This pipeline computes the correlation between significantly recurrent gene mutations and selected clinical features.
Testing the association between mutation status of 143 genes and 8 clinical features across 473 patients, 25 significant findings detected with Q value < 0.25.
-
IDH1 mutation correlated to 'Time to Death' and 'YEARS_TO_BIRTH'.
-
TP53 mutation correlated to 'YEARS_TO_BIRTH', 'HISTOLOGICAL_TYPE', and 'RADIATIONS_RADIATION_REGIMENINDICATION'.
-
ATRX mutation correlated to 'YEARS_TO_BIRTH' and 'HISTOLOGICAL_TYPE'.
-
CIC mutation correlated to 'Time to Death' and 'HISTOLOGICAL_TYPE'.
-
NOTCH1 mutation correlated to 'HISTOLOGICAL_TYPE'.
-
EGFR mutation correlated to 'Time to Death' and 'YEARS_TO_BIRTH'.
-
IDH2 mutation correlated to 'HISTOLOGICAL_TYPE'.
-
NF1 mutation correlated to 'Time to Death' and 'HISTOLOGICAL_TYPE'.
-
PTEN mutation correlated to 'Time to Death', 'YEARS_TO_BIRTH', and 'HISTOLOGICAL_TYPE'.
-
FUBP1 mutation correlated to 'YEARS_TO_BIRTH' and 'HISTOLOGICAL_TYPE'.
-
ZNF292 mutation correlated to 'YEARS_TO_BIRTH'.
-
PLCG1 mutation correlated to 'Time to Death'.
-
TMEM184A mutation correlated to 'Time to Death'.
-
HCFC1 mutation correlated to 'GENDER'.
-
TNFSF14 mutation correlated to 'Time to Death'.
Table 1. Get Full Table Overview of the association between mutation status of 143 genes and 8 clinical features. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 25 significant findings detected.
|
Clinical Features |
Time to Death |
YEARS TO BIRTH |
GENDER |
KARNOFSKY PERFORMANCE SCORE |
HISTOLOGICAL TYPE |
RADIATIONS RADIATION REGIMENINDICATION |
RACE | ETHNICITY | ||
| nMutated (%) | nWild-Type | logrank test | Wilcoxon-test | Fisher's exact test | Wilcoxon-test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| TP53 | 226 (48%) | 247 |
0.03 (0.621) |
1.74e-11 (6.65e-09) |
0.228 (0.923) |
0.188 (0.833) |
1e-05 (0.00104) |
0.00536 (0.245) |
0.0532 (0.771) |
0.633 (1.00) |
| PTEN | 23 (5%) | 450 |
2.57e-05 (0.00226) |
0.00441 (0.219) |
0.399 (1.00) |
0.509 (1.00) |
0.00126 (0.0861) |
0.419 (1.00) |
0.254 (0.945) |
1 (1.00) |
| IDH1 | 367 (78%) | 106 |
6.57e-14 (7.52e-11) |
4.24e-07 (6.92e-05) |
0.74 (1.00) |
0.0146 (0.47) |
0.00655 (0.268) |
0.679 (1.00) |
0.0831 (0.826) |
0.567 (1.00) |
| ATRX | 181 (38%) | 292 |
0.147 (0.831) |
1.63e-11 (6.65e-09) |
0.703 (1.00) |
0.588 (1.00) |
1e-05 (0.00104) |
0.012 (0.428) |
0.428 (1.00) |
0.629 (1.00) |
| CIC | 100 (21%) | 373 |
0.00433 (0.219) |
0.0432 (0.756) |
0.497 (1.00) |
0.457 (1.00) |
1e-05 (0.00104) |
0.0869 (0.826) |
0.822 (1.00) |
0.775 (1.00) |
| EGFR | 30 (6%) | 443 |
6.3e-09 (1.44e-06) |
4.56e-09 (1.3e-06) |
0.85 (1.00) |
0.0242 (0.579) |
0.0248 (0.579) |
0.235 (0.923) |
0.368 (1.00) |
0.622 (1.00) |
| NF1 | 31 (7%) | 442 |
2.17e-05 (0.00207) |
0.0189 (0.54) |
0.581 (1.00) |
0.435 (1.00) |
0.0039 (0.215) |
0.096 (0.826) |
0.276 (0.971) |
0.353 (1.00) |
| FUBP1 | 46 (10%) | 427 |
0.262 (0.947) |
0.00493 (0.235) |
0.535 (1.00) |
0.268 (0.966) |
1e-05 (0.00104) |
1 (1.00) |
0.735 (1.00) |
0.24 (0.929) |
| NOTCH1 | 42 (9%) | 431 |
0.231 (0.923) |
0.0436 (0.756) |
0.627 (1.00) |
0.488 (1.00) |
3e-05 (0.00245) |
1 (1.00) |
0.431 (1.00) |
0.675 (1.00) |
| IDH2 | 19 (4%) | 454 |
0.104 (0.831) |
0.12 (0.831) |
0.815 (1.00) |
0.881 (1.00) |
0.00128 (0.0861) |
0.773 (1.00) |
0.645 (1.00) |
0.165 (0.832) |
| ZNF292 | 13 (3%) | 460 |
0.614 (1.00) |
0.00395 (0.215) |
0.258 (0.947) |
0.612 (1.00) |
1 (1.00) |
0.724 (1.00) |
0.157 (0.831) |
1 (1.00) |
| PLCG1 | 6 (1%) | 467 |
5.98e-08 (1.14e-05) |
0.0153 (0.47) |
0.411 (1.00) |
0.642 (1.00) |
0.347 (1.00) |
0.331 (1.00) |
0.276 (0.971) |
1 (1.00) |
| TMEM184A | 3 (1%) | 470 |
0.000413 (0.0315) |
0.154 (0.831) |
1 (1.00) |
1 (1.00) |
0.478 (1.00) |
0.147 (0.831) |
1 (1.00) |
|
| HCFC1 | 7 (1%) | 466 |
0.286 (0.989) |
0.58 (1.00) |
0.0029 (0.184) |
0.499 (1.00) |
0.0301 (0.621) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| TNFSF14 | 5 (1%) | 468 |
0.00346 (0.208) |
