value = 0.169 (Fisher's exact test), Q value = 1
Table S30. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| CDKN2A MUTATED | 2 | 5 | 5 | 1 |
| CDKN2A WILD-TYPE | 27 | 17 | 29 | 26 |
P value = 0.38 (Fisher's exact test), Q value = 1
Table S31. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| CDKN2A MUTATED | 4 | 12 | 5 | 5 |
| CDKN2A WILD-TYPE | 36 | 50 | 45 | 21 |
P value = 0.0891 (Fisher's exact test), Q value = 1
Table S32. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| CDKN2A MUTATED | 6 | 16 | 4 |
| CDKN2A WILD-TYPE | 62 | 58 | 32 |
P value = 0.664 (Fisher's exact test), Q value = 1
Table S33. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| CDKN2A MUTATED | 16 | 1 | 4 | 3 | 0 |
| CDKN2A WILD-TYPE | 77 | 13 | 17 | 21 | 10 |
P value = 0.819 (Fisher's exact test), Q value = 1
Table S34. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| CDKN2A MUTATED | 5 | 6 | 10 | 3 |
| CDKN2A WILD-TYPE | 30 | 45 | 44 | 19 |
P value = 0.843 (Fisher's exact test), Q value = 1
Table S35. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 16 | 19 | 13 |
| CDKN2A MUTATED | 1 | 2 | 2 |
| CDKN2A WILD-TYPE | 15 | 17 | 11 |
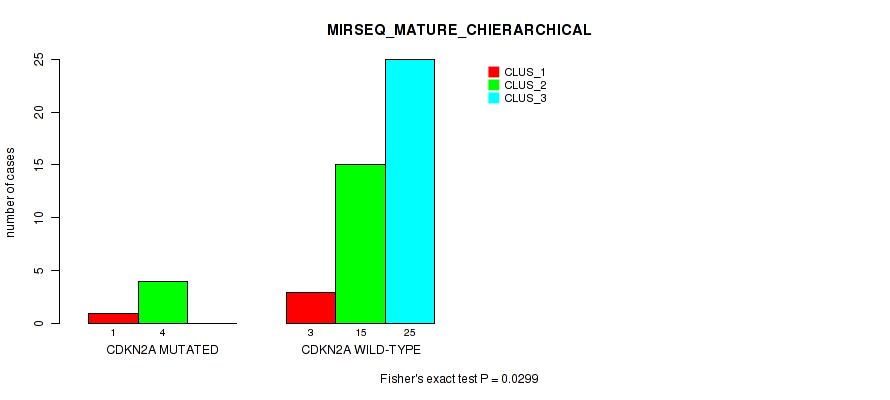
P value = 0.0299 (Fisher's exact test), Q value = 1
Table S36. Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 4 | 19 | 25 |
| CDKN2A MUTATED | 1 | 4 | 0 |
| CDKN2A WILD-TYPE | 3 | 15 | 25 |
Figure S18. Get High-res Image Gene #3: 'CDKN2A MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
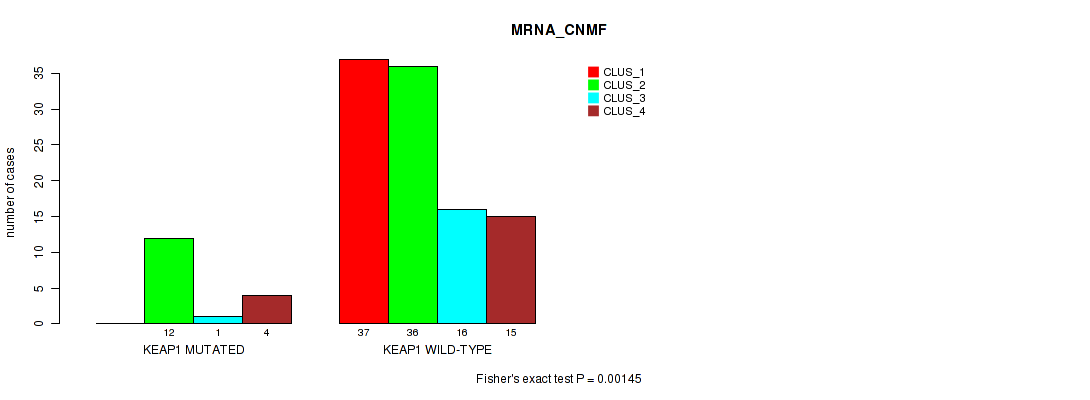
P value = 0.00145 (Fisher's exact test), Q value = 0.26
Table S37. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| KEAP1 MUTATED | 0 | 12 | 1 | 4 |
| KEAP1 WILD-TYPE | 37 | 36 | 16 | 15 |
Figure S19. Get High-res Image Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
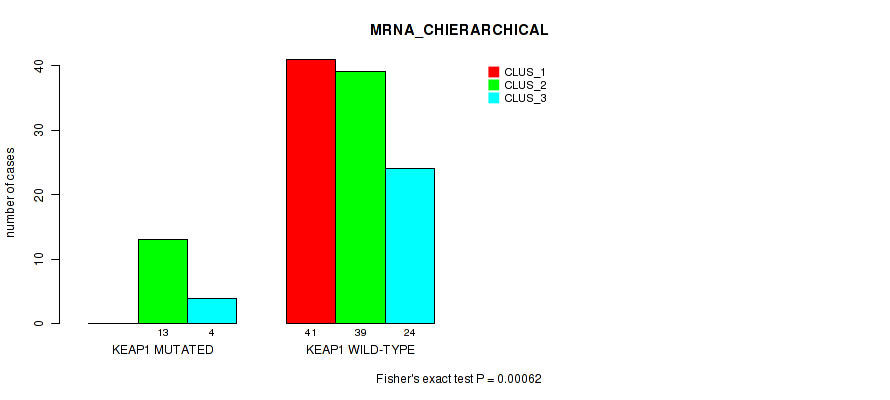
P value = 0.00062 (Fisher's exact test), Q value = 0.11
Table S38. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| KEAP1 MUTATED | 0 | 13 | 4 |
| KEAP1 WILD-TYPE | 41 | 39 | 24 |
Figure S20. Get High-res Image Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
P value = 0.119 (Fisher's exact test), Q value = 1
Table S39. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| KEAP1 MUTATED | 4 | 13 | 5 |
| KEAP1 WILD-TYPE | 54 | 56 | 46 |
P value = 0.0754 (Fisher's exact test), Q value = 1
Table S40. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| KEAP1 MUTATED | 0 | 4 | 3 |
| KEAP1 WILD-TYPE | 27 | 25 | 15 |
P value = 0.202 (Fisher's exact test), Q value = 1
Table S41. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| KEAP1 MUTATED | 5 | 2 | 4 |
| KEAP1 WILD-TYPE | 29 | 45 | 27 |
P value = 0.148 (Fisher's exact test), Q value = 1
Table S42. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| KEAP1 MUTATED | 4 | 1 | 1 | 5 |
| KEAP1 WILD-TYPE | 25 | 21 | 33 | 22 |
P value = 0.187 (Fisher's exact test), Q value = 1
Table S43. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| KEAP1 MUTATED | 2 | 12 | 5 | 3 |
| KEAP1 WILD-TYPE | 38 | 50 | 45 | 23 |
P value = 0.0297 (Fisher's exact test), Q value = 1
Table S44. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| KEAP1 MUTATED | 3 | 13 | 6 |
| KEAP1 WILD-TYPE | 65 | 61 | 30 |
Figure S21. Get High-res Image Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
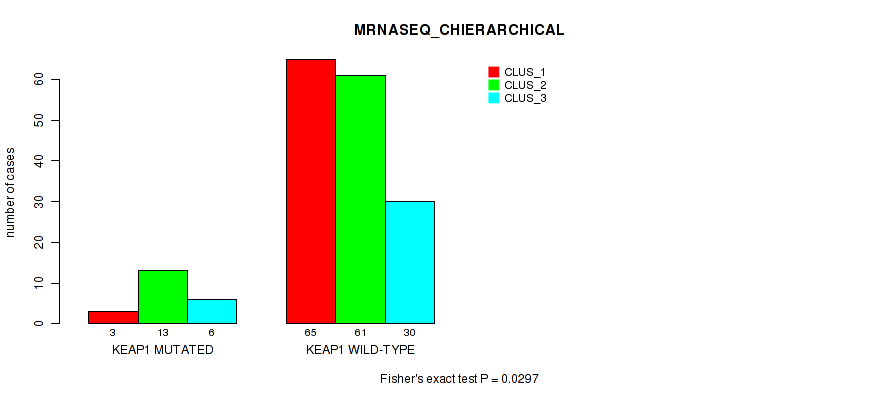
P value = 0.181 (Fisher's exact test), Q value = 1
Table S45. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| KEAP1 MUTATED | 10 | 3 | 0 | 3 | 2 |
| KEAP1 WILD-TYPE | 83 | 11 | 21 | 21 | 8 |
P value = 0.447 (Fisher's exact test), Q value = 1
Table S46. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| KEAP1 MUTATED | 5 | 3 | 8 | 2 |
| KEAP1 WILD-TYPE | 30 | 48 | 46 | 20 |
P value = 0.481 (Fisher's exact test), Q value = 1
Table S47. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 16 | 19 | 13 |
| KEAP1 MUTATED | 0 | 2 | 1 |
| KEAP1 WILD-TYPE | 16 | 17 | 12 |
P value = 0.669 (Fisher's exact test), Q value = 1
Table S48. Gene #4: 'KEAP1 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 4 | 19 | 25 |
| KEAP1 MUTATED | 0 | 2 | 1 |
