This pipeline computes the correlation between significant copy number variation (cnv focal) genes and molecular subtypes.
Testing the association between copy number variation 28 focal events and 8 molecular subtypes across 162 patients, 33 significant findings detected with P value < 0.05 and Q value < 0.25.
-
amp_17q21.31 cnv correlated to 'MRNASEQ_CHIERARCHICAL'.
-
del_1p12 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
del_3p24.1 cnv correlated to 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
del_3q26.1 cnv correlated to 'CN_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
del_11p15.4 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
del_11q22.1 cnv correlated to 'CN_CNMF', 'MRNASEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
del_17p13.2 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
del_17q11.2 cnv correlated to 'CN_CNMF'.
-
del_22q13.31 cnv correlated to 'CN_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
del_xp21.1 cnv correlated to 'CN_CNMF' and 'MIRSEQ_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 28 focal events and 8 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 33 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| del 11p15 4 | 58 (36%) | 104 |
1e-05 (0.00224) |
2e-05 (0.00422) |
0.00057 (0.112) |
1e-05 (0.00224) |
0.00013 (0.0266) |
1e-05 (0.00224) |
1e-05 (0.00224) |
0.00038 (0.0768) |
| del 3p24 1 | 64 (40%) | 98 |
0.00156 (0.298) |
0.00744 (1.00) |
5e-05 (0.0103) |
3e-05 (0.00621) |
1e-05 (0.00224) |
0.00022 (0.0447) |
1e-05 (0.00224) |
2e-05 (0.00422) |
| del 1p12 | 113 (70%) | 49 |
1e-05 (0.00224) |
2e-05 (0.00422) |
1e-05 (0.00224) |
1e-05 (0.00224) |
0.00066 (0.129) |
0.0167 (1.00) |
0.0074 (1.00) |
0.0153 (1.00) |
| del 11q22 1 | 48 (30%) | 114 |
1e-05 (0.00224) |
0.209 (1.00) |
0.00066 (0.129) |
0.0101 (1.00) |
0.00346 (0.65) |
0.0007 (0.136) |
0.00038 (0.0768) |
0.0311 (1.00) |
| del 3q26 1 | 93 (57%) | 69 |
2e-05 (0.00422) |
0.517 (1.00) |
0.0238 (1.00) |
1e-05 (0.00224) |
0.0192 (1.00) |
0.00951 (1.00) |
0.0115 (1.00) |
0.0108 (1.00) |
| del 17p13 2 | 64 (40%) | 98 |
0.00017 (0.0347) |
0.00042 (0.084) |
0.016 (1.00) |
0.00619 (1.00) |
0.0272 (1.00) |
0.00788 (1.00) |
0.171 (1.00) |
0.0169 (1.00) |
| del 22q13 31 | 65 (40%) | 97 |
1e-05 (0.00224) |
0.0203 (1.00) |
0.0203 (1.00) |
0.00047 (0.0935) |
0.011 (1.00) |
0.0267 (1.00) |
0.0162 (1.00) |
0.0192 (1.00) |