0.0554 (0.79) |
1 (1.00) |
0.097 (0.826) |
1 (1.00) |
0.588 (1.00) |
1 (1.00) |
1 (1.00) |
| PIK3R1 | 20 (4%) | 453 |
0.14 (0.831) |
0.1 (0.831) |
0.253 (0.945) |
0.684 (1.00) |
0.479 (1.00) |
0.0842 (0.826) |
0.119 (0.831) |
1 (1.00) |
| ARID1A | 19 (4%) | 454 |
0.0364 (0.686) |
0.695 (1.00) |
0.348 (1.00) |
0.526 (1.00) |
0.347 (1.00) |
0.773 (1.00) |
1 (1.00) |
0.557 (1.00) |
| PIK3CA | 42 (9%) | 431 |
0.563 (1.00) |
0.0459 (0.771) |
1 (1.00) |
0.116 (0.831) |
0.0163 (0.48) |
0.838 (1.00) |
0.839 (1.00) |
1 (1.00) |
| STK19 | 10 (2%) | 463 |
0.426 (1.00) |
0.312 (1.00) |
0.113 (0.831) |
0.176 (0.832) |
0.0674 (0.826) |
0.695 (1.00) |
1 (1.00) |
0.0591 (0.795) |
| NIPBL | 17 (4%) | 456 |
0.00599 (0.254) |
0.201 (0.853) |
0.32 (1.00) |
0.415 (1.00) |
0.0743 (0.826) |
0.545 (1.00) |
0.0799 (0.826) |
0.151 (0.831) |
| CREBZF | 6 (1%) | 467 |
0.659 (1.00) |
0.116 (0.831) |
0.411 (1.00) |
0.112 (0.831) |
0.113 (0.831) |
1 (1.00) |
0.273 (0.971) |
1 (1.00) |
| TCF12 | 15 (3%) | 458 |
0.88 (1.00) |
0.232 (0.923) |
0.598 (1.00) |
0.773 (1.00) |
0.945 (1.00) |
0.323 (1.00) |
1 (1.00) |
1 (1.00) |
| TRERF1 | 6 (1%) | 467 |
0.697 (1.00) |
0.285 (0.989) |
0.0909 (0.826) |
0.627 (1.00) |
0.147 (0.831) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| SRPX | 3 (1%) | 470 |
0.232 (0.923) |
0.0998 (0.831) |
0.0831 (0.826) |
0.787 (1.00) |
0.478 (1.00) |
1 (1.00) |
1 (1.00) |
|
| SMARCA4 | 24 (5%) | 449 |
0.0234 (0.579) |
0.387 (1.00) |
0.399 (1.00) |
0.117 (0.831) |
0.932 (1.00) |
0.107 (0.831) |
0.131 (0.831) |
0.576 (1.00) |
| EMG1 | 7 (1%) | 466 |
0.467 (1.00) |
0.0651 (0.82) |
0.704 (1.00) |
0.462 (1.00) |
0.561 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| FAM47C | 18 (4%) | 455 |
0.177 (0.832) |
0.571 (1.00) |
0.339 (1.00) |
0.877 (1.00) |
0.623 (1.00) |
0.762 (1.00) |
1 (1.00) |
0.517 (1.00) |
| TNRC18 | 10 (2%) | 463 |
0.75 (1.00) |
0.648 (1.00) |
0.198 (0.843) |
0.812 (1.00) |
0.0655 (0.82) |
0.695 (1.00) |
1 (1.00) |
1 (1.00) |
| DNMT3A | 10 (2%) | 463 |
0.0629 (0.82) |
0.486 (1.00) |
1 (1.00) |
0.804 (1.00) |
0.304 (1.00) |
0.695 (1.00) |
0.416 (1.00) |
0.346 (1.00) |
| HTRA2 | 5 (1%) | 468 |
0.348 (1.00) |
0.028 (0.621) |
1 (1.00) |
0.378 (1.00) |
0.588 (1.00) |
1 (1.00) |
0.19 (0.833) |
|
| ZNF709 | 4 (1%) | 469 |
0.0638 (0.82) |
0.597 (1.00) |
0.135 (0.831) |
0.302 (1.00) |
0.0246 (0.579) |
0.193 (0.833) |
0.155 (0.831) |
|
| RBPJ | 7 (1%) | 466 |
0.936 (1.00) |
0.0559 (0.79) |
1 (1.00) |
0.29 (0.995) |
0.306 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| NRAS | 4 (1%) | 469 |
0.289 (0.993) |
0.892 (1.00) |
1 (1.00) |
0.505 (1.00) |
0.0424 (0.756) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
| PTPN11 | 7 (1%) | 466 |
0.208 (0.869) |
0.342 (1.00) |
1 (1.00) |
0.428 (1.00) |
0.272 (0.971) |
0.626 (1.00) |
0.313 (1.00) |
1 (1.00) |
| GAGE2D | 5 (1%) | 468 |
0.648 (1.00) |
0.55 (1.00) |
0.392 (1.00) |
1 (1.00) |
1 (1.00) |
0.237 (0.923) |
0.19 (0.833) |
|
| ARID2 | 11 (2%) | 462 |
0.407 (1.00) |
0.114 (0.831) |
1 (1.00) |
0.942 (1.00) |
0.187 (0.833) |
0.454 (1.00) |
0.236 (0.923) |
1 (1.00) |
| ZMIZ1 | 8 (2%) | 465 |
0.0472 (0.771) |
0.904 (1.00) |
0.306 (1.00) |
0.868 (1.00) |
0.825 (1.00) |
1 (1.00) |
0.145 (0.831) |
0.0297 (0.621) |
| PDGFRA | 8 (2%) | 465 |
0.118 (0.831) |
0.7 (1.00) |
0.734 (1.00) |
0.119 (0.831) |
0.141 (0.831) |
0.0499 (0.771) |
1 (1.00) |
0.287 (0.989) |
| ZNF512B | 4 (1%) | 469 |
0.152 (0.831) |
0.592 (1.00) |
1 (1.00) |
0.681 (1.00) |
0.172 (0.832) |
1 (1.00) |
1 (1.00) |
|
| PPL | 5 (1%) | 468 |
0.0746 (0.826) |
0.824 (1.00) |
0.658 (1.00) |
0.965 (1.00) |
0.453 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| PRX | 10 (2%) | 463 |
0.924 (1.00) |
0.468 (1.00) |
0.345 (1.00) |
0.755 (1.00) |
0.718 (1.00) |
0.695 (1.00) |
1 (1.00) |
0.346 (1.00) |
| NKD2 | 4 (1%) | 469 |
0.552 (1.00) |
0.692 (1.00) |
1 (1.00) |
0.376 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| IRS4 | 7 (1%) | 466 |
0.225 (0.915) |
0.494 (1.00) |
1 (1.00) |