| KEAP1 WILD-TYPE | 4 | 17 | 24 |
P value = 0.381 (Fisher's exact test), Q value = 1
Table S49. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| PTEN MUTATED | 4 | 2 | 0 | 2 |
| PTEN WILD-TYPE | 33 | 46 | 17 | 17 |
P value = 0.323 (Fisher's exact test), Q value = 1
Table S50. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| PTEN MUTATED | 5 | 2 | 1 |
| PTEN WILD-TYPE | 36 | 50 | 27 |
P value = 0.412 (Fisher's exact test), Q value = 1
Table S51. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| PTEN MUTATED | 7 | 4 | 3 |
| PTEN WILD-TYPE | 51 | 65 | 48 |
P value = 0.135 (Fisher's exact test), Q value = 1
Table S52. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| PTEN MUTATED | 5 | 3 | 0 |
| PTEN WILD-TYPE | 22 | 26 | 18 |
P value = 0.722 (Fisher's exact test), Q value = 1
Table S53. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| PTEN MUTATED | 3 | 4 | 1 |
| PTEN WILD-TYPE | 31 | 43 | 30 |
P value = 0.961 (Fisher's exact test), Q value = 1
Table S54. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| PTEN MUTATED | 2 | 2 | 2 | 2 |
| PTEN WILD-TYPE | 27 | 20 | 32 | 25 |
P value = 0.594 (Fisher's exact test), Q value = 1
Table S55. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| PTEN MUTATED | 4 | 3 | 4 | 3 |
| PTEN WILD-TYPE | 36 | 59 | 46 | 23 |
P value = 0.511 (Fisher's exact test), Q value = 1
Table S56. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| PTEN MUTATED | 6 | 4 | 4 |
| PTEN WILD-TYPE | 62 | 70 | 32 |
P value = 0.573 (Fisher's exact test), Q value = 1
Table S57. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| PTEN MUTATED | 7 | 0 | 3 | 1 | 1 |
| PTEN WILD-TYPE | 86 | 14 | 18 | 23 | 9 |
P value = 0.513 (Fisher's exact test), Q value = 1
Table S58. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| PTEN MUTATED | 1 | 3 | 6 | 2 |
| PTEN WILD-TYPE | 34 | 48 | 48 | 20 |
P value = 0.199 (Fisher's exact test), Q value = 1
Table S59. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 16 | 19 | 13 |
| PTEN MUTATED | 1 | 4 | 0 |
| PTEN WILD-TYPE | 15 | 15 | 13 |
P value = 1 (Fisher's exact test), Q value = 1
Table S60. Gene #5: 'PTEN MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 4 | 19 | 25 |
| PTEN MUTATED | 0 | 2 | 3 |
| PTEN WILD-TYPE | 4 | 17 | 22 |
P value = 0.69 (Fisher's exact test), Q value = 1
Table S61. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| PIK3CA MUTATED | 4 | 6 | 3 | 4 |
| PIK3CA WILD-TYPE | 33 | 42 | 14 | 15 |
P value = 0.754 (Fisher's exact test), Q value = 1
Table S62. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| PIK3CA MUTATED | 7 | 6 | 4 |
| PIK3CA WILD-TYPE | 34 | 46 | 24 |
P value = 0.291 (Fisher's exact test), Q value = 1
Table S63. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| PIK3CA MUTATED | 10 | 7 | 10 |
| PIK3CA WILD-TYPE | 48 | 62 | 41 |
P value = 0.546 (Fisher's exact test), Q value = 1
Table S64. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| PIK3CA MUTATED | 4 | 3 | 4 |
| PIK3CA WILD-TYPE | 23 | 26 | 14 |
P value = 1 (Fisher's exact test), Q value = 1
Table S65. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| PIK3CA MUTATED | 5 | 7 | 5 |
| PIK3CA WILD-TYPE | 29 | 40 | 26 |
P value = 0.666 (Fisher's exact test), Q value = 1
Table S66. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| PIK3CA MUTATED | 5 | 2 | 7 | 3 |
| PIK3CA WILD-TYPE | 24 | 20 | 27 | 24 |
P value = 0.339 (Fisher's exact test), Q value = 1
Table S67. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| PIK3CA MUTATED | 8 | 7 | 10 | 2 |
| PIK3CA WILD-TYPE | 32 | 55 | 40 | 24 |
P value = 0.586 (Fisher's exact test), Q value = 1
Table S68. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| PIK3CA MUTATED | 11 | 9 | 7 |
| PIK3CA WILD-TYPE | 57 | 65 | 29 |
P value = 0.364 (Fisher's exact test), Q value = 1
Table S69. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| PIK3CA MUTATED | 15 | 4 | 3 | 2 | 0 |
| PIK3CA WILD-TYPE | 78 | 10 | 18 | 22 | 10 |
P value = 0.379 (Fisher's exact test), Q value = 1
Table S70. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| PIK3CA MUTATED | 3 | 11 | 8 | 2 |