| del xp21 1 | 46 (28%) | 116 |
0.00099 (0.19) |
0.00593 (1.00) |
0.0336 (1.00) |
0.00388 (0.722) |
0.00072 (0.139) |
0.0451 (1.00) |
0.185 (1.00) |
0.0335 (1.00) |
| amp 17q21 31 | 16 (10%) | 146 |
0.628 (1.00) |
0.89 (1.00) |
0.536 (1.00) |
0.00053 (0.105) |
0.753 (1.00) |
0.143 (1.00) |
0.0663 (1.00) |
0.455 (1.00) |
| del 17q11 2 | 42 (26%) | 120 |
1e-05 (0.00224) |
0.00371 (0.694) |
0.0242 (1.00) |
0.00289 (0.546) |
0.103 (1.00) |
0.0713 (1.00) |
0.368 (1.00) |
0.14 (1.00) |
| amp 1q21 3 | 30 (19%) | 132 |
0.00488 (0.903) |
0.609 (1.00) |
0.637 (1.00) |
0.941 (1.00) |
0.902 (1.00) |
0.447 (1.00) |
0.693 (1.00) |
0.825 (1.00) |
| amp 4q25 | 6 (4%) | 156 |
0.882 (1.00) |
0.761 (1.00) |
0.13 (1.00) |
0.388 (1.00) |
0.766 (1.00) |
0.624 (1.00) |
0.293 (1.00) |
0.504 (1.00) |
| amp 4q31 1 | 13 (8%) | 149 |
0.00548 (1.00) |
0.0661 (1.00) |
0.613 (1.00) |
0.00781 (1.00) |
0.405 (1.00) |
0.228 (1.00) |
0.115 (1.00) |
0.323 (1.00) |
| amp 11p15 2 | 8 (5%) | 154 |
0.0936 (1.00) |
0.0782 (1.00) |
0.332 (1.00) |
0.104 (1.00) |
0.41 (1.00) |
0.399 (1.00) |
0.458 (1.00) |
0.81 (1.00) |
| amp 12q13 3 | 16 (10%) | 146 |
0.0449 (1.00) |
0.139 (1.00) |
0.61 (1.00) |
0.62 (1.00) |
0.0466 (1.00) |
0.274 (1.00) |
0.129 (1.00) |
0.323 (1.00) |
| amp 14q24 3 | 10 (6%) | 152 |
0.0298 (1.00) |
0.199 (1.00) |
0.852 (1.00) |
0.173 (1.00) |
0.457 (1.00) |
0.711 (1.00) |
0.443 (1.00) |
0.858 (1.00) |
| del 1q42 13 | 18 (11%) | 144 |
0.0219 (1.00) |
0.136 (1.00) |
0.025 (1.00) |
0.178 (1.00) |
0.333 (1.00) |
0.536 (1.00) |
0.0729 (1.00) |
0.247 (1.00) |
| del 4q28 3 | 10 (6%) | 152 |
0.0734 (1.00) |
0.242 (1.00) |
0.0182 (1.00) |
0.338 (1.00) |
0.457 (1.00) |
0.715 (1.00) |
0.325 (1.00) |
0.321 (1.00) |
| del 5q15 | 12 (7%) | 150 |
0.596 (1.00) |
1 (1.00) |
0.975 (1.00) |
1 (1.00) |
0.928 (1.00) |
0.187 (1.00) |
0.244 (1.00) |
0.294 (1.00) |
| del 6p12 3 | 10 (6%) | 152 |
0.0113 (1.00) |
0.266 (1.00) |
0.0205 (1.00) |
0.0864 (1.00) |
0.197 (1.00) |
0.163 (1.00) |
0.929 (1.00) |
0.0513 (1.00) |
| del 6q16 1 | 24 (15%) | 138 |
0.136 (1.00) |
0.0765 (1.00) |
0.0263 (1.00) |
0.00747 (1.00) |
0.0152 (1.00) |
0.0057 (1.00) |
0.0829 (1.00) |
0.0959 (1.00) |
| del 8p22 | 25 (15%) | 137 |
0.00167 (0.317) |
0.0833 (1.00) |
0.00636 (1.00) |
0.00903 (1.00) |
0.209 (1.00) |
0.239 (1.00) |
0.027 (1.00) |
0.243 (1.00) |
| del 8q23 3 | 13 (8%) | 149 |
0.0326 (1.00) |
0.868 (1.00) |
0.275 (1.00) |