0.552 (1.00) |
0.113 (0.831) |
0.626 (1.00) |
1 (1.00) |
0.256 (0.945) |
| SPTAN1 | 5 (1%) | 468 |
0.0867 (0.826) |
0.452 (1.00) |
1 (1.00) |
0.704 (1.00) |
0.224 (0.915) |
0.251 (0.945) |
1 (1.00) |
0.19 (0.833) |
| CIB1 | 4 (1%) | 469 |
0.42 (1.00) |
0.996 (1.00) |
1 (1.00) |
0.868 (1.00) |
0.839 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| EIF1AX | 4 (1%) | 469 |
0.346 (1.00) |
0.93 (1.00) |
0.135 (0.831) |
0.94 (1.00) |
0.193 (0.833) |
0.172 (0.832) |
1 (1.00) |
1 (1.00) |
| SCN4A | 6 (1%) | 467 |
0.283 (0.988) |
0.261 (0.947) |
0.411 (1.00) |
0.255 (0.945) |
1 (1.00) |
0.0909 (0.826) |
1 (1.00) |
1 (1.00) |
| DLC1 | 5 (1%) | 468 |
0.882 (1.00) |
0.983 (1.00) |
0.658 (1.00) |
0.549 (1.00) |
0.0528 (0.771) |
1 (1.00) |
1 (1.00) |
|
| TMEM216 | 3 (1%) | 470 |
0.78 (1.00) |
0.832 (1.00) |
0.0831 (0.826) |
0.474 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| MYT1 | 6 (1%) | 467 |
0.372 (1.00) |
0.403 (1.00) |
1 (1.00) |
0.875 (1.00) |
0.147 (0.831) |
0.331 (1.00) |
1 (1.00) |
1 (1.00) |
| SMARCB1 | 4 (1%) | 469 |
0.432 (1.00) |
0.807 (1.00) |
0.635 (1.00) |
0.868 (1.00) |
0.839 (1.00) |
0.0246 (0.579) |
1 (1.00) |
1 (1.00) |
| RIPK4 | 8 (2%) | 465 |
0.845 (1.00) |
0.325 (1.00) |
0.734 (1.00) |
0.959 (1.00) |
0.754 (1.00) |
0.656 (1.00) |
0.0728 (0.826) |
1 (1.00) |
| NEU2 | 4 (1%) | 469 |
0.72 (1.00) |
0.138 (0.831) |
0.635 (1.00) |
0.68 (1.00) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
|
| PTPRH | 8 (2%) | 465 |
0.883 (1.00) |
0.0587 (0.795) |
0.734 (1.00) |
0.213 (0.88) |
0.431 (1.00) |
0.19 (0.833) |
1 (1.00) |
1 (1.00) |
| MAPK15 | 4 (1%) | 469 |
0.0057 (0.251) |
0.489 (1.00) |
0.635 (1.00) |
0.459 (1.00) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
|
| TPX2 | 3 (1%) | 470 |
0.14 (0.831) |
0.411 (1.00) |
1 (1.00) |
0.629 (1.00) |
0.0982 (0.826) |
1 (1.00) |
1 (1.00) |
|
| SLC12A7 | 5 (1%) | 468 |
0.0967 (0.826) |
0.57 (1.00) |
0.658 (1.00) |
0.581 (1.00) |
0.647 (1.00) |
0.0528 (0.771) |
0.235 (0.923) |
1 (1.00) |
| PLXNA3 | 8 (2%) | 465 |
0.13 (0.831) |
0.295 (1.00) |
0.146 (0.831) |
0.672 (1.00) |
1 (1.00) |
0.0499 (0.771) |
0.0486 (0.771) |
1 (1.00) |
| RET | 6 (1%) | 467 |
0.921 (1.00) |
0.76 (1.00) |
0.411 (1.00) |
0.419 (1.00) |
0.037 (0.686) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| ZNF148 | 4 (1%) | 469 |
0.14 (0.831) |
0.661 (1.00) |
1 (1.00) |
0.455 (1.00) |
0.58 (1.00) |
0.0818 (0.826) |
1 (1.00) |
|
| HDAC2 | 4 (1%) | 469 |
0.518 (1.00) |
0.199 (0.845) |
1 (1.00) |
0.456 (1.00) |
0.0246 (0.579) |
1 (1.00) |
1 (1.00) |
|
| RBBP6 | 6 (1%) | 467 |
0.273 (0.971) |
0.324 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.125 (0.831) |
1 (1.00) |
|
| MED9 | 3 (1%) | 470 |
0.196 (0.84) |
0.86 (1.00) |
0.26 (0.947) |
0.0152 (0.47) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| DDX5 | 5 (1%) | 468 |
0.156 (0.831) |
0.852 (1.00) |
1 (1.00) |
0.263 (0.948) |
0.647 (1.00) |
0.251 (0.945) |
1 (1.00) |
0.155 (0.831) |
| ZC3H11A | 5 (1%) | 468 |
0.131 (0.831) |
0.162 (0.832) |
0.392 (1.00) |
0.179 (0.833) |
0.155 (0.831) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| SLC6A3 | 9 (2%) | 464 |
0.856 (1.00) |
0.383 (1.00) |
0.514 (1.00) |
0.759 (1.00) |
0.689 (1.00) |
1 (1.00) |
0.256 (0.945) |
|
| TPTE2 | 5 (1%) | 468 |
0.412 (1.00) |
0.543 (1.00) |
0.658 (1.00) |
0.177 (0.832) |
0.0509 (0.771) |
0.588 (1.00) |
1 (1.00) |
1 (1.00) |
| C4BPA | 4 (1%) | 469 |
0.494 (1.00) |
0.717 (1.00) |
0.323 (1.00) |
0.65 (1.00) |
0.142 (0.831) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
| MATK | 5 (1%) | 468 |
0.431 (1.00) |
0.446 (1.00) |
0.392 (1.00) |
1 (1.00) |
0.0528 (0.771) |
1 (1.00) |
1 (1.00) |
|
| SMOC1 | 3 (1%) | 470 |
0.506 (1.00) |
0.447 (1.00) |
1 (1.00) |
0.94 (1.00) |
0.362 (1.00) |
0.0982 (0.826) |
1 (1.00) |
1 (1.00) |
| NAP1L2 | 4 (1%) | 469 |
0.285 (0.989) |
0.181 (0.833) |
1 (1.00) |
0.3 (1.00) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
|
| CD99L2 | 3 (1%) | 470 |
0.168 (0.832) |
0.308 (1.00) |