| PIK3CA WILD-TYPE | 32 | 40 | 46 | 20 |
P value = 0.724 (Fisher's exact test), Q value = 1
Table S71. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 16 | 19 | 13 |
| PIK3CA MUTATED | 2 | 4 | 3 |
| PIK3CA WILD-TYPE | 14 | 15 | 10 |
P value = 0.753 (Fisher's exact test), Q value = 1
Table S72. Gene #6: 'PIK3CA MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 4 | 19 | 25 |
| PIK3CA MUTATED | 1 | 4 | 4 |
| PIK3CA WILD-TYPE | 3 | 15 | 21 |
P value = 0.168 (Fisher's exact test), Q value = 1
Table S73. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| MLL2 MUTATED | 4 | 10 | 6 | 5 |
| MLL2 WILD-TYPE | 33 | 38 | 11 | 14 |
P value = 0.464 (Fisher's exact test), Q value = 1
Table S74. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| MLL2 MUTATED | 6 | 13 | 6 |
| MLL2 WILD-TYPE | 35 | 39 | 22 |
P value = 0.646 (Fisher's exact test), Q value = 1
Table S75. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| MLL2 MUTATED | 9 | 15 | 11 |
| MLL2 WILD-TYPE | 49 | 54 | 40 |
P value = 0.314 (Fisher's exact test), Q value = 1
Table S76. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| MLL2 MUTATED | 6 | 6 | 1 |
| MLL2 WILD-TYPE | 21 | 23 | 17 |
P value = 0.697 (Fisher's exact test), Q value = 1
Table S77. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| MLL2 MUTATED | 6 | 9 | 8 |
| MLL2 WILD-TYPE | 28 | 38 | 23 |
P value = 0.363 (Fisher's exact test), Q value = 1
Table S78. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| MLL2 MUTATED | 6 | 7 | 7 | 3 |
| MLL2 WILD-TYPE | 23 | 15 | 27 | 24 |
P value = 0.324 (Fisher's exact test), Q value = 1
Table S79. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| MLL2 MUTATED | 4 | 15 | 11 | 5 |
| MLL2 WILD-TYPE | 36 | 47 | 39 | 21 |
P value = 0.856 (Fisher's exact test), Q value = 1
Table S80. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| MLL2 MUTATED | 12 | 16 | 7 |
| MLL2 WILD-TYPE | 56 | 58 | 29 |
P value = 0.93 (Fisher's exact test), Q value = 1
Table S81. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| MLL2 MUTATED | 17 | 2 | 5 | 4 | 1 |
| MLL2 WILD-TYPE | 76 | 12 | 16 | 20 | 9 |
P value = 0.941 (Fisher's exact test), Q value = 1
Table S82. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| MLL2 MUTATED | 5 | 10 | 10 | 4 |
| MLL2 WILD-TYPE | 30 | 41 | 44 | 18 |
P value = 0.743 (Fisher's exact test), Q value = 1
Table S83. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 16 | 19 | 13 |
| MLL2 MUTATED | 4 | 3 | 3 |
| MLL2 WILD-TYPE | 12 | 16 | 10 |
P value = 1 (Fisher's exact test), Q value = 1
Table S84. Gene #7: 'MLL2 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 4 | 19 | 25 |
| MLL2 MUTATED | 1 | 4 | 5 |
| MLL2 WILD-TYPE | 3 | 15 | 20 |
P value = 0.313 (Fisher's exact test), Q value = 1
Table S85. Gene #8: 'RB1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| RB1 MUTATED | 2 | 5 | 0 | 3 |
| RB1 WILD-TYPE | 35 | 43 | 17 | 16 |
P value = 0.646 (Fisher's exact test), Q value = 1
Table S86. Gene #8: 'RB1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| RB1 MUTATED | 2 | 5 | 3 |
| RB1 WILD-TYPE | 39 | 47 | 25 |
P value = 0.524 (Fisher's exact test), Q value = 1
Table S87. Gene #8: 'RB1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| RB1 MUTATED | 4 | 3 | 5 |
| RB1 WILD-TYPE | 54 | 66 | 46 |
P value = 0.107 (Fisher's exact test), Q value = 1
Table S88. Gene #8: 'RB1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| RB1 MUTATED | 0 | 4 | 1 |
| RB1 WILD-TYPE | 27 | 25 | 17 |
P value = 0.656 (Fisher's exact test), Q value = 1
Table S89. Gene #8: 'RB1 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| RB1 MUTATED | 3 | 2 | 1 |
| RB1 WILD-TYPE | 31 | 45 | 30 |
P value = 0.48 (Fisher's exact test), Q value = 1
Table S90. Gene #8: 'RB1 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| RB1 MUTATED | 1 | 0 | 2 | 3 |
| RB1 WILD-TYPE | 28 | 22 | 32 | 24 |
P value = 0.775 (Fisher's exact test), Q value = 1
Table S91. Gene #8: 'RB1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| RB1 MUTATED | 2 | 4 | 3 | 3 |
| RB1 WILD-TYPE | 38 | 58 | 47 | 23 |
P value = 0.241 (Fisher's exact test), Q value = 1