0.518 (1.00) |
0.0718 (1.00) |
0.465 (1.00) |
0.929 (1.00) |
0.297 (1.00) |
| del 9p24 2 | 15 (9%) | 147 |
0.288 (1.00) |
0.34 (1.00) |
0.0914 (1.00) |
0.177 (1.00) |
0.481 (1.00) |
0.0779 (1.00) |
0.482 (1.00) |
0.305 (1.00) |
| del 9q21 12 | 14 (9%) | 148 |
0.00968 (1.00) |
0.549 (1.00) |
0.369 (1.00) |
0.389 (1.00) |
0.726 (1.00) |
0.111 (1.00) |
0.585 (1.00) |
0.343 (1.00) |
| del 12q21 33 | 8 (5%) | 154 |
0.0599 (1.00) |
0.892 (1.00) |
0.913 (1.00) |
0.877 (1.00) |
0.592 (1.00) |
0.697 (1.00) |
0.835 (1.00) |
1 (1.00) |
| del 13q22 3 | 8 (5%) | 154 |
0.249 (1.00) |
0.893 (1.00) |
0.272 (1.00) |
0.429 (1.00) |
1 (1.00) |
0.611 (1.00) |
0.719 (1.00) |
0.856 (1.00) |
| del 16q21 | 5 (3%) | 157 |
0.0662 (1.00) |
0.115 (1.00) |
0.551 (1.00) |
0.0261 (1.00) |
0.212 (1.00) |
0.59 (1.00) |
0.0901 (1.00) |
0.828 (1.00) |
P value = 0.00053 (Fisher's exact test), Q value = 0.1
Table S1. Gene #7: 'amp_17q21.31' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 34 | 64 | 43 | 21 |
| AMP PEAK 7(17Q21.31) MUTATED | 10 | 5 | 1 | 0 |
| AMP PEAK 7(17Q21.31) WILD-TYPE | 24 | 59 | 42 | 21 |
Figure S1. Get High-res Image Gene #7: 'amp_17q21.31' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
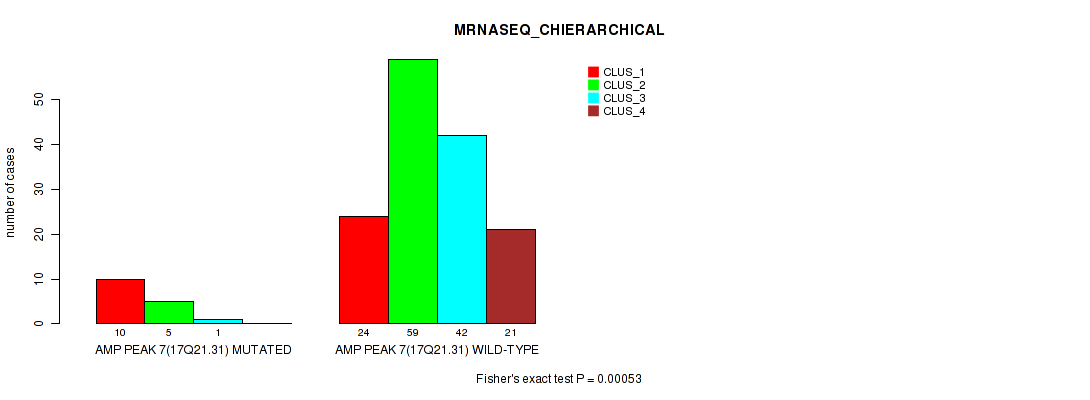
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S2. Gene #8: 'del_1p12' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 60 | 63 | 39 |
| DEL PEAK 1(1P12) MUTATED | 54 | 27 | 32 |
| DEL PEAK 1(1P12) WILD-TYPE | 6 | 36 | 7 |
Figure S2. Get High-res Image Gene #8: 'del_1p12' versus Molecular Subtype #1: 'CN_CNMF'
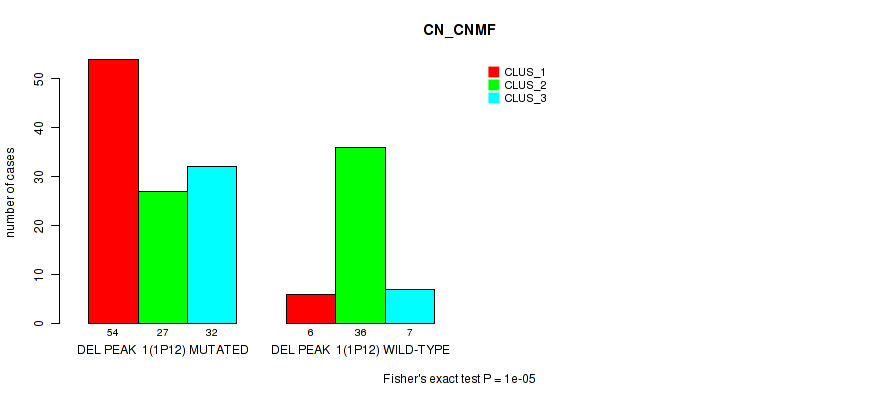
P value = 2e-05 (Fisher's exact test), Q value = 0.0042
Table S3. Gene #8: 'del_1p12' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 40 | 78 | 44 |
| DEL PEAK 1(1P12) MUTATED | 24 | 68 | 21 |
| DEL PEAK 1(1P12) WILD-TYPE | 16 | 10 | 23 |
Figure S3. Get High-res Image Gene #8: 'del_1p12' versus Molecular Subtype #2: 'METHLYATION_CNMF'
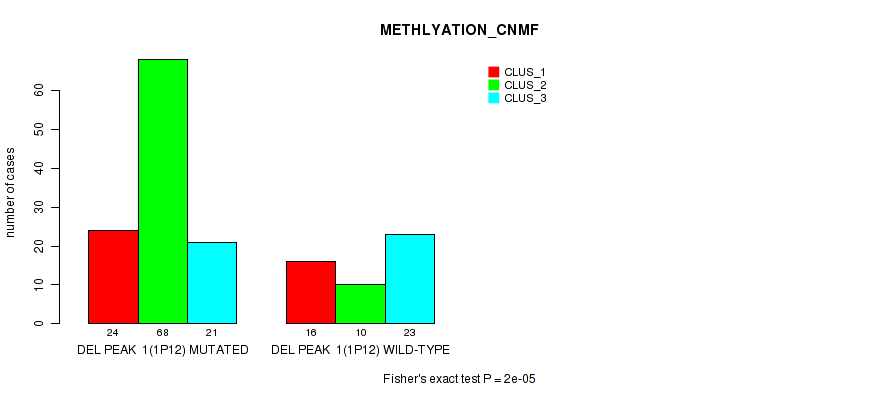
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S4. Gene #8: 'del_1p12' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 64 | 44 | 21 |
| DEL PEAK 1(1P12) MUTATED | 24 | 58 | 21 | 10 |
| DEL PEAK 1(1P12) WILD-TYPE | 9 | 6 | 23 | 11 |
Figure S4. Get High-res Image Gene #8: 'del_1p12' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
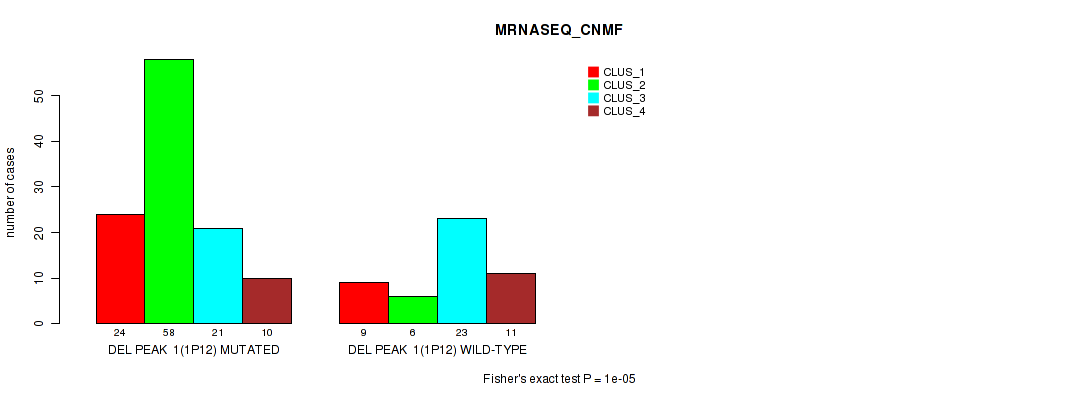