0.0831 (0.826) |
0.787 (1.00) |
0.478 (1.00) |
1 (1.00) |
1 (1.00) |
|
| SELE | 6 (1%) | 467 |
0.824 (1.00) |
0.168 (0.832) |
0.411 (1.00) |
0.483 (1.00) |
1 (1.00) |
0.331 (1.00) |
1 (1.00) |
1 (1.00) |
| FHOD1 | 6 (1%) | 467 |
0.245 (0.945) |
0.175 (0.832) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| HTR3A | 7 (1%) | 466 |
0.581 (1.00) |
0.545 (1.00) |
0.047 (0.771) |
1 (1.00) |
1 (1.00) |
0.0079 (0.292) |
1 (1.00) |
|
| RB1 | 5 (1%) | 468 |
0.968 (1.00) |
0.389 (1.00) |
0.173 (0.832) |
0.222 (0.915) |
0.0528 (0.771) |
0.235 (0.923) |
1 (1.00) |
|
| FGD6 | 4 (1%) | 469 |
0.771 (1.00) |
0.96 (1.00) |
0.0361 (0.686) |
0.428 (1.00) |
0.565 (1.00) |
0.0246 (0.579) |
1 (1.00) |
1 (1.00) |
| CD72 | 3 (1%) | 470 |
0.483 (1.00) |
0.968 (1.00) |
0.584 (1.00) |
0.428 (1.00) |
0.0659 (0.82) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| NKPD1 | 3 (1%) | 470 |
0.158 (0.831) |
0.0823 (0.826) |
0.584 (1.00) |
0.628 (1.00) |
1 (1.00) |
1 (1.00) |
0.119 (0.831) |
|
| MTMR1 | 4 (1%) | 469 |
0.472 (1.00) |
0.171 (0.832) |
0.323 (1.00) |
0.681 (1.00) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
|
| GPR133 | 3 (1%) | 470 |
0.128 (0.831) |
0.584 (1.00) |
1 (1.00) |
0.478 (1.00) |
1 (1.00) |
0.119 (0.831) |
||
| CNOT4 | 5 (1%) | 468 |
0.131 (0.831) |
0.666 (1.00) |
1 (1.00) |
0.119 (0.831) |
0.648 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| ZBTB20 | 19 (4%) | 454 |
0.203 (0.856) |
0.53 (1.00) |
0.64 (1.00) |
0.938 (1.00) |
0.176 (0.832) |
0.552 (1.00) |
0.193 (0.833) |
0.537 (1.00) |
| ROBO3 | 4 (1%) | 469 |
0.646 (1.00) |
0.808 (1.00) |
0.323 (1.00) |
0.147 (0.831) |
0.681 (1.00) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
| PDE8A | 6 (1%) | 467 |
0.62 (1.00) |
0.0304 (0.621) |
1 (1.00) |
0.455 (1.00) |
0.331 (1.00) |
1 (1.00) |
1 (1.00) |
|
| EEF1D | 3 (1%) | 470 |
0.36 (1.00) |
0.101 (0.831) |
0.584 (1.00) |
1 (1.00) |
0.00717 (0.283) |
1 (1.00) |
1 (1.00) |
|
| MET | 7 (1%) | 466 |
0.259 (0.947) |
0.446 (1.00) |
1 (1.00) |
0.554 (1.00) |
0.562 (1.00) |
0.137 (0.831) |
1 (1.00) |
1 (1.00) |
| CUL4B | 10 (2%) | 463 |
0.341 (1.00) |
0.815 (1.00) |
0.113 (0.831) |
0.817 (1.00) |
0.304 (1.00) |
1 (1.00) |
0.212 (0.88) |
1 (1.00) |
| SNX31 | 4 (1%) | 469 |
0.0939 (0.826) |
0.465 (1.00) |
0.323 (1.00) |
0.839 (1.00) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
|
| PHF3 | 8 (2%) | 465 |
0.903 (1.00) |
0.617 (1.00) |
0.734 (1.00) |
0.213 (0.88) |
0.539 (1.00) |
0.19 (0.833) |
0.35 (1.00) |
0.256 (0.945) |
| G6PC | 5 (1%) | 468 |
0.868 (1.00) |
0.112 (0.831) |
0.392 (1.00) |
0.377 (1.00) |
0.251 (0.945) |
1 (1.00) |
0.19 (0.833) |
|
| FOXQ1 | 3 (1%) | 470 |
0.5 (1.00) |
0.252 (0.945) |
0.584 (1.00) |
1 (1.00) |
0.0982 (0.826) |
1 (1.00) |
1 (1.00) |
|
| AEBP1 | 4 (1%) | 469 |
0.429 (1.00) |
0.317 (1.00) |
1 (1.00) |
0.457 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| SETD2 | 8 (2%) | 465 |
0.826 (1.00) |
0.0314 (0.63) |
0.734 (1.00) |
0.565 (1.00) |
0.752 (1.00) |
0.19 (0.833) |
0.349 (1.00) |
1 (1.00) |
| DST | 11 (2%) | 462 |
0.0775 (0.826) |
0.397 (1.00) |
0.763 (1.00) |
0.875 (1.00) |
0.0925 (0.826) |
1 (1.00) |
1 (1.00) |
0.0705 (0.826) |
| USP6 | 5 (1%) | 468 |
0.0788 (0.826) |
0.237 (0.923) |
1 (1.00) |
0.356 (1.00) |
0.379 (1.00) |
0.251 (0.945) |
0.235 (0.923) |
1 (1.00) |
| TPM1 | 5 (1%) | 468 |
0.127 (0.831) |
0.381 (1.00) |
1 (1.00) |
0.851 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| GLYAT | 4 (1%) | 469 |
0.0789 (0.826) |
0.583 (1.00) |
0.635 (1.00) |
0.839 (1.00) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
|
| DIAPH1 | 4 (1%) | 469 |
0.146 (0.831) |
0.489 (1.00) |
1 (1.00) |
0.142 (0.831) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
|
| ZMYM2 | 6 (1%) | 467 |
0.283 (0.988) |
0.309 (1.00) |
0.7 (1.00) |
0.704 (1.00) |
0.776 (1.00) |
0.331 (1.00) |
1 (1.00) |
1 (1.00) |
| MST1 | 4 (1%) | 469 |
0.12 (0.831) |
0.321 (1.00) |
0.0361 (0.686) |
0.94 (1.00) |
0.681 (1.00) |
0.58 (1.00) |
1 (1.00) |
0.155 (0.831) |