Table S92. Gene #8: 'RB1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| RB1 MUTATED | 2 | 6 | 4 |
| RB1 WILD-TYPE | 66 | 68 | 32 |
P value = 0.322 (Fisher's exact test), Q value = 1
Table S93. Gene #8: 'RB1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| RB1 MUTATED | 7 | 1 | 0 | 2 | 2 |
| RB1 WILD-TYPE | 86 | 13 | 21 | 22 | 8 |
P value = 0.754 (Fisher's exact test), Q value = 1
Table S94. Gene #8: 'RB1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| RB1 MUTATED | 2 | 3 | 6 | 1 |
| RB1 WILD-TYPE | 33 | 48 | 48 | 21 |
P value = 0.489 (Fisher's exact test), Q value = 1
Table S95. Gene #9: 'IBTK MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| IBTK MUTATED | 1 | 2 | 0 | 2 |
| IBTK WILD-TYPE | 36 | 46 | 17 | 17 |
P value = 0.847 (Fisher's exact test), Q value = 1
Table S96. Gene #9: 'IBTK MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| IBTK MUTATED | 1 | 3 | 1 |
| IBTK WILD-TYPE | 40 | 49 | 27 |
P value = 0.739 (Fisher's exact test), Q value = 1
Table S97. Gene #9: 'IBTK MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| IBTK MUTATED | 1 | 3 | 1 |
| IBTK WILD-TYPE | 57 | 66 | 50 |
P value = 0.807 (Fisher's exact test), Q value = 1
Table S98. Gene #9: 'IBTK MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| IBTK MUTATED | 1 | 3 | 1 | 0 |
| IBTK WILD-TYPE | 39 | 59 | 49 | 26 |
P value = 0.842 (Fisher's exact test), Q value = 1
Table S99. Gene #9: 'IBTK MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| IBTK MUTATED | 1 | 3 | 1 |
| IBTK WILD-TYPE | 67 | 71 | 35 |
P value = 1 (Fisher's exact test), Q value = 1
Table S100. Gene #9: 'IBTK MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| IBTK MUTATED | 4 | 0 | 0 | 1 | 0 |
| IBTK WILD-TYPE | 89 | 14 | 21 | 23 | 10 |
P value = 0.608 (Fisher's exact test), Q value = 1
Table S101. Gene #9: 'IBTK MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| IBTK MUTATED | 1 | 3 | 1 | 0 |
| IBTK WILD-TYPE | 34 | 48 | 53 | 22 |
P value = 0.653 (Fisher's exact test), Q value = 1
Table S102. Gene #10: 'CYP11B1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| CYP11B1 MUTATED | 3 | 5 | 3 | 1 |
| CYP11B1 WILD-TYPE | 34 | 43 | 14 | 18 |
P value = 0.677 (Fisher's exact test), Q value = 1
Table S103. Gene #10: 'CYP11B1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| CYP11B1 MUTATED | 3 | 5 | 4 |
| CYP11B1 WILD-TYPE | 38 | 47 | 24 |
P value = 0.659 (Fisher's exact test), Q value = 1
Table S104. Gene #10: 'CYP11B1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| CYP11B1 MUTATED | 4 | 5 | 6 |
| CYP11B1 WILD-TYPE | 54 | 64 | 45 |
P value = 0.125 (Fisher's exact test), Q value = 1
Table S105. Gene #10: 'CYP11B1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| CYP11B1 MUTATED | 3 | 0 | 1 |
| CYP11B1 WILD-TYPE | 24 | 29 | 17 |
P value = 0.24 (Fisher's exact test), Q value = 1
Table S106. Gene #10: 'CYP11B1 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| CYP11B1 MUTATED | 5 | 2 | 2 |
| CYP11B1 WILD-TYPE | 29 | 45 | 29 |
P value = 0.0944 (Fisher's exact test), Q value = 1
Table S107. Gene #10: 'CYP11B1 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| CYP11B1 MUTATED | 4 | 0 | 1 | 4 |
| CYP11B1 WILD-TYPE | 25 | 22 | 33 | 23 |
P value = 0.57 (Fisher's exact test), Q value = 1
Table S108. Gene #10: 'CYP11B1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| CYP11B1 MUTATED | 2 | 4 | 6 | 3 |
| CYP11B1 WILD-TYPE | 38 | 58 | 44 | 23 |
P value = 0.368 (Fisher's exact test), Q value = 1
Table S109. Gene #10: 'CYP11B1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| CYP11B1 MUTATED | 4 | 6 | 5 |
| CYP11B1 WILD-TYPE | 64 | 68 | 31 |
P value = 0.562 (Fisher's exact test), Q value = 1
Table S110. Gene #10: 'CYP11B1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| CYP11B1 MUTATED | 6 | 2 | 1 | 3 | 1 |
| CYP11B1 WILD-TYPE | 87 | 12 | 20 | 21 | 9 |
P value = 0.977 (Fisher's exact test), Q value = 1
Table S111. Gene #10: 'CYP11B1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| CYP11B1 MUTATED | 3 | 4 | 5 | 1 |
| CYP11B1 WILD-TYPE | 32 | 47 | 49 | 21 |
P value = 0.816 (Fisher's exact test), Q value = 1