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S5. Gene #8: 'del_1p12' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 34 | 64 | 43 | 21 |
| DEL PEAK 1(1P12) MUTATED | 19 | 58 | 27 | 9 |
| DEL PEAK 1(1P12) WILD-TYPE | 15 | 6 | 16 | 12 |
Figure S5. Get High-res Image Gene #8: 'del_1p12' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
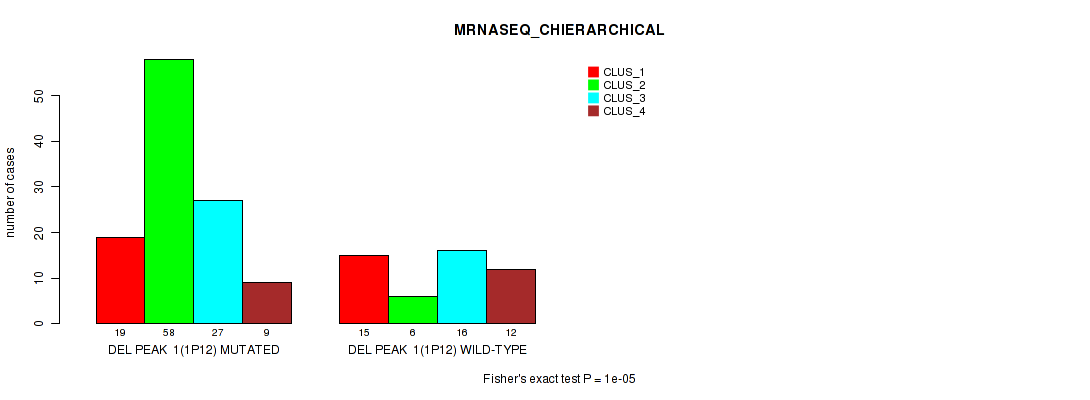
P value = 0.00066 (Fisher's exact test), Q value = 0.13
Table S6. Gene #8: 'del_1p12' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 58 | 39 |
| DEL PEAK 1(1P12) MUTATED | 39 | 51 | 23 |
| DEL PEAK 1(1P12) WILD-TYPE | 26 | 7 | 16 |
Figure S6. Get High-res Image Gene #8: 'del_1p12' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
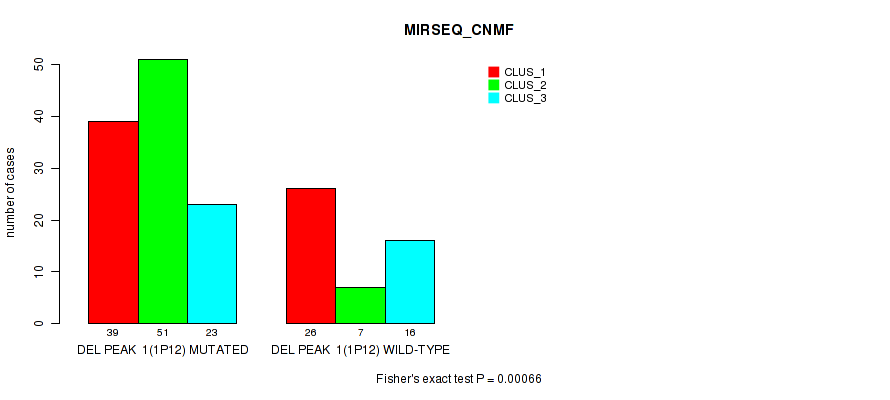
P value = 5e-05 (Fisher's exact test), Q value = 0.01
Table S7. Gene #10: 'del_3p24.1' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 64 | 44 | 21 |
| DEL PEAK 3(3P24.1) MUTATED | 18 | 15 | 27 | 4 |
| DEL PEAK 3(3P24.1) WILD-TYPE | 15 | 49 | 17 | 17 |
Figure S7. Get High-res Image Gene #10: 'del_3p24.1' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
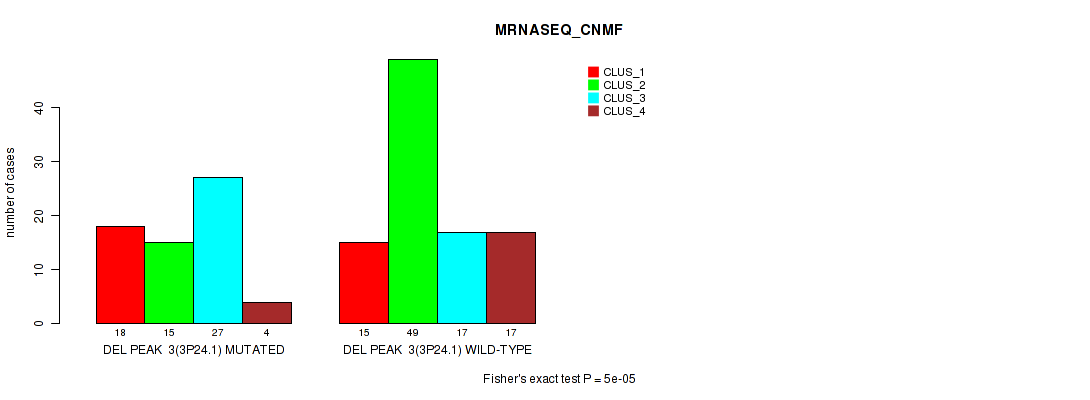
P value = 3e-05 (Fisher's exact test), Q value = 0.0062
Table S8. Gene #10: 'del_3p24.1' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 34 | 64 | 43 | 21 |
| DEL PEAK 3(3P24.1) MUTATED | 12 | 17 | 31 | 4 |
| DEL PEAK 3(3P24.1) WILD-TYPE | 22 | 47 | 12 | 17 |
Figure S8. Get High-res Image Gene #10: 'del_3p24.1' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
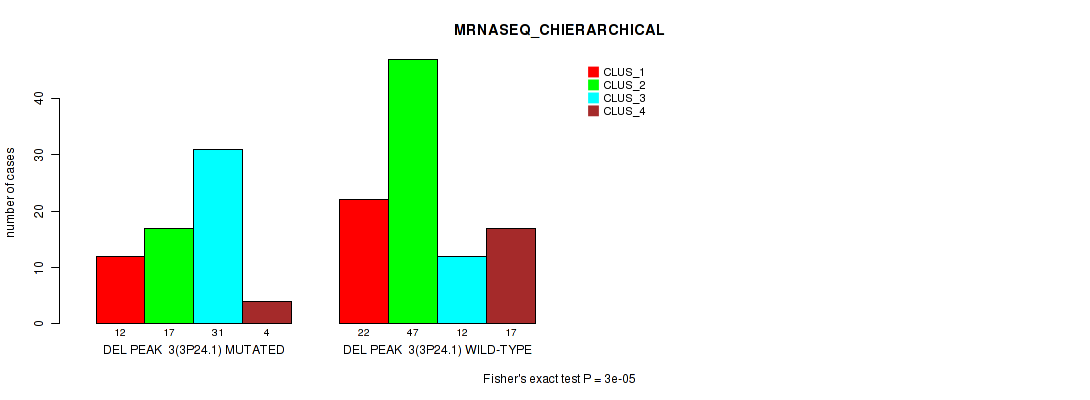
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S9. Gene #10: 'del_3p24.1' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 58 | 39 |
| DEL PEAK 3(3P24.1) MUTATED | 39 | 19 | 6 |
| DEL PEAK 3(3P24.1) WILD-TYPE | 26 | 39 | 33 |
Figure S9. Get High-res Image Gene #10: 'del_3p24.1' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
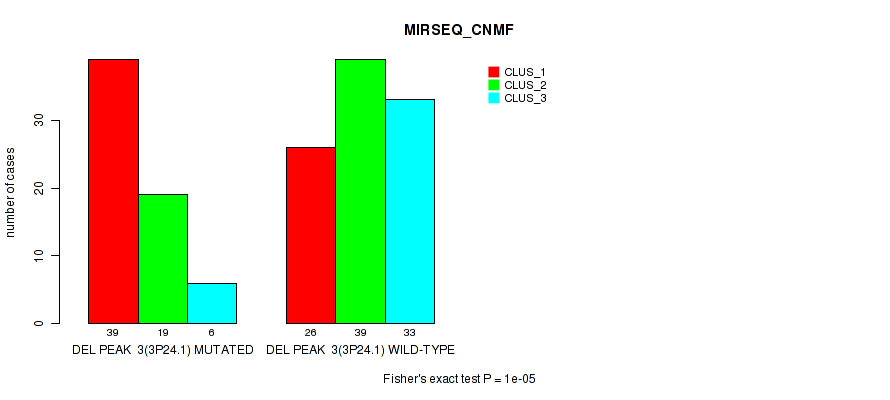
P value = 0.00022 (Fisher's exact test), Q value = 0.045
Table S10. Gene #10: 'del_3p24.1' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 66 | 54 | 18 | 24 |
| DEL PEAK 3(3P24.1) MUTATED | 39 | 15 | 6 | 4 |
| DEL PEAK 3(3P24.1) WILD-TYPE | 27 | 39 | 12 | 20 |
Figure S10. Get High-res Image Gene #10: 'del_3p24.1' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
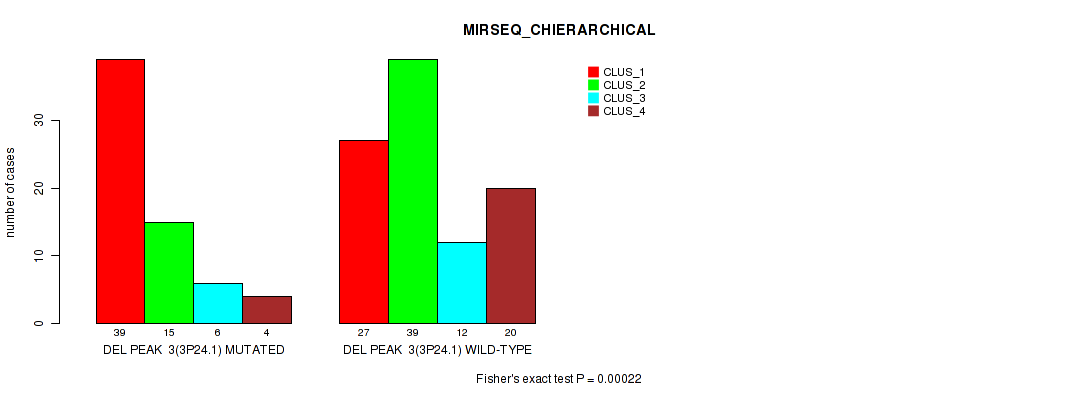