| DNAH7 | 10 (2%) | 463 |
0.776 (1.00) |
0.0571 (0.795) |
1 (1.00) |
0.104 (0.831) |
0.477 (1.00) |
0.417 (1.00) |
1 (1.00) |
1 (1.00) |
| ENGASE | 3 (1%) | 470 |
0.344 (1.00) |
0.668 (1.00) |
0.584 (1.00) |
0.628 (1.00) |
0.478 (1.00) |
1 (1.00) |
1 (1.00) |
|
| MYH8 | 11 (2%) | 462 |
0.0635 (0.82) |
0.716 (1.00) |
0.224 (0.915) |
0.519 (1.00) |
0.186 (0.833) |
0.454 (1.00) |
0.0808 (0.826) |
0.373 (1.00) |
| VSIG4 | 6 (1%) | 467 |
0.75 (1.00) |
0.947 (1.00) |
0.411 (1.00) |
1 (1.00) |
0.602 (1.00) |
1 (1.00) |
1 (1.00) |
|
| KRT3 | 4 (1%) | 469 |
0.0709 (0.826) |
0.865 (1.00) |
0.635 (1.00) |
0.428 (1.00) |
0.681 (1.00) |
0.172 (0.832) |
1 (1.00) |
0.155 (0.831) |
| STAG2 | 4 (1%) | 469 |
0.104 (0.831) |
0.659 (1.00) |
0.323 (1.00) |
0.139 (0.831) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
|
| MUC17 | 24 (5%) | 449 |
0.699 (1.00) |
0.15 (0.831) |
0.3 (1.00) |
0.653 (1.00) |
0.233 (0.923) |
1 (1.00) |
0.269 (0.966) |
0.614 (1.00) |
| ABCA7 | 9 (2%) | 464 |
0.98 (1.00) |
0.163 (0.832) |
0.738 (1.00) |
0.415 (1.00) |
0.487 (1.00) |
0.689 (1.00) |
1 (1.00) |
1 (1.00) |
| FOXK1 | 4 (1%) | 469 |
0.0924 (0.826) |
0.336 (1.00) |
0.323 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
0.155 (0.831) |
|
| SLFN11 | 3 (1%) | 470 |
0.175 (0.832) |
0.0681 (0.826) |
1 (1.00) |
1 (1.00) |
0.478 (1.00) |
1 (1.00) |
1 (1.00) |
|
| MX2 | 4 (1%) | 469 |
0.323 (1.00) |
0.0297 (0.621) |
1 (1.00) |
0.683 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| SLC6A4 | 4 (1%) | 469 |
0.158 (0.831) |
0.449 (1.00) |
0.323 (1.00) |
0.14 (0.831) |
0.172 (0.832) |
1 (1.00) |
1 (1.00) |
|
| KRT15 | 6 (1%) | 467 |
0.695 (1.00) |
0.117 (0.831) |
0.7 (1.00) |
0.34 (1.00) |
0.773 (1.00) |
0.602 (1.00) |
0.277 (0.971) |
1 (1.00) |
| F8 | 8 (2%) | 465 |
0.382 (1.00) |
0.116 (0.831) |
0.024 (0.579) |
0.205 (0.859) |
0.54 (1.00) |
0.19 (0.833) |
0.0468 (0.771) |
1 (1.00) |
| HOOK1 | 3 (1%) | 470 |
0.125 (0.831) |
0.904 (1.00) |
0.584 (1.00) |
0.628 (1.00) |
0.0982 (0.826) |
1 (1.00) |
1 (1.00) |
|
| FMR1 | 5 (1%) | 468 |
0.104 (0.831) |
0.182 (0.833) |
1 (1.00) |
0.428 (1.00) |
0.0966 (0.826) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| SON | 8 (2%) | 465 |
0.97 (1.00) |
0.204 (0.859) |
1 (1.00) |
0.425 (1.00) |
0.433 (1.00) |
1 (1.00) |
1 (1.00) |
0.256 (0.945) |
| TTC30B | 6 (1%) | 467 |
0.164 (0.832) |
0.154 (0.831) |
0.7 (1.00) |
0.187 (0.833) |
0.587 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| FANCD2 | 7 (1%) | 466 |
0.356 (1.00) |
0.0974 (0.826) |
0.704 (1.00) |
0.349 (1.00) |
0.626 (1.00) |
1 (1.00) |
0.256 (0.945) |
|
| RGL2 | 4 (1%) | 469 |
0.126 (0.831) |
0.194 (0.833) |
1 (1.00) |
0.123 (0.831) |
0.139 (0.831) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
| NR1H3 | 4 (1%) | 469 |
0.131 (0.831) |
0.497 (1.00) |
0.323 (1.00) |
0.141 (0.831) |
0.172 (0.832) |
1 (1.00) |
0.155 (0.831) |
|
| TYRP1 | 4 (1%) | 469 |
0.485 (1.00) |
0.503 (1.00) |
1 (1.00) |
0.139 (0.831) |
0.172 (0.832) |
1 (1.00) |
1 (1.00) |
|
| SLC25A5 | 4 (1%) | 469 |
0.0156 (0.47) |
0.581 (1.00) |
0.323 (1.00) |
0.423 (1.00) |
1 (1.00) |
0.58 (1.00) |
0.193 (0.833) |
1 (1.00) |
| AGBL1 | 4 (1%) | 469 |
0.65 (1.00) |
0.362 (1.00) |
0.635 (1.00) |
0.193 (0.833) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| AMPD1 | 4 (1%) | 469 |
0.903 (1.00) |
0.745 (1.00) |
1 (1.00) |
0.683 (1.00) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
|
| NGEF | 5 (1%) | 468 |
0.361 (1.00) |
0.904 (1.00) |
0.658 (1.00) |
0.41 (1.00) |
0.305 (1.00) |
1 (1.00) |
1 (1.00) |
0.0148 (0.47) |
| YIPF1 | 3 (1%) | 470 |
0.17 (0.832) |
0.951 (1.00) |
0.0831 (0.826) |
0.629 (1.00) |
0.478 (1.00) |
1 (1.00) |
1 (1.00) |
|
| PDHA1 | 5 (1%) | 468 |
0.967 (1.00) |
0.188 (0.833) |
0.0156 (0.47) |
0.483 (1.00) |
0.304 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
| ASXL2 | 3 (1%) | 470 |
0.163 (0.832) |
0.415 (1.00) |
0.584 (1.00) |
0.788 (1.00) |
0.0982 (0.826) |
1 (1.00) |
1 (1.00) |
|
| HLA-DRA | 3 (1%) | 470 |
0.448 (1.00) |
0.792 (1.00) |