Table S112. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| NOTCH1 MUTATED | 1 | 4 | 1 | 1 |
| NOTCH1 WILD-TYPE | 36 | 44 | 16 | 18 |
P value = 0.603 (Fisher's exact test), Q value = 1
Table S113. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| NOTCH1 MUTATED | 1 | 4 | 2 |
| NOTCH1 WILD-TYPE | 40 | 48 | 26 |
P value = 0.468 (Fisher's exact test), Q value = 1
Table S114. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| NOTCH1 MUTATED | 3 | 5 | 6 |
| NOTCH1 WILD-TYPE | 55 | 64 | 45 |
P value = 0.0345 (Fisher's exact test), Q value = 1
Table S115. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| NOTCH1 MUTATED | 0 | 4 | 4 |
| NOTCH1 WILD-TYPE | 27 | 25 | 14 |
Figure S22. Get High-res Image Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
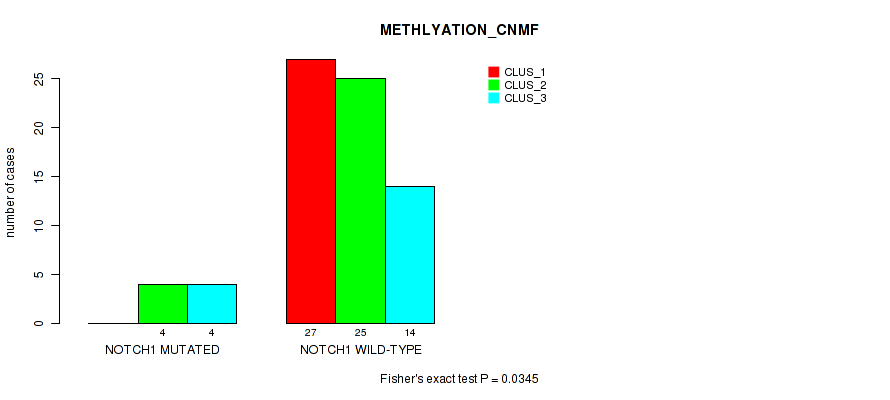
P value = 0.59 (Fisher's exact test), Q value = 1
Table S116. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| NOTCH1 MUTATED | 1 | 4 | 3 |
| NOTCH1 WILD-TYPE | 33 | 43 | 28 |
P value = 0.628 (Fisher's exact test), Q value = 1
Table S117. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| NOTCH1 MUTATED | 2 | 3 | 2 | 1 |
| NOTCH1 WILD-TYPE | 27 | 19 | 32 | 26 |
P value = 0.756 (Fisher's exact test), Q value = 1
Table S118. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| NOTCH1 MUTATED | 2 | 6 | 5 | 1 |
| NOTCH1 WILD-TYPE | 38 | 56 | 45 | 25 |
P value = 0.392 (Fisher's exact test), Q value = 1
Table S119. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| NOTCH1 MUTATED | 3 | 8 | 3 |
| NOTCH1 WILD-TYPE | 65 | 66 | 33 |
P value = 0.349 (Fisher's exact test), Q value = 1
Table S120. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| NOTCH1 MUTATED | 7 | 2 | 0 | 3 | 1 |
| NOTCH1 WILD-TYPE | 86 | 12 | 21 | 21 | 9 |
P value = 0.32 (Fisher's exact test), Q value = 1
Table S121. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| NOTCH1 MUTATED | 4 | 3 | 6 | 0 |
| NOTCH1 WILD-TYPE | 31 | 48 | 48 | 22 |
P value = 0.0103 (Fisher's exact test), Q value = 1
Table S122. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 16 | 19 | 13 |
| NOTCH1 MUTATED | 1 | 0 | 4 |
| NOTCH1 WILD-TYPE | 15 | 19 | 9 |
Figure S23. Get High-res Image Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
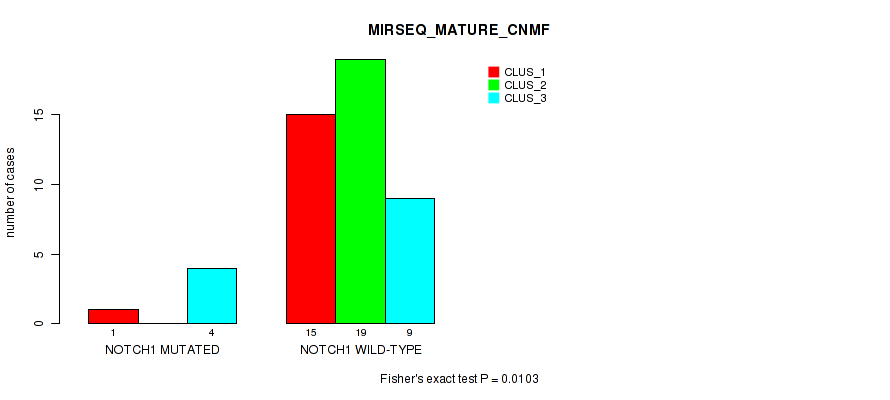
P value = 0.0296 (Fisher's exact test), Q value = 1
Table S123. Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 4 | 19 | 25 |
| NOTCH1 MUTATED | 1 | 4 | 0 |
| NOTCH1 WILD-TYPE | 3 | 15 | 25 |
Figure S24. Get High-res Image Gene #11: 'NOTCH1 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
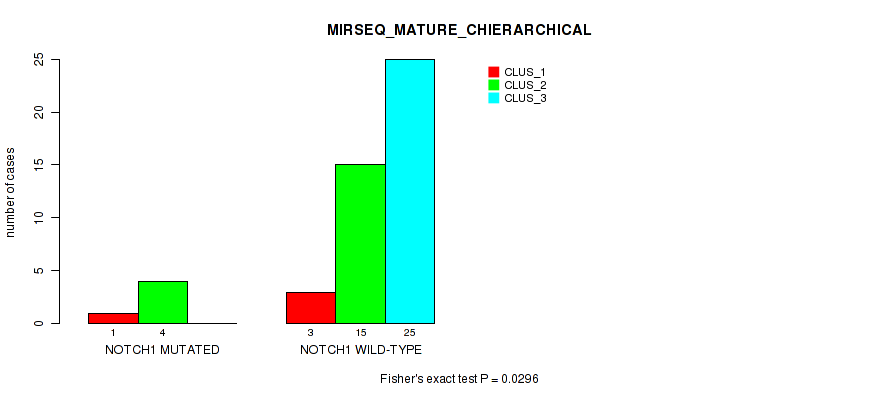
P value = 0.0136 (Fisher's exact test), Q value = 1
Table S124. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| SLC28A1 MUTATED | 0 | 1 | 3 | 0 |
| SLC28A1 WILD-TYPE | 37 | 47 | 14 | 19 |
Figure S25. Get High-res Image Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
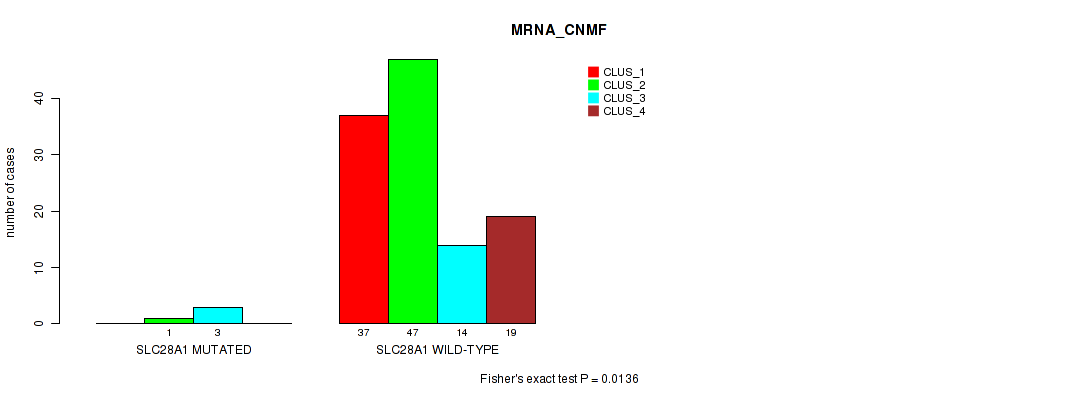
P value = 0.0501 (Fisher's exact test), Q value = 1
Table S125. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| SLC28A1 MUTATED | 0 | 1 | 3 |
| SLC28A1 WILD-TYPE | 41 | 51 | 25 |
P value = 0.0793 (Fisher's exact test), Q value = 1
Table S126. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| SLC28A1 MUTATED | 6 | 1 | 2 |
| SLC28A1 WILD-TYPE | 52 | 68 | 49 |
P value = 0.134 (Fisher's exact test), Q value = 1
Table S127. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| SLC28A1 MUTATED | 3 | 0 | 2 |
| SLC28A1 WILD-TYPE | 24 | 29 | 16 |
P value = 0.32 (Fisher's exact test), Q value = 1
Table S128. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| SLC28A1 MUTATED | 2 | 4 | 0 |
| SLC28A1 WILD-TYPE | 32 | 43 | 31 |
P value = 0.523 (Fisher's exact test), Q value = 1
Table S129. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| SLC28A1 MUTATED | 2 | 1 | 3 | 0 |
| SLC28A1 WILD-TYPE | 27 | 21 | 31 | 27 |
P value = 0.0367 (Fisher's exact test), Q value = 1
Table S130. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| SLC28A1 MUTATED | 3 | 0 | 5 | 1 |
| SLC28A1 WILD-TYPE | 37 | 62 | 45 | 25 |
Figure S26. Get High-res Image Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