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S11. Gene #10: 'del_3p24.1' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 34 | 58 | 11 | 35 | 23 |
| DEL PEAK 3(3P24.1) MUTATED | 12 | 15 | 2 | 29 | 6 |
| DEL PEAK 3(3P24.1) WILD-TYPE | 22 | 43 | 9 | 6 | 17 |
Figure S11. Get High-res Image Gene #10: 'del_3p24.1' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
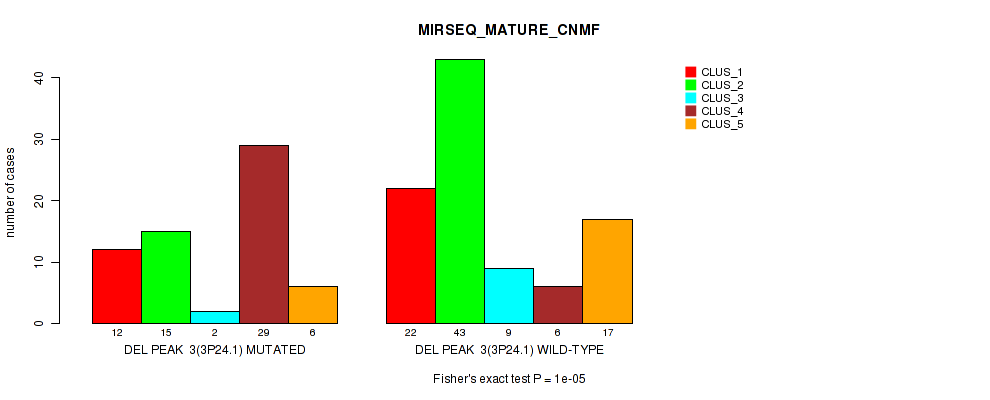
P value = 2e-05 (Fisher's exact test), Q value = 0.0042
Table S12. Gene #10: 'del_3p24.1' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 71 | 53 | 18 | 19 |
| DEL PEAK 3(3P24.1) MUTATED | 18 | 36 | 6 | 4 |
| DEL PEAK 3(3P24.1) WILD-TYPE | 53 | 17 | 12 | 15 |
Figure S12. Get High-res Image Gene #10: 'del_3p24.1' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
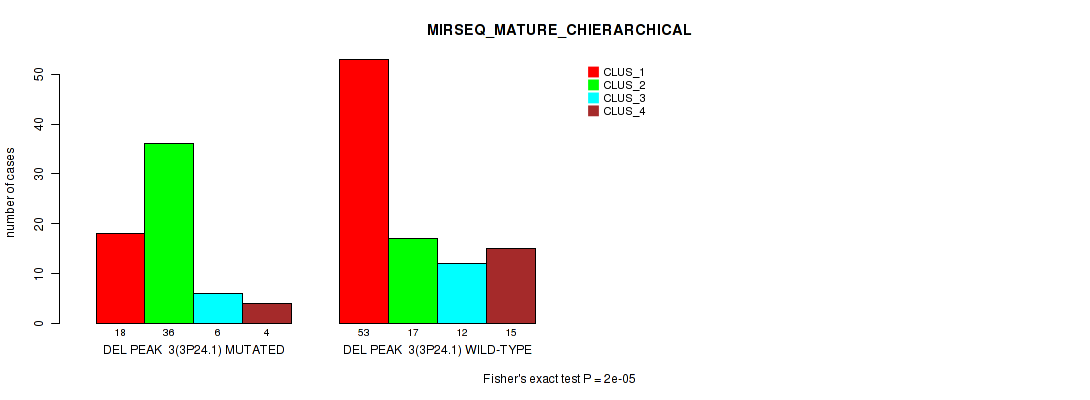
P value = 2e-05 (Fisher's exact test), Q value = 0.0042
Table S13. Gene #11: 'del_3q26.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 60 | 63 | 39 |
| DEL PEAK 4(3Q26.1) MUTATED | 40 | 22 | 31 |
| DEL PEAK 4(3Q26.1) WILD-TYPE | 20 | 41 | 8 |
Figure S13. Get High-res Image Gene #11: 'del_3q26.1' versus Molecular Subtype #1: 'CN_CNMF'
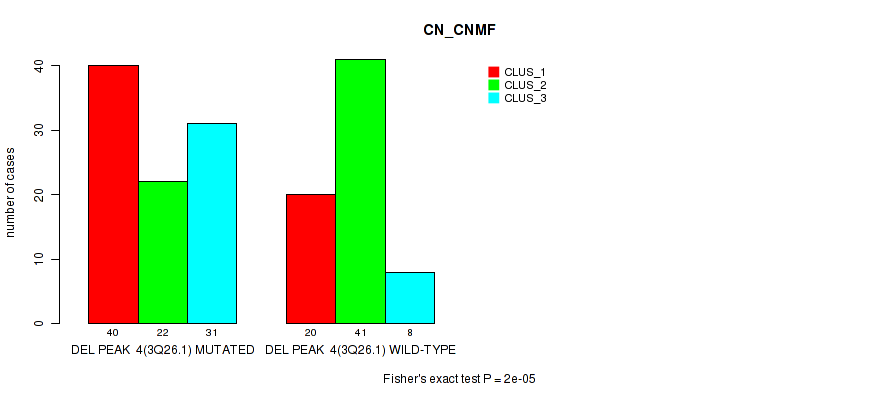
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S14. Gene #11: 'del_3q26.1' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 34 | 64 | 43 | 21 |
| DEL PEAK 4(3Q26.1) MUTATED | 6 | 47 | 33 | 7 |
| DEL PEAK 4(3Q26.1) WILD-TYPE | 28 | 17 | 10 | 14 |
Figure S14. Get High-res Image Gene #11: 'del_3q26.1' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
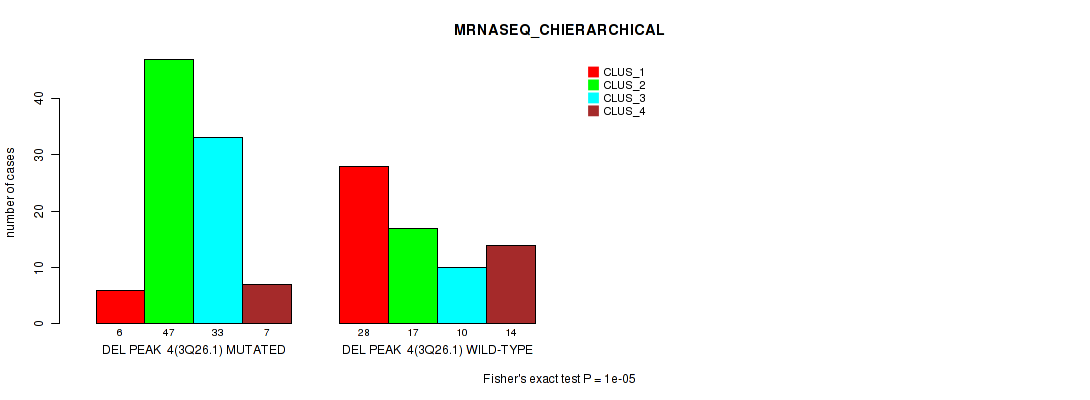
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S15. Gene #20: 'del_11p15.4' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 60 | 63 | 39 |
| DEL PEAK 13(11P15.4) MUTATED | 2 | 39 | 17 |
| DEL PEAK 13(11P15.4) WILD-TYPE | 58 | 24 | 22 |
Figure S15. Get High-res Image Gene #20: 'del_11p15.4' versus Molecular Subtype #1: 'CN_CNMF'
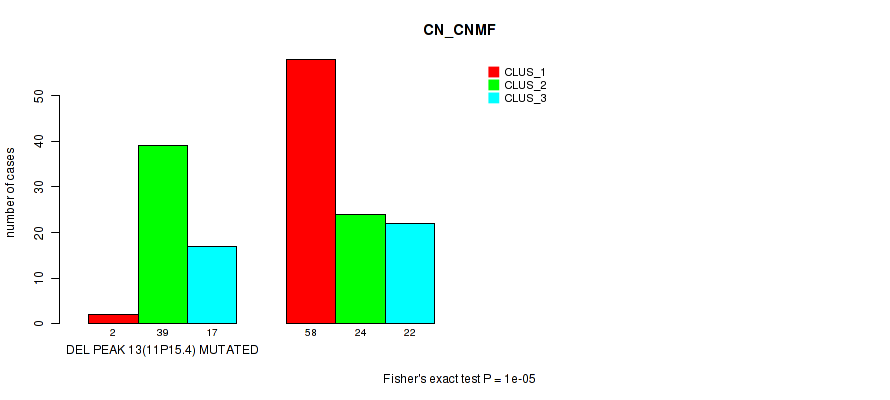
P value = 2e-05 (Fisher's exact test), Q value = 0.0042
Table S16. Gene #20: 'del_11p15.4' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 40 | 78 | 44 |
| DEL PEAK 13(11P15.4) MUTATED | 23 | 14 | 21 |
| DEL PEAK 13(11P15.4) WILD-TYPE | 17 | 64 | 23 |
Figure S16. Get High-res Image Gene #20: 'del_11p15.4' versus Molecular Subtype #2: 'METHLYATION_CNMF'
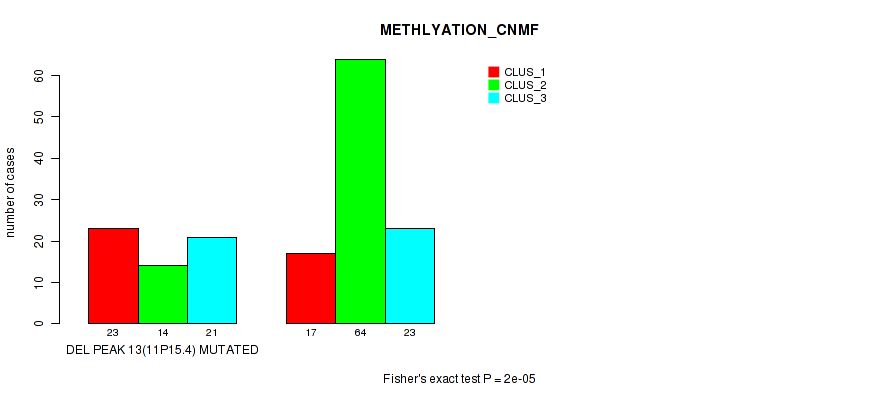
P value = 0.00057 (Fisher's exact test), Q value = 0.11