0.0831 (0.826) |
0.0659 (0.82) |
0.0982 (0.826) |
0.149 (0.831) |
1 (1.00) |
|
| ALPP | 4 (1%) | 469 |
0.387 (1.00) |
0.913 (1.00) |
0.323 (1.00) |
0.85 (1.00) |
0.298 (1.00) |
0.58 (1.00) |
0.0213 (0.579) |
0.0806 (0.826) |
| CDH3 | 3 (1%) | 470 |
0.429 (1.00) |
0.621 (1.00) |
0.0831 (0.826) |
0.789 (1.00) |
0.478 (1.00) |
1 (1.00) |
1 (1.00) |
|
| NADSYN1 | 3 (1%) | 470 |
0.17 (0.832) |
0.825 (1.00) |
0.584 (1.00) |
0.362 (1.00) |
0.478 (1.00) |
1 (1.00) |
1 (1.00) |
|
| LHFPL1 | 4 (1%) | 469 |
0.963 (1.00) |
0.0266 (0.609) |
0.635 (1.00) |
1 (1.00) |
0.58 (1.00) |
1 (1.00) |
1 (1.00) |
|
| CXCR2 | 4 (1%) | 469 |
0.823 (1.00) |
0.825 (1.00) |
0.635 (1.00) |
0.682 (1.00) |
0.172 (0.832) |
1 (1.00) |
1 (1.00) |
|
| SEC14L4 | 3 (1%) | 470 |
0.638 (1.00) |
0.0378 (0.686) |
0.26 (0.947) |
0.627 (1.00) |
0.0982 (0.826) |
1 (1.00) |
1 (1.00) |
|
| ANKRD17 | 6 (1%) | 467 |
0.154 (0.831) |
0.485 (1.00) |
0.7 (1.00) |
0.177 (0.832) |
1 (1.00) |
0.331 (1.00) |
0.276 (0.971) |
1 (1.00) |
| MAST3 | 5 (1%) | 468 |
0.476 (1.00) |
0.689 (1.00) |
0.658 (1.00) |
0.0377 (0.686) |
0.452 (1.00) |
1 (1.00) |
0.235 (0.923) |
1 (1.00) |
| AMMECR1 | 3 (1%) | 470 |
0.66 (1.00) |
0.937 (1.00) |
0.584 (1.00) |
0.868 (1.00) |
0.628 (1.00) |
0.0982 (0.826) |
1 (1.00) |
1 (1.00) |
| NEO1 | 7 (1%) | 466 |
0.0579 (0.795) |
0.14 (0.831) |
0.248 (0.945) |
0.559 (1.00) |
1 (1.00) |
1 (1.00) |
1 (1.00) |
|
| IGSF8 | 3 (1%) | 470 |
0.157 (0.831) |
0.382 (1.00) |
0.584 (1.00) |
0.628 (1.00) |
0.0982 (0.826) |
1 (1.00) |
1 (1.00) |
|
| RGS3 | 5 (1%) | 468 |
0.335 (1.00) |
0.487 (1.00) |
0.658 (1.00) |
0.303 (1.00) |
0.00772 (0.292) |
1 (1.00) |
1 (1.00) |
0.19 (0.833) |
P value = 6.57e-14 (logrank test), Q value = 7.5e-11
Table S1. Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 471 | 93 | 0.0 - 211.2 (18.6) |
| IDH1 MUTATED | 365 | 53 | 0.0 - 182.3 (20.1) |
| IDH1 WILD-TYPE | 106 | 40 | 0.1 - 211.2 (14.6) |
Figure S1. Get High-res Image Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
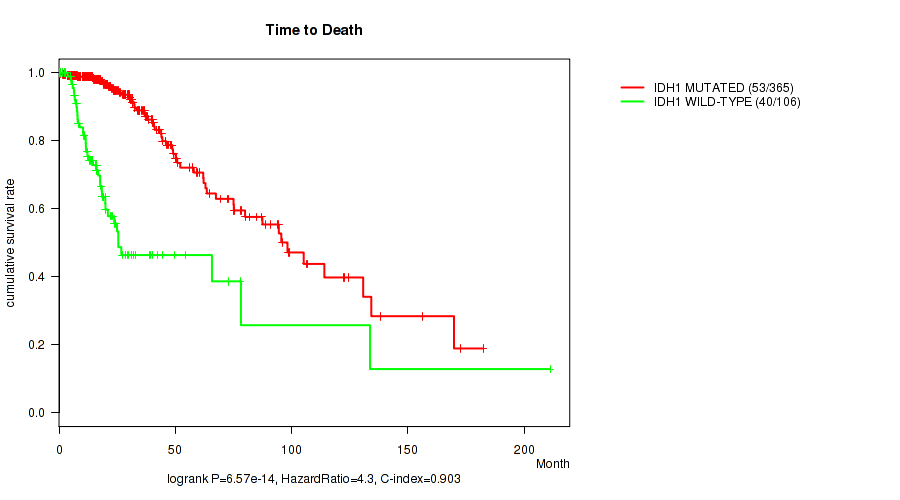
P value = 4.24e-07 (Wilcoxon-test), Q value = 6.9e-05
Table S2. Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 472 | 43.0 (13.5) |
| IDH1 MUTATED | 366 | 41.2 (12.5) |
| IDH1 WILD-TYPE | 106 | 49.2 (14.8) |
Figure S2. Get High-res Image Gene #1: 'IDH1 MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
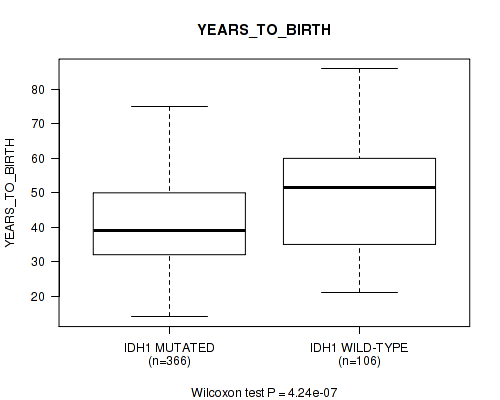
P value = 1.74e-11 (Wilcoxon-test), Q value = 6.6e-09
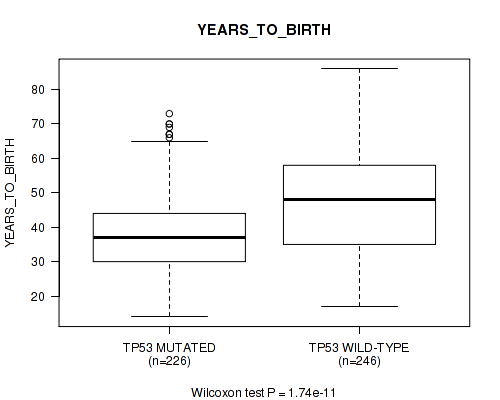
Table S3. Gene #2: 'TP53 MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 472 | 43.0 (13.5) |
| TP53 MUTATED | 226 | 38.6 (11.5) |
| TP53 WILD-TYPE | 246 | 47.0 (13.9) |
Figure S3. Get High-res Image Gene #2: 'TP53 MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
P value = 1e-05 (Fisher's exact test), Q value = 0.001
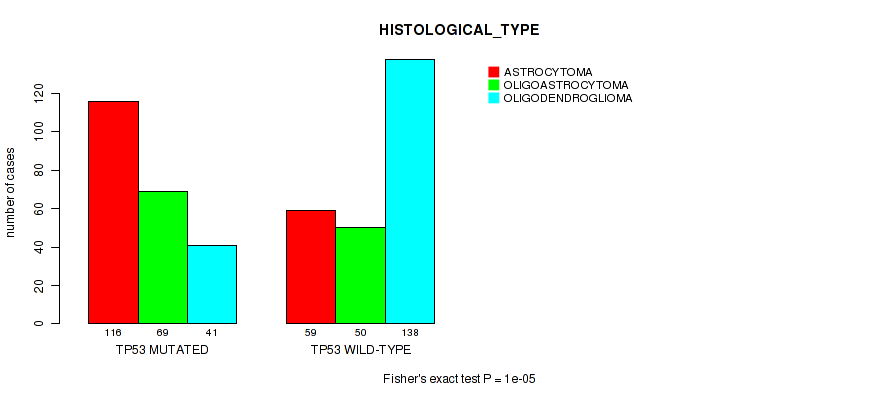
Table S4. Gene #2: 'TP53 MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 175 | 119 | 179 |
| TP53 MUTATED | 116 | 69 | 41 |
| TP53 WILD-TYPE | 59 | 50 | 138 |
Figure S4. Get High-res Image Gene #2: 'TP53 MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
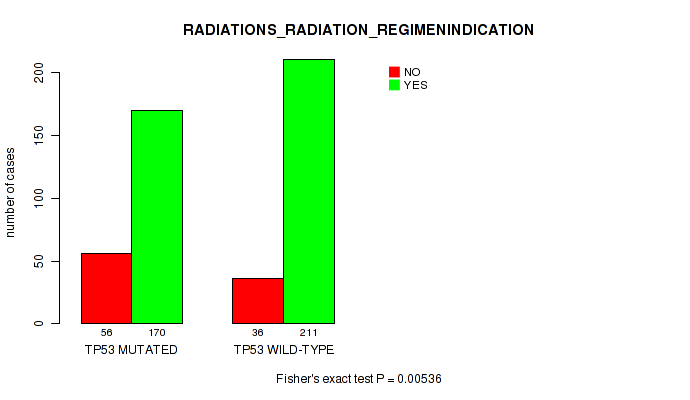
P value = 0.00536 (Fisher's exact test), Q value = 0.25
Table S5. Gene #2: 'TP53 MUTATION STATUS' versus Clinical Feature #6: 'RADIATIONS_RADIATION_REGIMENINDICATION'
| nPatients | NO | YES |
|---|---|---|
| ALL | 92 | 381 |
| TP53 MUTATED | 56 | 170 |
| TP53 WILD-TYPE | 36 | 211 |
Figure S5. Get High-res Image Gene #2: 'TP53 MUTATION STATUS' versus Clinical Feature #6: 'RADIATIONS_RADIATION_REGIMENINDICATION'
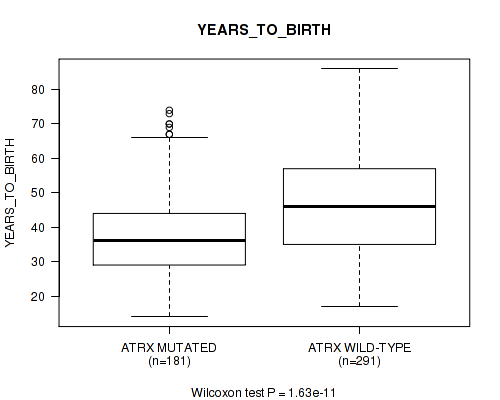
P value = 1.63e-11 (Wilcoxon-test), Q value = 6.6e-09
Table S6. Gene #3: 'ATRX MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 472 | 43.0 (13.5) |
| ATRX MUTATED | 181 | 37.9 (12.1) |
| ATRX WILD-TYPE | 291 | 46.1 (13.3) |
Figure S6. Get High-res Image Gene #3: 'ATRX MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