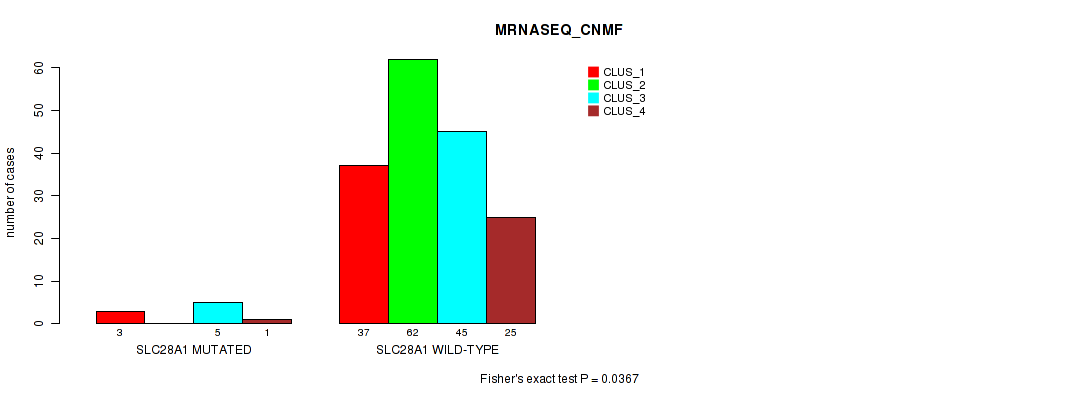
P value = 0.0281 (Fisher's exact test), Q value = 1
Table S131. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| SLC28A1 MUTATED | 3 | 1 | 5 |
| SLC28A1 WILD-TYPE | 65 | 73 | 31 |
Figure S27. Get High-res Image Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
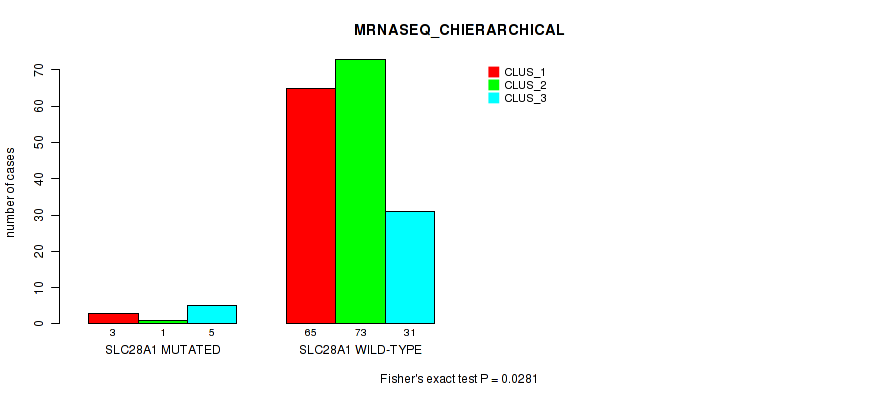
P value = 0.185 (Fisher's exact test), Q value = 1
Table S132. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| SLC28A1 MUTATED | 4 | 1 | 3 | 0 | 0 |
| SLC28A1 WILD-TYPE | 89 | 13 | 18 | 24 | 10 |
P value = 0.0169 (Fisher's exact test), Q value = 1
Table S133. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| SLC28A1 MUTATED | 0 | 1 | 3 | 4 |
| SLC28A1 WILD-TYPE | 35 | 50 | 51 | 18 |
Figure S28. Get High-res Image Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
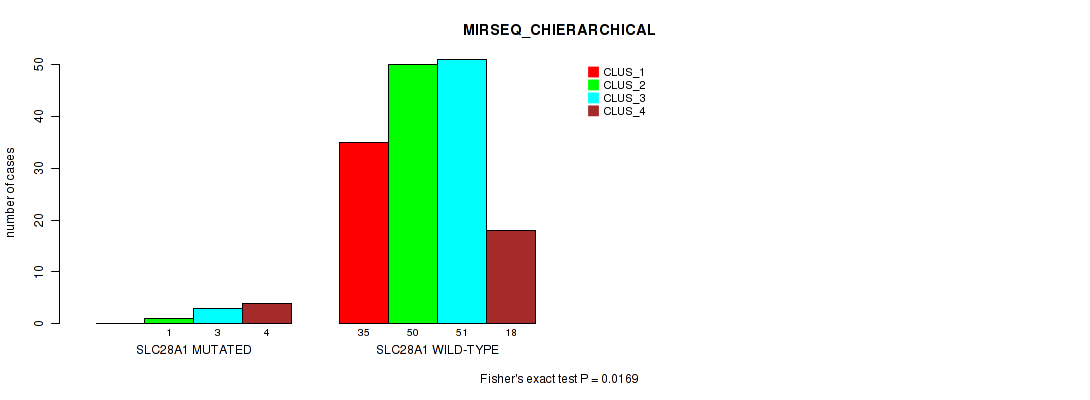
P value = 0.819 (Fisher's exact test), Q value = 1
Table S134. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 16 | 19 | 13 |
| SLC28A1 MUTATED | 2 | 1 | 1 |
| SLC28A1 WILD-TYPE | 14 | 18 | 12 |
P value = 0.183 (Fisher's exact test), Q value = 1
Table S135. Gene #12: 'SLC28A1 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 4 | 19 | 25 |
| SLC28A1 MUTATED | 0 | 0 | 4 |
| SLC28A1 WILD-TYPE | 4 | 19 | 21 |
P value = 0.496 (Fisher's exact test), Q value = 1
Table S136. Gene #13: 'ASB5 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| ASB5 MUTATED | 1 | 4 | 2 | 2 |
| ASB5 WILD-TYPE | 36 | 44 | 15 | 17 |
P value = 0.55 (Fisher's exact test), Q value = 1
Table S137. Gene #13: 'ASB5 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| ASB5 MUTATED | 2 | 4 | 3 |
| ASB5 WILD-TYPE | 39 | 48 | 25 |
P value = 0.263 (Fisher's exact test), Q value = 1
Table S138. Gene #13: 'ASB5 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| ASB5 MUTATED | 2 | 6 | 1 |
| ASB5 WILD-TYPE | 56 | 63 | 50 |
P value = 0.472 (Fisher's exact test), Q value = 1
Table S139. Gene #13: 'ASB5 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| ASB5 MUTATED | 2 | 1 | 0 |
| ASB5 WILD-TYPE | 32 | 46 | 31 |
P value = 1 (Fisher's exact test), Q value = 1
Table S140. Gene #13: 'ASB5 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| ASB5 MUTATED | 1 | 0 | 1 | 1 |
| ASB5 WILD-TYPE | 28 | 22 | 33 | 26 |
P value = 0.48 (Fisher's exact test), Q value = 1
Table S141. Gene #13: 'ASB5 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| ASB5 MUTATED | 1 | 4 | 4 | 0 |
| ASB5 WILD-TYPE | 39 | 58 | 46 | 26 |
P value = 0.497 (Fisher's exact test), Q value = 1
Table S142. Gene #13: 'ASB5 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| ASB5 MUTATED | 2 | 4 | 3 |
| ASB5 WILD-TYPE | 66 | 70 | 33 |
P value = 0.252 (Fisher's exact test), Q value = 1
Table S143. Gene #13: 'ASB5 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| ASB5 MUTATED | 5 | 2 | 0 | 0 | 0 |
| ASB5 WILD-TYPE | 88 | 12 | 21 | 24 | 10 |
P value = 0.337 (Fisher's exact test), Q value = 1
Table S144. Gene #13: 'ASB5 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| ASB5 MUTATED | 0 | 4 | 2 | 1 |
| ASB5 WILD-TYPE | 35 | 47 | 52 | 21 |
P value = 0.768 (Fisher's exact test), Q value = 1
Table S145. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| CPS1 MUTATED | 5 | 8 | 4 | 2 |
| CPS1 WILD-TYPE | 32 | 40 | 13 | 17 |
P value = 0.235 (Fisher's exact test), Q value = 1
Table S146. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| CPS1 MUTATED | 4 | 8 | 7 |
| CPS1 WILD-TYPE | 37 | 44 | 21 |
P value = 0.267 (Fisher's exact test), Q value = 1
Table S147. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| CPS1 MUTATED | 5 | 10 | 10 |
| CPS1 WILD-TYPE | 53 | 59 | 41 |
P value = 0.892 (Fisher's exact test), Q value = 1
Table S148. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| CPS1 MUTATED | 3 | 3 | 1 |
| CPS1 WILD-TYPE | 24 | 26 | 17 |
P value = 0.731 (Fisher's exact test), Q value = 1
Table S149. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| CPS1 MUTATED | 4 | 8 | 3 |
| CPS1 WILD-TYPE | 30 | 39 | 28 |
P value = 0.498 (Fisher's exact test), Q value = 1