Table S17. Gene #20: 'del_11p15.4' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 64 | 44 | 21 |
| DEL PEAK 13(11P15.4) MUTATED | 13 | 13 | 26 | 6 |
| DEL PEAK 13(11P15.4) WILD-TYPE | 20 | 51 | 18 | 15 |
Figure S17. Get High-res Image Gene #20: 'del_11p15.4' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
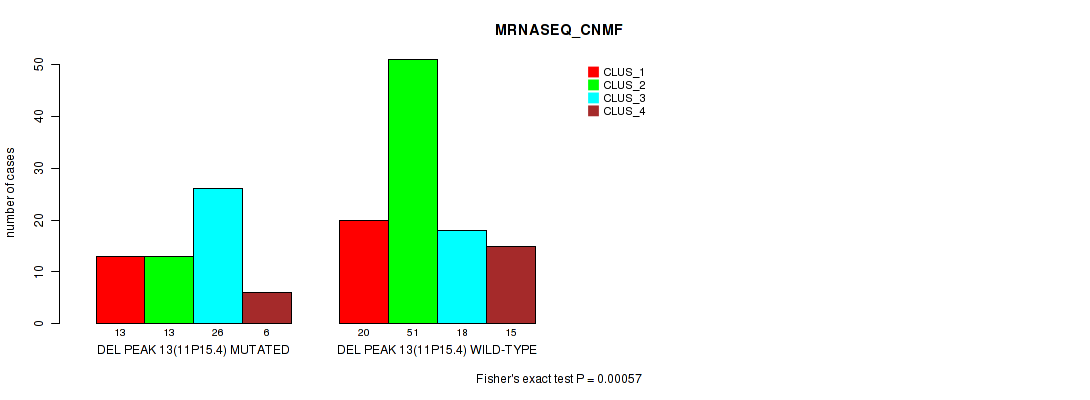
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S18. Gene #20: 'del_11p15.4' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 34 | 64 | 43 | 21 |
| DEL PEAK 13(11P15.4) MUTATED | 11 | 10 | 31 | 6 |
| DEL PEAK 13(11P15.4) WILD-TYPE | 23 | 54 | 12 | 15 |
Figure S18. Get High-res Image Gene #20: 'del_11p15.4' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
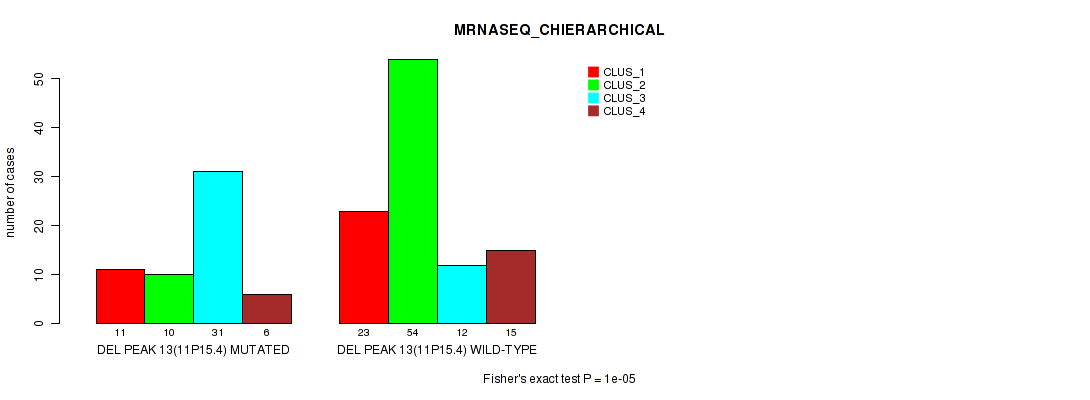
P value = 0.00013 (Fisher's exact test), Q value = 0.027
Table S19. Gene #20: 'del_11p15.4' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 58 | 39 |
| DEL PEAK 13(11P15.4) MUTATED | 36 | 12 | 10 |
| DEL PEAK 13(11P15.4) WILD-TYPE | 29 | 46 | 29 |
Figure S19. Get High-res Image Gene #20: 'del_11p15.4' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
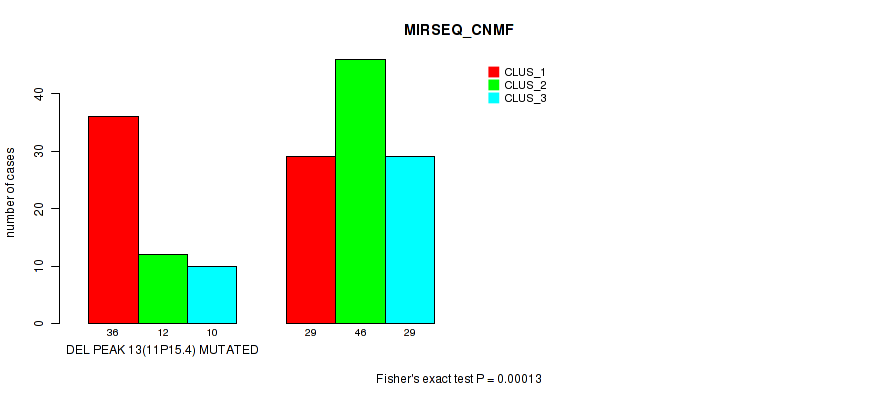
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S20. Gene #20: 'del_11p15.4' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 66 | 54 | 18 | 24 |
| DEL PEAK 13(11P15.4) MUTATED | 39 | 8 | 5 | 6 |
| DEL PEAK 13(11P15.4) WILD-TYPE | 27 | 46 | 13 | 18 |
Figure S20. Get High-res Image Gene #20: 'del_11p15.4' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
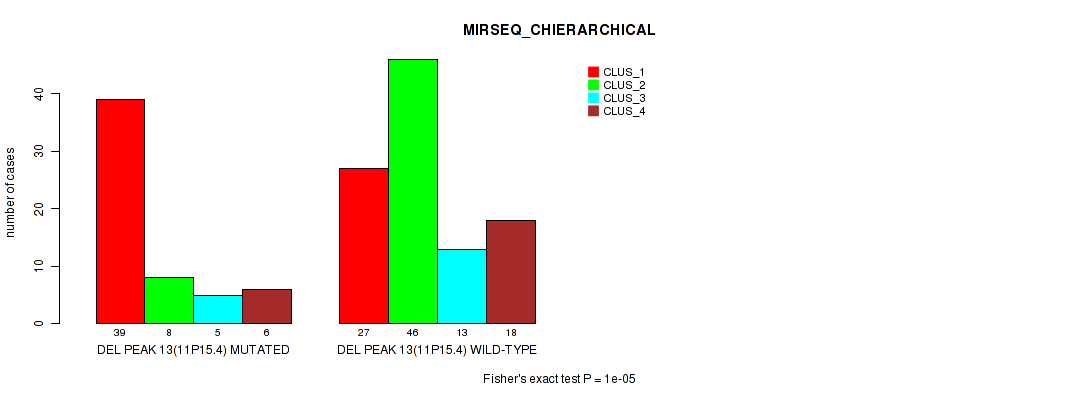
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S21. Gene #20: 'del_11p15.4' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 34 | 58 | 11 | 35 | 23 |
| DEL PEAK 13(11P15.4) MUTATED | 12 | 8 | 2 | 24 | 11 |
| DEL PEAK 13(11P15.4) WILD-TYPE | 22 | 50 | 9 | 11 | 12 |
Figure S21. Get High-res Image Gene #20: 'del_11p15.4' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
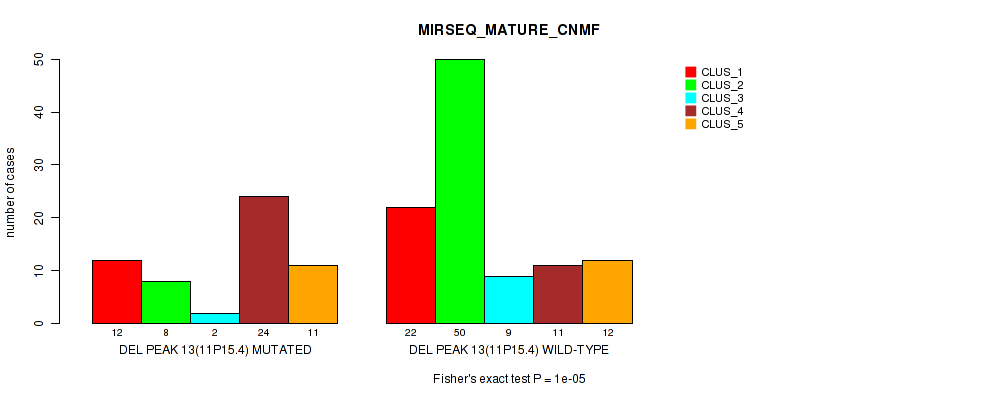
P value = 0.00038 (Fisher's exact test), Q value = 0.077
Table S22. Gene #20: 'del_11p15.4' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 71 | 53 | 18 | 19 |
| DEL PEAK 13(11P15.4) MUTATED | 16 | 31 | 5 | 5 |
| DEL PEAK 13(11P15.4) WILD-TYPE | 55 | 22 | 13 | 14 |
Figure S22. Get High-res Image Gene #20: 'del_11p15.4' versus Molecular Subtype #8: 'MIRSEQ_MATURE_CHIERARCHICAL'