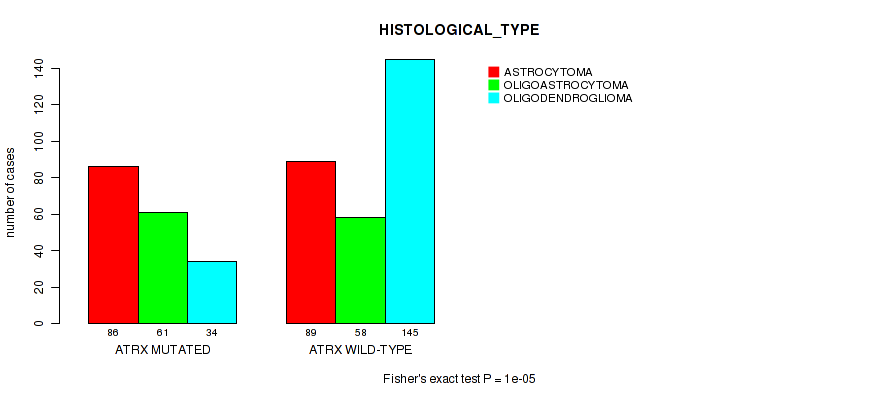
P value = 1e-05 (Fisher's exact test), Q value = 0.001
Table S7. Gene #3: 'ATRX MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 175 | 119 | 179 |
| ATRX MUTATED | 86 | 61 | 34 |
| ATRX WILD-TYPE | 89 | 58 | 145 |
Figure S7. Get High-res Image Gene #3: 'ATRX MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
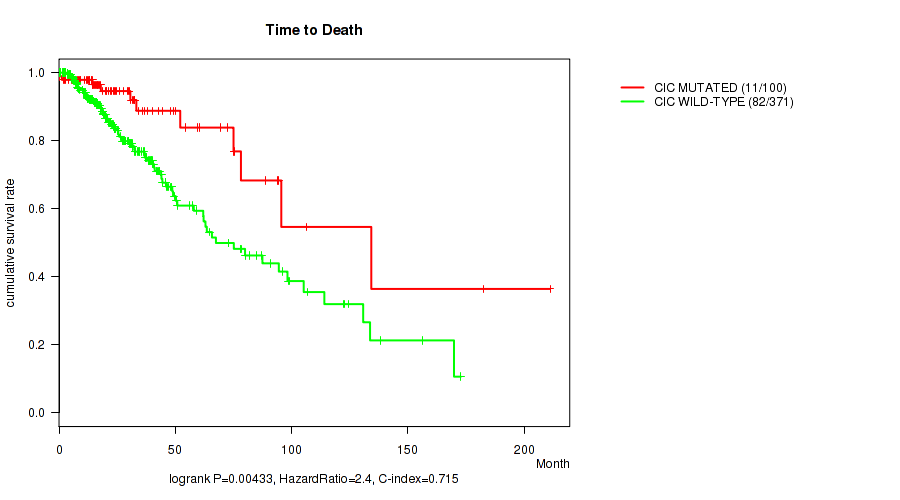
P value = 0.00433 (logrank test), Q value = 0.22
Table S8. Gene #4: 'CIC MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 471 | 93 | 0.0 - 211.2 (18.6) |
| CIC MUTATED | 100 | 11 | 0.1 - 211.2 (20.2) |
| CIC WILD-TYPE | 371 | 82 | 0.0 - 172.8 (18.5) |
Figure S8. Get High-res Image Gene #4: 'CIC MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
P value = 1e-05 (Fisher's exact test), Q value = 0.001
Table S9. Gene #4: 'CIC MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 175 | 119 | 179 |
| CIC MUTATED | 3 | 20 | 77 |
| CIC WILD-TYPE | 172 | 99 | 102 |
Figure S9. Get High-res Image Gene #4: 'CIC MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
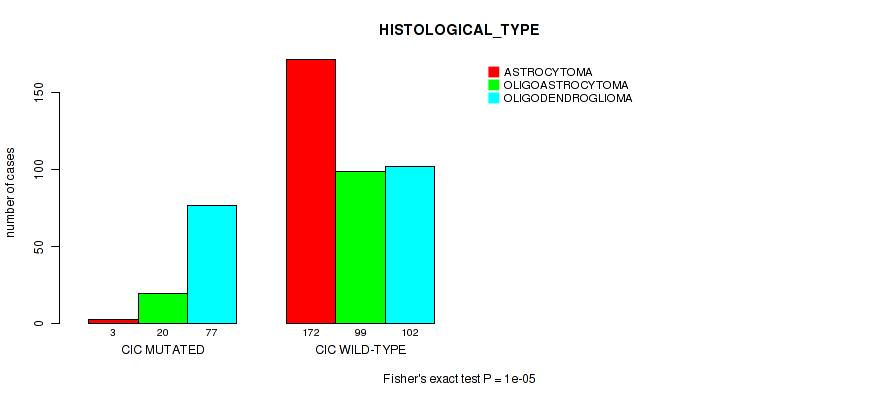
P value = 3e-05 (Fisher's exact test), Q value = 0.0025
Table S10. Gene #5: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 175 | 119 | 179 |
| NOTCH1 MUTATED | 6 | 7 | 29 |
| NOTCH1 WILD-TYPE | 169 | 112 | 150 |
Figure S10. Get High-res Image Gene #5: 'NOTCH1 MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
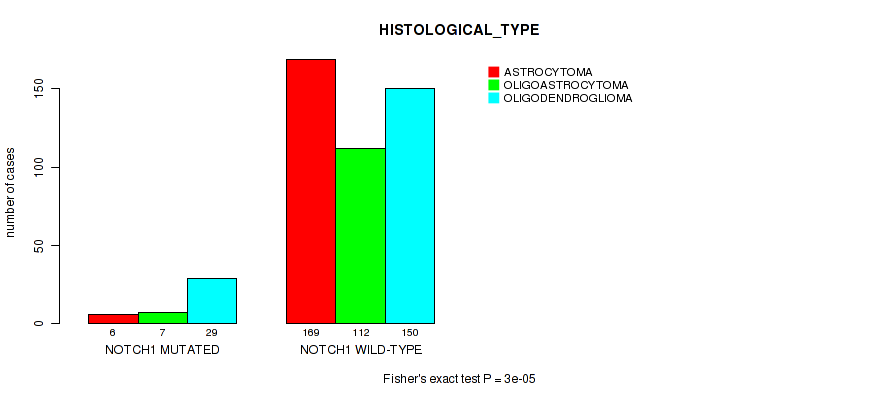
P value = 6.3e-09 (logrank test), Q value = 1.4e-06
Table S11. Gene #6: 'EGFR MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 471 | 93 | 0.0 - 211.2 (18.6) |
| EGFR MUTATED | 30 | 14 | 0.5 - 211.2 (11.1) |
| EGFR WILD-TYPE | 441 | 79 | 0.0 - 182.3 (19.2) |
Figure S11. Get High-res Image Gene #6: 'EGFR MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
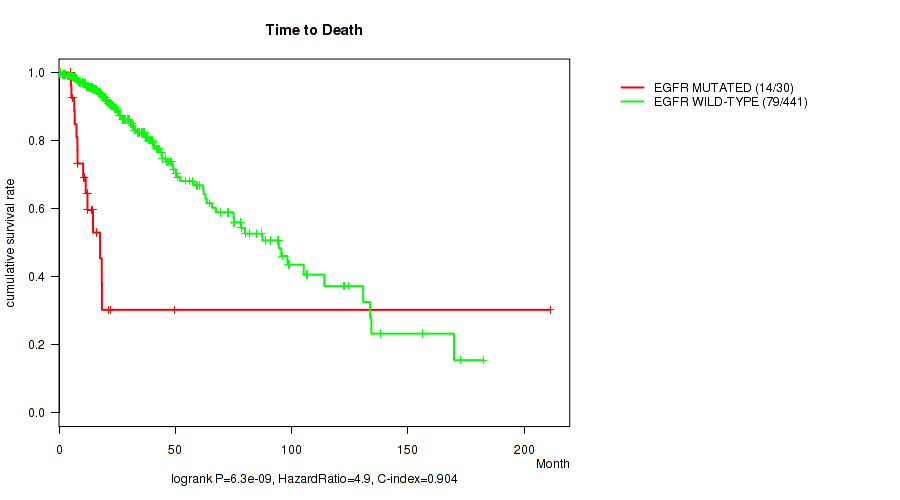
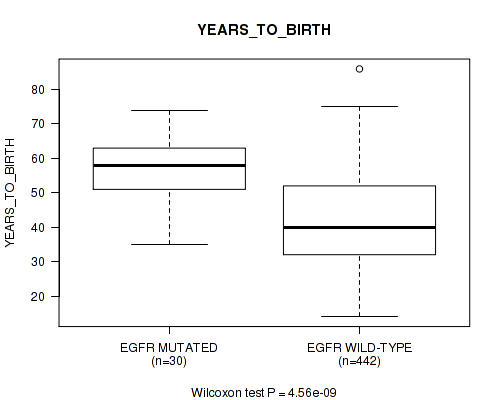
P value = 4.56e-09 (Wilcoxon-test), Q value = 1.3e-06
Table S12. Gene #6: 'EGFR MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 472 | 43.0 (13.5) |
| EGFR MUTATED | 30 | 57.3 (9.7) |
| EGFR WILD-TYPE | 442 | 42.0 (13.1) |
Figure S12. Get High-res Image Gene #6: 'EGFR MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
P value = 0.00128 (Fisher's exact test), Q value = 0.086
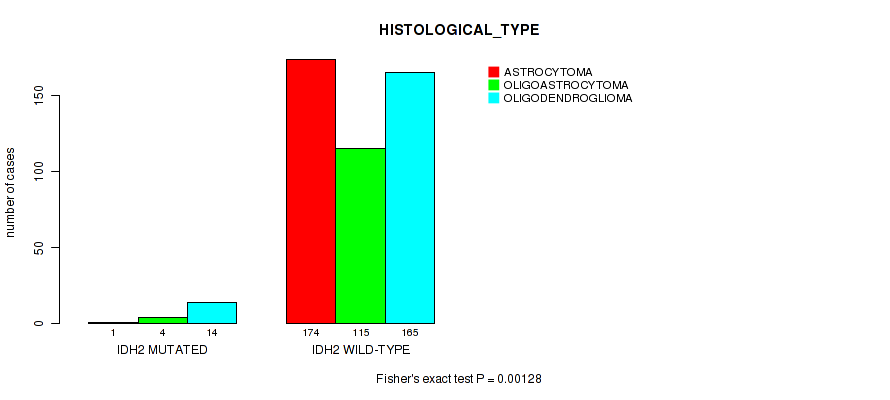
Table S13. Gene #8: 'IDH2 MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 175 | 119 | 179 |
| IDH2 MUTATED | 1 | 4 | 14 |
| IDH2 WILD-TYPE | 174 | 115 | 165 |
Figure S13. Get High-res Image Gene #8: 'IDH2 MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
P value = 2.17e-05 (logrank test), Q value = 0.0021
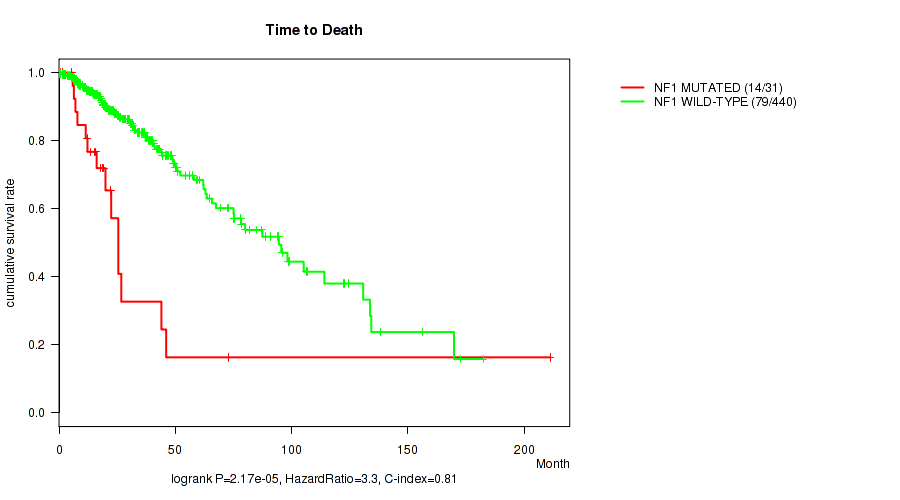
Table S14. Gene #9: 'NF1 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 471 | 93 | 0.0 - 211.2 (18.6) |
| NF1 MUTATED | 31 | 14 | 0.2 - 211.2 (16.2) |
| NF1 WILD-TYPE | 440 | 79 | 0.0 - 182.3 (18.7) |
Figure S14. Get High-res Image Gene #9: 'NF1 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
P value = 0.0039 (Fisher's exact test), Q value = 0.21
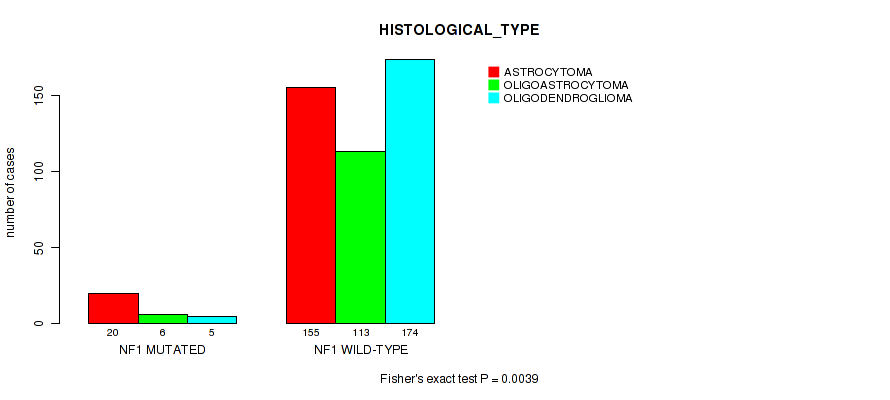
Table S15. Gene #9: 'NF1 MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 175 | 119 | 179 |
| NF1 MUTATED | 20 | 6 | 5 |
| NF1 WILD-TYPE | 155 | 113 | 174 |
Figure S15. Get High-res Image Gene #9: 'NF1 MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
P value = 2.57e-05 (logrank test), Q value = 0.0023
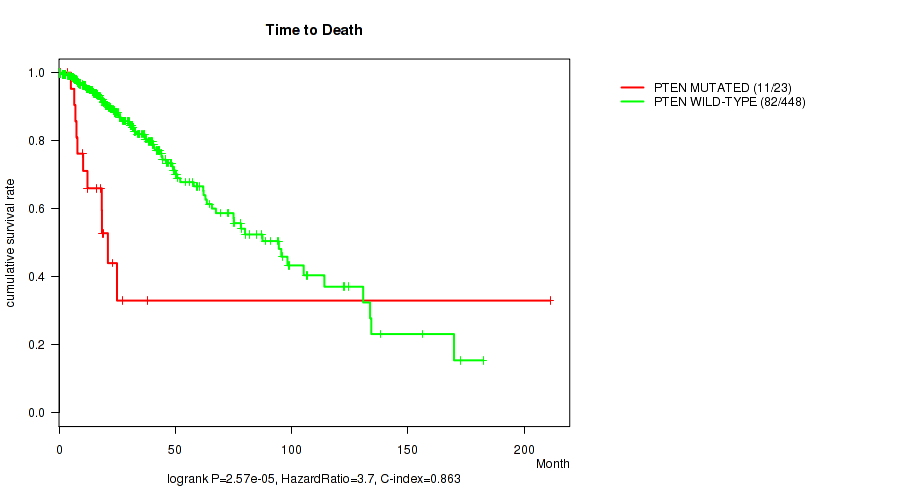
Table S16. Gene #10: 'PTEN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 471 | 93 | 0.0 - 211.2 (18.6) |
| PTEN MUTATED | 23 | 11 | 0.5 - 211.2 (16.2) |
| PTEN WILD-TYPE | 448 | 82 | 0.0 - 182.3 (18.8) |
Figure S16. Get High-res Image Gene #10: 'PTEN MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
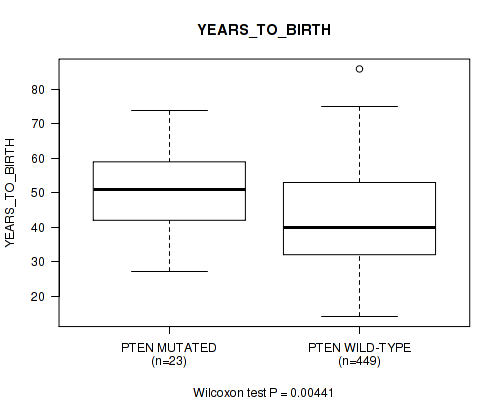
P value = 0.00441 (Wilcoxon-test), Q value = 0.22
Table S17. Gene #10: 'PTEN MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 472 | 43.0 (13.5) |