Table S150. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| CPS1 MUTATED | 4 | 2 | 7 | 2 |
| CPS1 WILD-TYPE | 25 | 20 | 27 | 25 |
P value = 0.99 (Fisher's exact test), Q value = 1
Table S151. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| CPS1 MUTATED | 5 | 9 | 7 | 4 |
| CPS1 WILD-TYPE | 35 | 53 | 43 | 22 |
P value = 0.726 (Fisher's exact test), Q value = 1
Table S152. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| CPS1 MUTATED | 8 | 12 | 5 |
| CPS1 WILD-TYPE | 60 | 62 | 31 |
P value = 0.0379 (Fisher's exact test), Q value = 1
Table S153. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| CPS1 MUTATED | 7 | 0 | 5 | 5 | 2 |
| CPS1 WILD-TYPE | 86 | 14 | 16 | 19 | 8 |
Figure S29. Get High-res Image Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
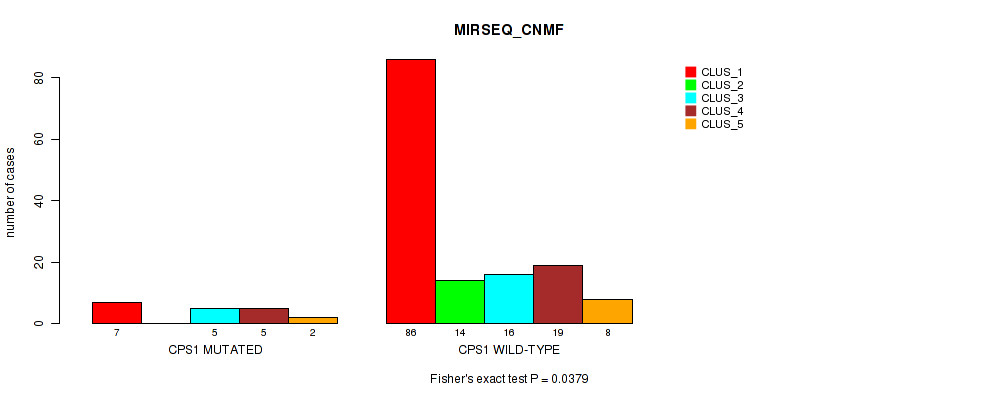
P value = 0.279 (Fisher's exact test), Q value = 1
Table S154. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| CPS1 MUTATED | 6 | 3 | 6 | 4 |
| CPS1 WILD-TYPE | 29 | 48 | 48 | 18 |
P value = 1 (Fisher's exact test), Q value = 1
Table S155. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #11: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 16 | 19 | 13 |
| CPS1 MUTATED | 2 | 2 | 1 |
| CPS1 WILD-TYPE | 14 | 17 | 12 |
P value = 1 (Fisher's exact test), Q value = 1
Table S156. Gene #14: 'CPS1 MUTATION STATUS' versus Molecular Subtype #12: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 4 | 19 | 25 |
| CPS1 MUTATED | 0 | 2 | 3 |
| CPS1 WILD-TYPE | 4 | 17 | 22 |
P value = 0.15 (Fisher's exact test), Q value = 1
Table S157. Gene #15: 'EP300 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| EP300 MUTATED | 5 | 2 | 0 | 0 |
| EP300 WILD-TYPE | 32 | 46 | 17 | 19 |
P value = 0.118 (Fisher's exact test), Q value = 1
Table S158. Gene #15: 'EP300 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| EP300 MUTATED | 5 | 2 | 0 |
| EP300 WILD-TYPE | 36 | 50 | 28 |
P value = 0.437 (Fisher's exact test), Q value = 1
Table S159. Gene #15: 'EP300 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| EP300 MUTATED | 2 | 2 | 4 |
| EP300 WILD-TYPE | 56 | 67 | 47 |
P value = 0.357 (Fisher's exact test), Q value = 1
Table S160. Gene #15: 'EP300 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| EP300 MUTATED | 0 | 1 | 2 |
| EP300 WILD-TYPE | 34 | 46 | 29 |
P value = 1 (Fisher's exact test), Q value = 1
Table S161. Gene #15: 'EP300 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| EP300 MUTATED | 1 | 0 | 1 | 1 |
| EP300 WILD-TYPE | 28 | 22 | 33 | 26 |
P value = 0.0811 (Fisher's exact test), Q value = 1
Table S162. Gene #15: 'EP300 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| EP300 MUTATED | 3 | 0 | 3 | 2 |
| EP300 WILD-TYPE | 37 | 62 | 47 | 24 |
P value = 0.083 (Fisher's exact test), Q value = 1
Table S163. Gene #15: 'EP300 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| EP300 MUTATED | 6 | 1 | 1 |
| EP300 WILD-TYPE | 62 | 73 | 35 |
P value = 0.57 (Fisher's exact test), Q value = 1
Table S164. Gene #15: 'EP300 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| EP300 MUTATED | 6 | 1 | 0 | 0 | 0 |
| EP300 WILD-TYPE | 87 | 13 | 21 | 24 | 10 |
P value = 0.822 (Fisher's exact test), Q value = 1
Table S165. Gene #15: 'EP300 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| EP300 MUTATED | 1 | 3 | 3 | 0 |
| EP300 WILD-TYPE | 34 | 48 | 51 | 22 |
P value = 0.313 (Fisher's exact test), Q value = 1
Table S166. Gene #16: 'ARID1A MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| ARID1A MUTATED | 1 | 4 | 1 | 3 |
| ARID1A WILD-TYPE | 36 | 44 | 16 | 16 |
P value = 0.552 (Fisher's exact test), Q value = 1
Table S167. Gene #16: 'ARID1A MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| ARID1A MUTATED | 2 | 4 | 3 |
| ARID1A WILD-TYPE | 39 | 48 | 25 |
P value = 0.452 (Fisher's exact test), Q value = 1
Table S168. Gene #16: 'ARID1A MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| ARID1A MUTATED | 6 | 4 | 2 |
| ARID1A WILD-TYPE | 52 | 65 | 49 |
P value = 0.354 (Fisher's exact test), Q value = 1
Table S169. Gene #16: 'ARID1A MUTATION STATUS' versus Molecular Subtype #4: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 27 | 29 | 18 |