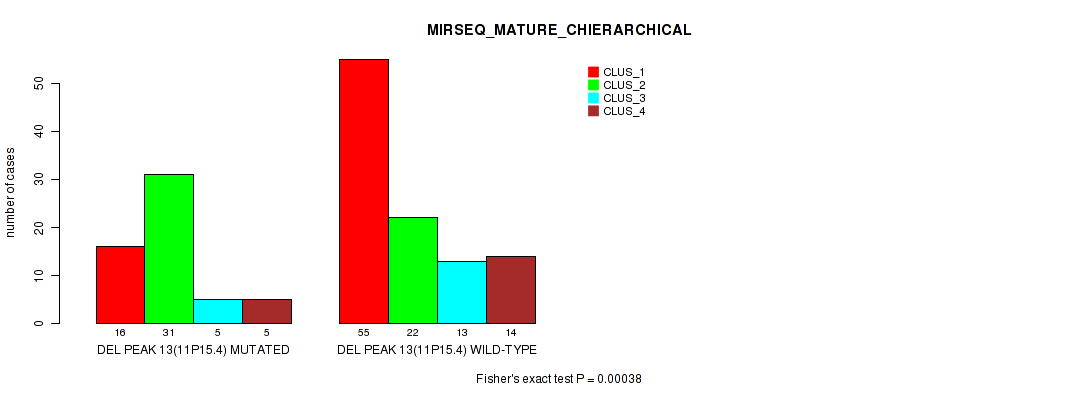
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S23. Gene #21: 'del_11q22.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 60 | 63 | 39 |
| DEL PEAK 14(11Q22.1) MUTATED | 4 | 26 | 18 |
| DEL PEAK 14(11Q22.1) WILD-TYPE | 56 | 37 | 21 |
Figure S23. Get High-res Image Gene #21: 'del_11q22.1' versus Molecular Subtype #1: 'CN_CNMF'
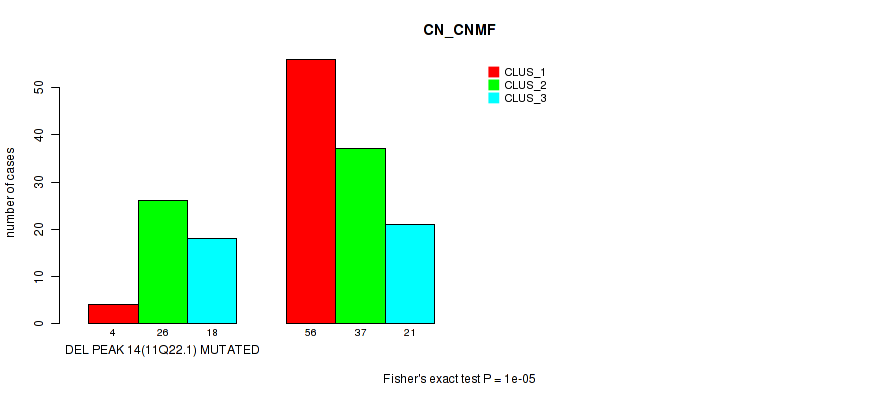
P value = 0.00066 (Fisher's exact test), Q value = 0.13
Table S24. Gene #21: 'del_11q22.1' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 64 | 44 | 21 |
| DEL PEAK 14(11Q22.1) MUTATED | 9 | 10 | 23 | 6 |
| DEL PEAK 14(11Q22.1) WILD-TYPE | 24 | 54 | 21 | 15 |
Figure S24. Get High-res Image Gene #21: 'del_11q22.1' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
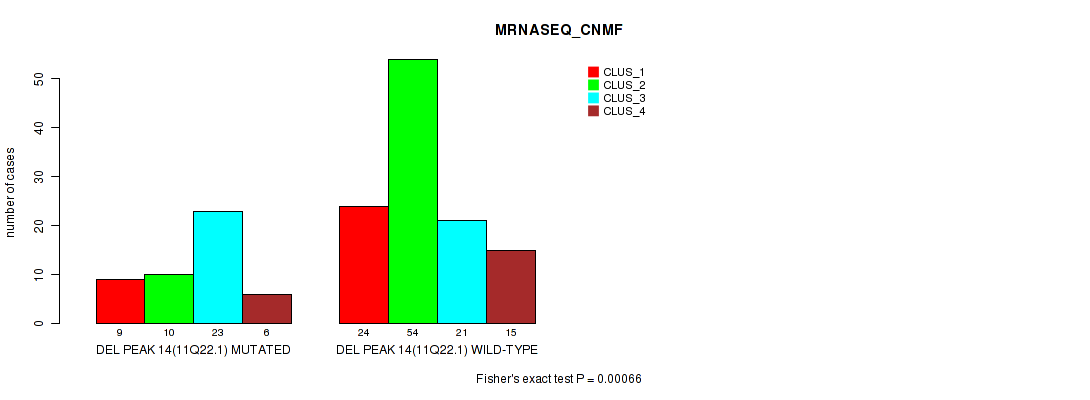
P value = 7e-04 (Fisher's exact test), Q value = 0.14
Table S25. Gene #21: 'del_11q22.1' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 66 | 54 | 18 | 24 |
| DEL PEAK 14(11Q22.1) MUTATED | 30 | 8 | 2 | 8 |
| DEL PEAK 14(11Q22.1) WILD-TYPE | 36 | 46 | 16 | 16 |
Figure S25. Get High-res Image Gene #21: 'del_11q22.1' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
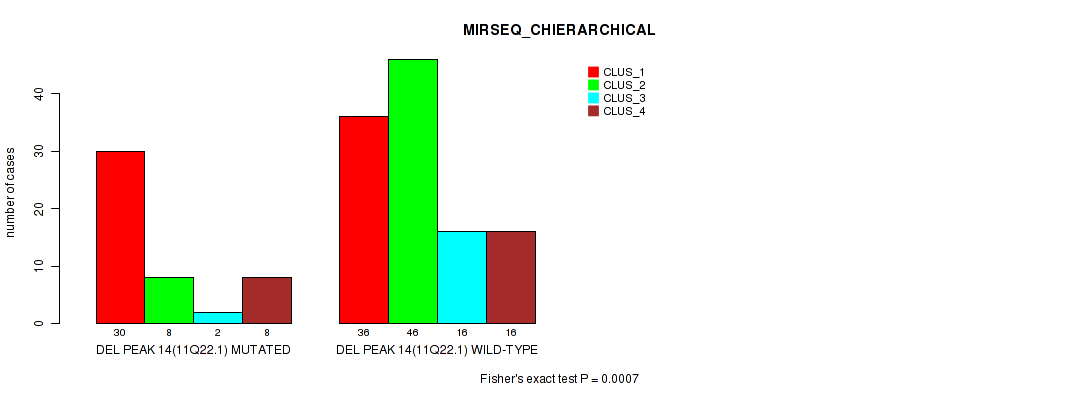
P value = 0.00038 (Fisher's exact test), Q value = 0.077
Table S26. Gene #21: 'del_11q22.1' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 34 | 58 | 11 | 35 | 23 |
| DEL PEAK 14(11Q22.1) MUTATED | 10 | 8 | 2 | 20 | 8 |
| DEL PEAK 14(11Q22.1) WILD-TYPE | 24 | 50 | 9 | 15 | 15 |
Figure S26. Get High-res Image Gene #21: 'del_11q22.1' versus Molecular Subtype #7: 'MIRSEQ_MATURE_CNMF'
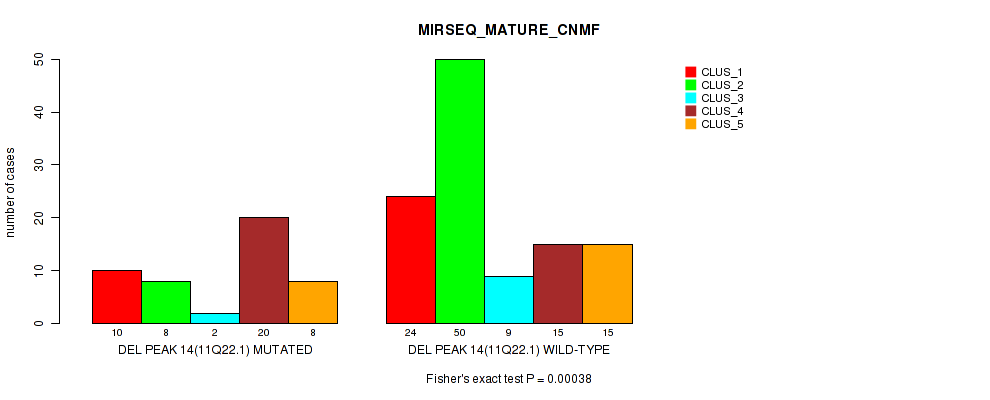
P value = 0.00017 (Fisher's exact test), Q value = 0.035
Table S27. Gene #25: 'del_17p13.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 60 | 63 | 39 |
| DEL PEAK 18(17P13.2) MUTATED | 22 | 16 | 26 |
| DEL PEAK 18(17P13.2) WILD-TYPE | 38 | 47 | 13 |
Figure S27. Get High-res Image Gene #25: 'del_17p13.2' versus Molecular Subtype #1: 'CN_CNMF'
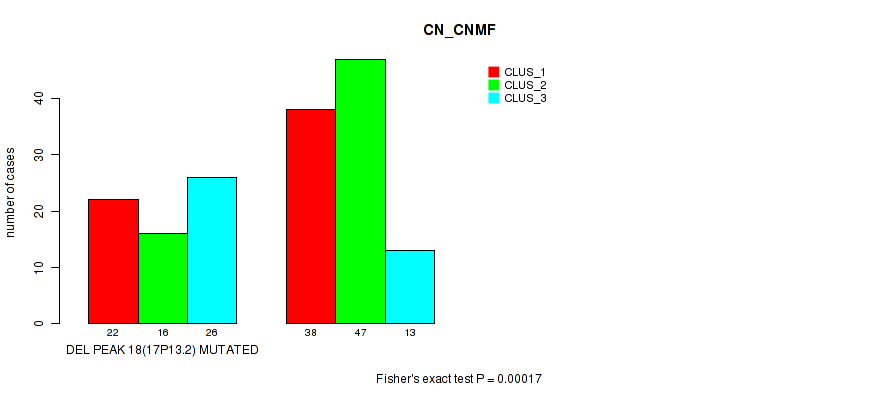
P value = 0.00042 (Fisher's exact test), Q value = 0.084
Table S28. Gene #25: 'del_17p13.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 40 | 78 | 44 |
| DEL PEAK 18(17P13.2) MUTATED | 9 | 43 | 12 |
| DEL PEAK 18(17P13.2) WILD-TYPE | 31 | 35 | 32 |
Figure S28. Get High-res Image Gene #25: 'del_17p13.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