| PTEN MUTATED | 23 | 50.4 (11.8) |
| PTEN WILD-TYPE | 449 | 42.6 (13.4) |
Figure S17. Get High-res Image Gene #10: 'PTEN MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
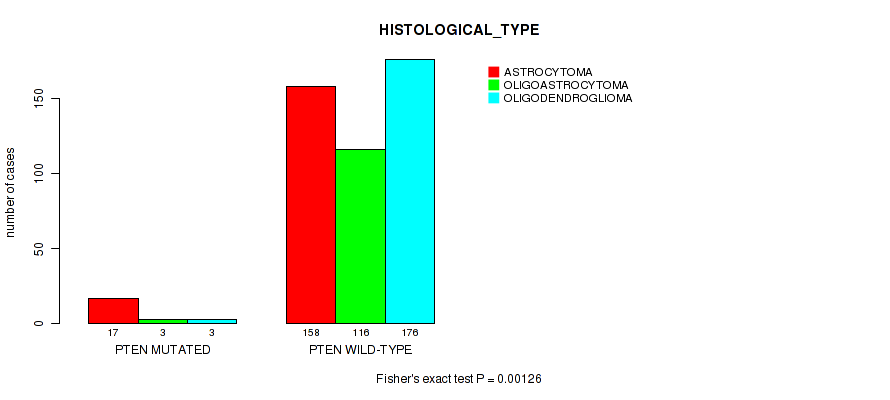
P value = 0.00126 (Fisher's exact test), Q value = 0.086
Table S18. Gene #10: 'PTEN MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 175 | 119 | 179 |
| PTEN MUTATED | 17 | 3 | 3 |
| PTEN WILD-TYPE | 158 | 116 | 176 |
Figure S18. Get High-res Image Gene #10: 'PTEN MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
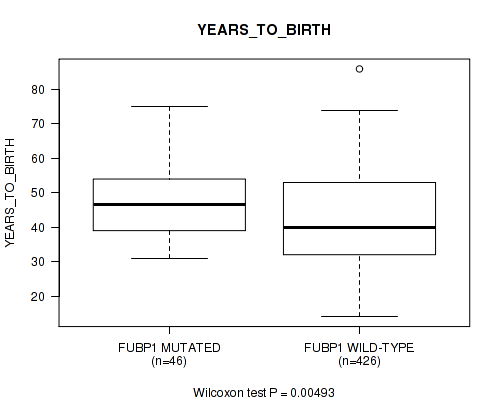
P value = 0.00493 (Wilcoxon-test), Q value = 0.23
Table S19. Gene #11: 'FUBP1 MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 472 | 43.0 (13.5) |
| FUBP1 MUTATED | 46 | 47.2 (9.6) |
| FUBP1 WILD-TYPE | 426 | 42.5 (13.7) |
Figure S19. Get High-res Image Gene #11: 'FUBP1 MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
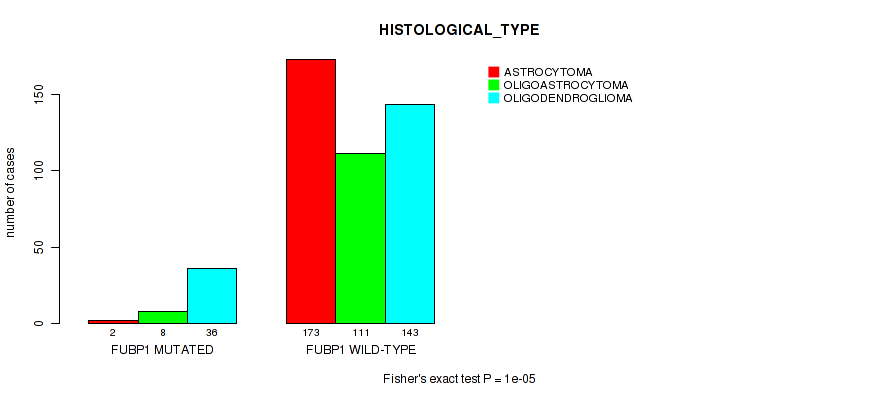
P value = 1e-05 (Fisher's exact test), Q value = 0.001
Table S20. Gene #11: 'FUBP1 MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
| nPatients | ASTROCYTOMA | OLIGOASTROCYTOMA | OLIGODENDROGLIOMA |
|---|---|---|---|
| ALL | 175 | 119 | 179 |
| FUBP1 MUTATED | 2 | 8 | 36 |
| FUBP1 WILD-TYPE | 173 | 111 | 143 |
Figure S20. Get High-res Image Gene #11: 'FUBP1 MUTATION STATUS' versus Clinical Feature #5: 'HISTOLOGICAL_TYPE'
P value = 0.00395 (Wilcoxon-test), Q value = 0.21
Table S21. Gene #29: 'ZNF292 MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 472 | 43.0 (13.5) |
| ZNF292 MUTATED | 13 | 53.7 (12.0) |
| ZNF292 WILD-TYPE | 459 | 42.7 (13.4) |
Figure S21. Get High-res Image Gene #29: 'ZNF292 MUTATION STATUS' versus Clinical Feature #2: 'YEARS_TO_BIRTH'
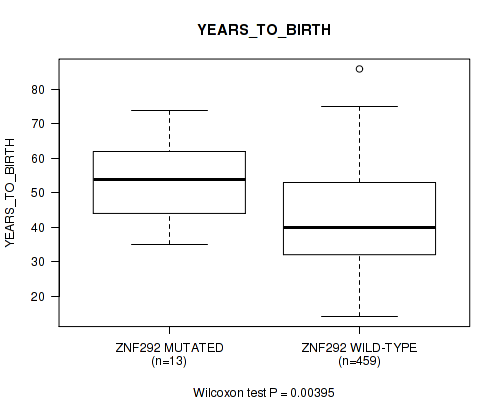
P value = 5.98e-08 (logrank test), Q value = 1.1e-05
Table S22. Gene #44: 'PLCG1 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 471 | 93 | 0.0 - 211.2 (18.6) |
| PLCG1 MUTATED | 6 | 4 | 6.7 - 23.7 (11.3) |
| PLCG1 WILD-TYPE | 465 | 89 | 0.0 - 211.2 (18.6) |
Figure S22. Get High-res Image Gene #44: 'PLCG1 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
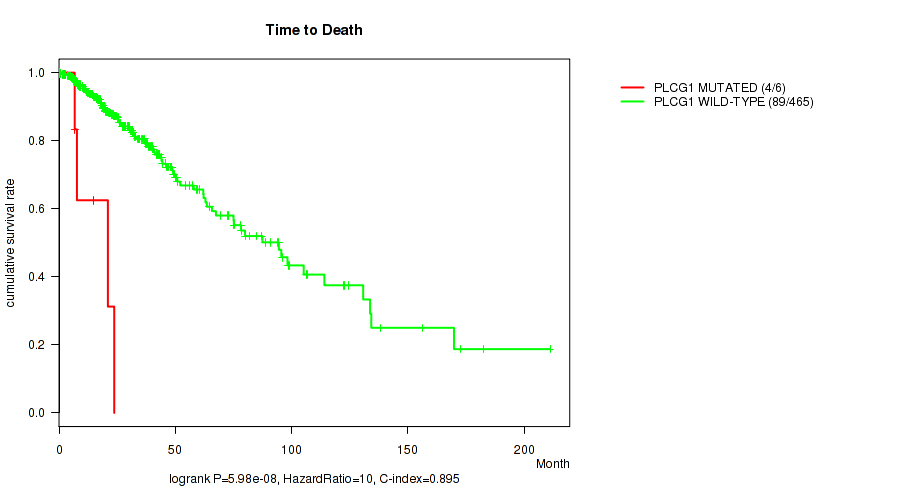
P value = 0.000413 (logrank test), Q value = 0.031
Table S23. Gene #71: 'TMEM184A MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 471 | 93 | 0.0 - 211.2 (18.6) |
| TMEM184A MUTATED | 3 | 1 | 0.3 - 9.2 (6.5) |
| TMEM184A WILD-TYPE | 468 | 92 | 0.0 - 211.2 (18.7) |
Figure S23. Get High-res Image Gene #71: 'TMEM184A MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
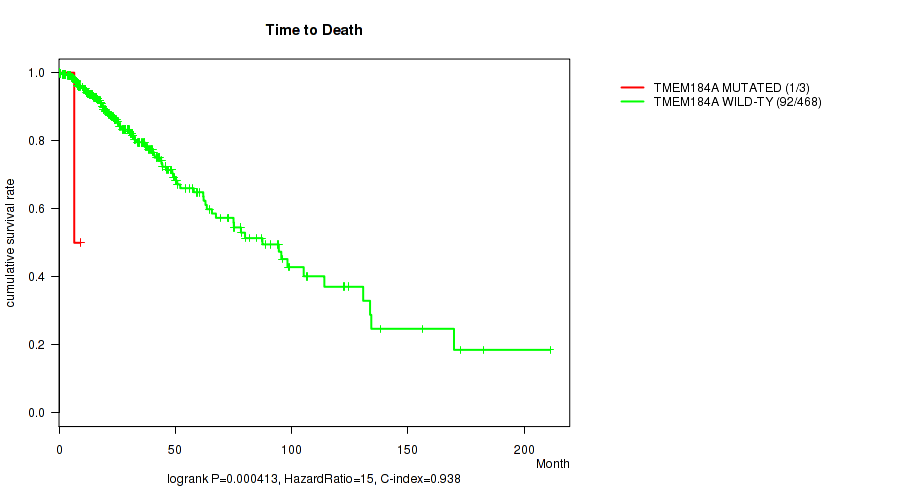
P value = 0.0029 (Fisher's exact test), Q value = 0.18
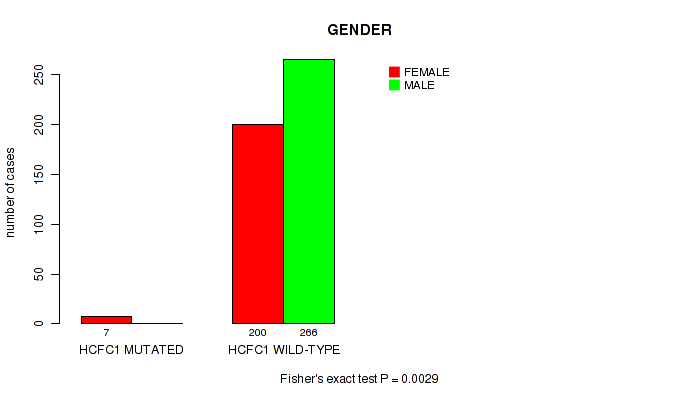
Table S24. Gene #99: 'HCFC1 MUTATION STATUS' versus Clinical Feature #3: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 207 | 266 |
| HCFC1 MUTATED | 7 | 0 |
| HCFC1 WILD-TYPE | 200 | 266 |
Figure S24. Get High-res Image Gene #99: 'HCFC1 MUTATION STATUS' versus Clinical Feature #3: 'GENDER'
P value = 0.00346 (logrank test), Q value = 0.21
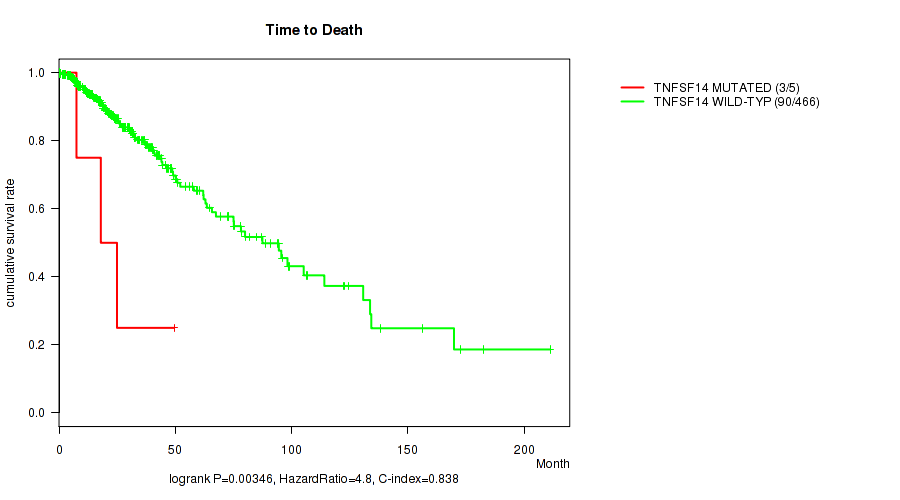
Table S25. Gene #101: 'TNFSF14 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 471 | 93 | 0.0 - 211.2 (18.6) |
| TNFSF14 MUTATED | 5 | 3 | 0.1 - 49.5 (18.0) |
| TNFSF14 WILD-TYPE | 466 | 90 | 0.0 - 211.2 (18.6) |
Figure S25. Get High-res Image Gene #101: 'TNFSF14 MUTATION STATUS' versus Clinical Feature #1: 'Time to Death'
-
Mutation data file = sample_sig_gene_table.txt from Mutsig_2CV pipeline
-
Processed Mutation data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/LGG-TP/15716584/transformed.cor.cli.txt
-
Clinical data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/Append_Data/LGG-TP/15082624/LGG-TP.merged_data.txt
-
Number of patients = 473
-
Number of significantly mutated genes = 143
-
Number of selected clinical features = 8
-
Exclude genes that fewer than K tumors have mutations, K = 3
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.