| ARID1A MUTATED | 3 | 1 | 0 |
| ARID1A WILD-TYPE | 24 | 28 | 18 |
P value = 0.495 (Fisher's exact test), Q value = 1
Table S170. Gene #16: 'ARID1A MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| ARID1A MUTATED | 1 | 2 | 3 |
| ARID1A WILD-TYPE | 33 | 45 | 28 |
P value = 0.601 (Fisher's exact test), Q value = 1
Table S171. Gene #16: 'ARID1A MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| ARID1A MUTATED | 1 | 1 | 1 | 3 |
| ARID1A WILD-TYPE | 28 | 21 | 33 | 24 |
P value = 0.111 (Fisher's exact test), Q value = 1
Table S172. Gene #16: 'ARID1A MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| ARID1A MUTATED | 0 | 5 | 6 | 1 |
| ARID1A WILD-TYPE | 40 | 57 | 44 | 25 |
P value = 0.862 (Fisher's exact test), Q value = 1
Table S173. Gene #16: 'ARID1A MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| ARID1A MUTATED | 4 | 5 | 3 |
| ARID1A WILD-TYPE | 64 | 69 | 33 |
P value = 0.53 (Fisher's exact test), Q value = 1
Table S174. Gene #16: 'ARID1A MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| ARID1A MUTATED | 4 | 1 | 2 | 2 | 1 |
| ARID1A WILD-TYPE | 89 | 13 | 19 | 22 | 9 |
P value = 0.692 (Fisher's exact test), Q value = 1
Table S175. Gene #16: 'ARID1A MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| ARID1A MUTATED | 3 | 2 | 3 | 2 |
| ARID1A WILD-TYPE | 32 | 49 | 51 | 20 |
P value = 0.493 (Fisher's exact test), Q value = 1
Table S176. Gene #17: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #1: 'MRNA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 37 | 48 | 17 | 19 |
| FBXW7 MUTATED | 1 | 4 | 2 | 2 |
| FBXW7 WILD-TYPE | 36 | 44 | 15 | 17 |
P value = 0.83 (Fisher's exact test), Q value = 1
Table S177. Gene #17: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #2: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 52 | 28 |
| FBXW7 MUTATED | 2 | 5 | 2 |
| FBXW7 WILD-TYPE | 39 | 47 | 26 |
P value = 0.07 (Fisher's exact test), Q value = 1
Table S178. Gene #17: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #3: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 58 | 69 | 51 |
| FBXW7 MUTATED | 4 | 6 | 0 |
| FBXW7 WILD-TYPE | 54 | 63 | 51 |
P value = 0.623 (Fisher's exact test), Q value = 1
Table S179. Gene #17: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #5: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 34 | 47 | 31 |
| FBXW7 MUTATED | 0 | 2 | 1 |
| FBXW7 WILD-TYPE | 34 | 45 | 30 |
P value = 0.788 (Fisher's exact test), Q value = 1
Table S180. Gene #17: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #6: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 29 | 22 | 34 | 27 |
| FBXW7 MUTATED | 0 | 1 | 1 | 1 |
| FBXW7 WILD-TYPE | 29 | 21 | 33 | 26 |
P value = 0.833 (Fisher's exact test), Q value = 1
Table S181. Gene #17: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #7: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 40 | 62 | 50 | 26 |
| FBXW7 MUTATED | 2 | 5 | 2 | 1 |
| FBXW7 WILD-TYPE | 38 | 57 | 48 | 25 |
P value = 0.526 (Fisher's exact test), Q value = 1
Table S182. Gene #17: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #8: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 68 | 74 | 36 |
| FBXW7 MUTATED | 3 | 6 | 1 |
| FBXW7 WILD-TYPE | 65 | 68 | 35 |
P value = 0.962 (Fisher's exact test), Q value = 1
Table S183. Gene #17: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #9: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 93 | 14 | 21 | 24 | 10 |
| FBXW7 MUTATED | 5 | 1 | 1 | 1 | 0 |
| FBXW7 WILD-TYPE | 88 | 13 | 20 | 23 | 10 |
P value = 0.0672 (Fisher's exact test), Q value = 1
Table S184. Gene #17: 'FBXW7 MUTATION STATUS' versus Molecular Subtype #10: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 35 | 51 | 54 | 22 |
| FBXW7 MUTATED | 4 | 3 | 0 | 1 |
| FBXW7 WILD-TYPE | 31 | 48 | 54 | 21 |
-
Mutation data file = transformed.cor.cli.txt
-
Molecular subtypes file = LUSC-TP.transferedmergedcluster.txt
-
Number of patients = 178
-
Number of significantly mutated genes = 17
-
Number of Molecular subtypes = 12
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.