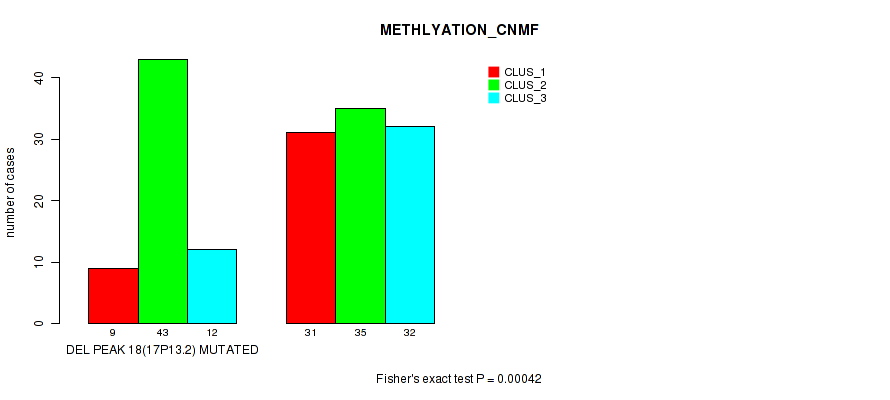
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S29. Gene #26: 'del_17q11.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 60 | 63 | 39 |
| DEL PEAK 19(17Q11.2) MUTATED | 13 | 4 | 25 |
| DEL PEAK 19(17Q11.2) WILD-TYPE | 47 | 59 | 14 |
Figure S29. Get High-res Image Gene #26: 'del_17q11.2' versus Molecular Subtype #1: 'CN_CNMF'
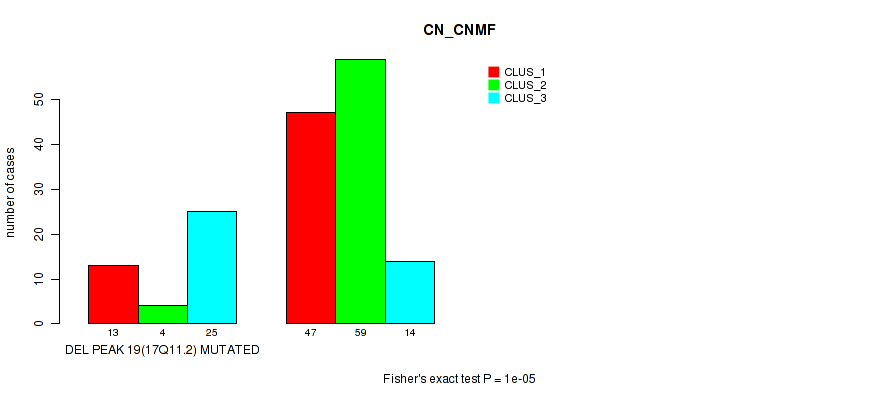
P value = 1e-05 (Fisher's exact test), Q value = 0.0022
Table S30. Gene #27: 'del_22q13.31' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 60 | 63 | 39 |
| DEL PEAK 20(22Q13.31) MUTATED | 40 | 9 | 16 |
| DEL PEAK 20(22Q13.31) WILD-TYPE | 20 | 54 | 23 |
Figure S30. Get High-res Image Gene #27: 'del_22q13.31' versus Molecular Subtype #1: 'CN_CNMF'
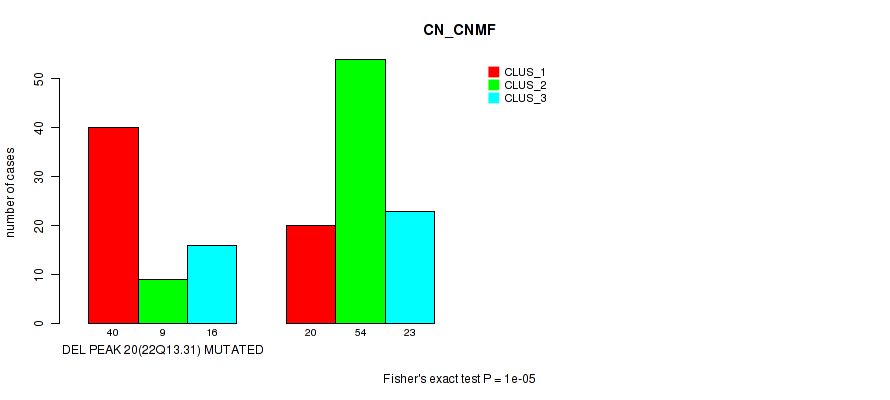
P value = 0.00047 (Fisher's exact test), Q value = 0.094
Table S31. Gene #27: 'del_22q13.31' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 34 | 64 | 43 | 21 |
| DEL PEAK 20(22Q13.31) MUTATED | 13 | 37 | 8 | 7 |
| DEL PEAK 20(22Q13.31) WILD-TYPE | 21 | 27 | 35 | 14 |
Figure S31. Get High-res Image Gene #27: 'del_22q13.31' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
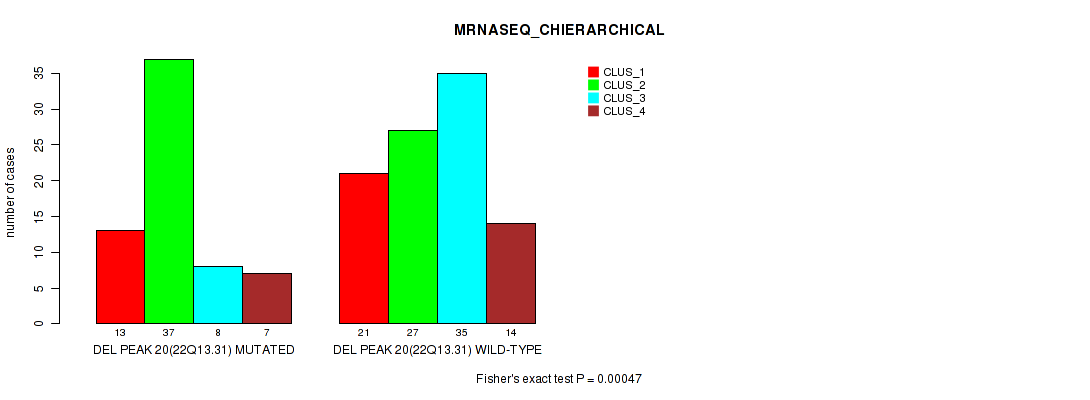
P value = 0.00099 (Fisher's exact test), Q value = 0.19
Table S32. Gene #28: 'del_xp21.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 60 | 63 | 39 |
| DEL PEAK 21(XP21.1) MUTATED | 21 | 8 | 17 |
| DEL PEAK 21(XP21.1) WILD-TYPE | 39 | 55 | 22 |
Figure S32. Get High-res Image Gene #28: 'del_xp21.1' versus Molecular Subtype #1: 'CN_CNMF'
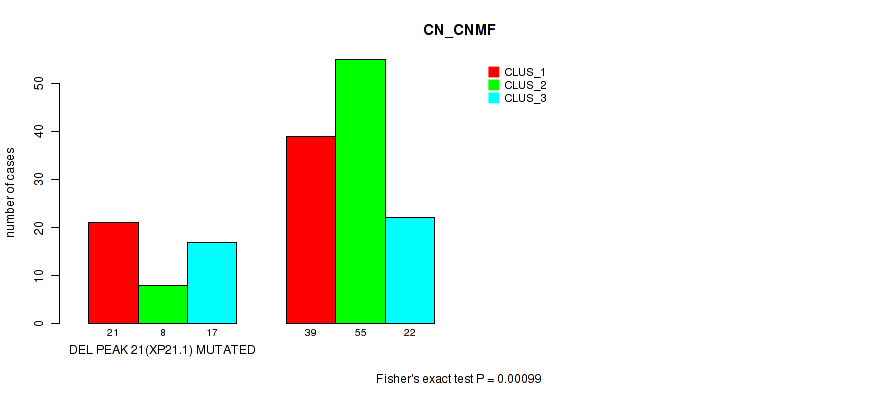
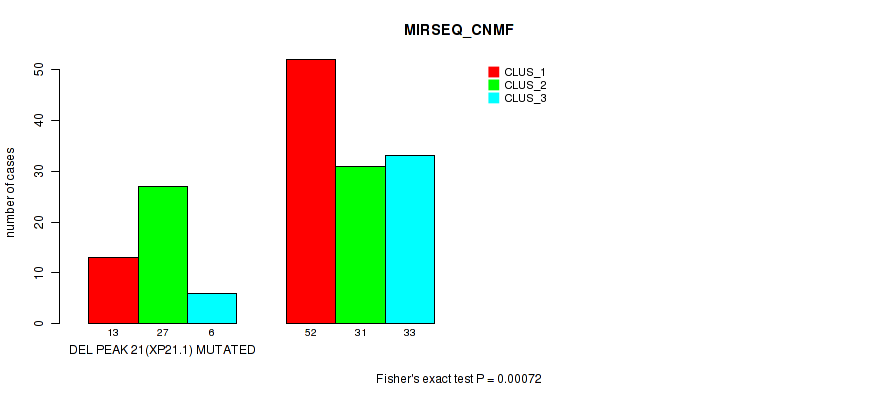
P value = 0.00072 (Fisher's exact test), Q value = 0.14
Table S33. Gene #28: 'del_xp21.1' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 58 | 39 |
| DEL PEAK 21(XP21.1) MUTATED | 13 | 27 | 6 |
| DEL PEAK 21(XP21.1) WILD-TYPE | 52 | 31 | 33 |
Figure S33. Get High-res Image Gene #28: 'del_xp21.1' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
-
Copy number data file = transformed.cor.cli.txt
-
Molecular subtype file = PCPG-TP.transferedmergedcluster.txt
-
Number of patients = 162
-
Number of significantly focal cnvs = 28
-
Number of molecular subtypes = 8
-
Exclude genes that fewer than K tumors have alterations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.