This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and molecular subtypes.
Testing the association between copy number variation 59 arm-level events and 10 molecular subtypes across 501 patients, 279 significant findings detected with P value < 0.05 and Q value < 0.25.
-
1p gain cnv correlated to 'CN_CNMF'.
-
1q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
3p gain cnv correlated to 'CN_CNMF'.
-
4p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
4q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
5p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
5q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
7p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
7q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
8p gain cnv correlated to 'CN_CNMF'.
-
8q gain cnv correlated to 'CN_CNMF'.
-
9p gain cnv correlated to 'CN_CNMF'.
-
9q gain cnv correlated to 'CN_CNMF'.
-
11p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
11q gain cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
12p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
12q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
13q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
14q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
16p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
16q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
17p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
17q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CHIERARCHICAL', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
18p gain cnv correlated to 'CN_CNMF'.
-
18q gain cnv correlated to 'CN_CNMF'.
-
19p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
19q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
20p gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
20q gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
21q gain cnv correlated to 'CN_CNMF'.
-
xp gain cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MRNASEQ_CNMF'.
-
xq gain cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
1p loss cnv correlated to 'CN_CNMF'.
-
1q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
2p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
2q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
3p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
3q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
6p loss cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
6q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'RPPA_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
8p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
8q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
9p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
9q loss cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
10p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
10q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
11p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
11q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
13q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
15q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
17p loss cnv correlated to 'CN_CNMF'.
-
18p loss cnv correlated to 'CN_CNMF'.
-
18q loss cnv correlated to 'CN_CNMF'.
-
19p loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
21q loss cnv correlated to 'CN_CNMF' and 'MIRSEQ_CHIERARCHICAL'.
-
22q loss cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', 'MIRSEQ_MATURE_CNMF', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
-
xp loss cnv correlated to 'MIRSEQ_MATURE_CNMF'.
-
xq loss cnv correlated to 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 59 arm-level events and 10 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 279 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| 1q gain | 26 (5%) | 475 |
1e-05 (0.000105) |
0.0161 (0.0413) |
0.00302 (0.0102) |
0.0645 (0.129) |
0.00144 (0.00548) |
4e-05 (0.000288) |
0.087 (0.167) |
0.0493 (0.105) |
0.0146 (0.0384) |
0.0154 (0.0401) |
| 7q gain | 20 (4%) | 481 |
1e-05 (0.000105) |
1e-05 (0.000105) |
0.572 (0.711) |
0.0132 (0.0355) |
0.00013 (0.000737) |
2e-05 (0.000182) |
5e-05 (0.000347) |
1e-05 (0.000105) |
0.612 (0.748) |
0.00089 (0.00365) |
| 17p gain | 16 (3%) | 485 |
1e-05 (0.000105) |
1e-05 (0.000105) |
0.537 (0.676) |
0.0278 (0.0642) |
1e-05 (0.000105) |
1e-05 (0.000105) |
2e-05 (0.000182) |
1e-05 (0.000105) |
0.362 (0.509) |
0.00027 (0.00132) |
| 17q gain | 18 (4%) | 483 |
1e-05 (0.000105) |
1e-05 (0.000105) |
0.636 (0.763) |
0.016 (0.0412) |
1e-05 (0.000105) |
1e-05 (0.000105) |
1e-05 (0.000105) |
1e-05 (0.000105) |
0.512 (0.653) |
0.00017 (0.00092) |
| 6q loss | 7 (1%) | 494 |
4e-05 (0.000288) |
0.00234 (0.00827) |
0.0124 (0.0341) |
0.575 (0.712) |
0.028 (0.0642) |
0.018 (0.044) |
0.00489 (0.0155) |
0.00064 (0.00278) |
1 (1.00) |
0.00979 (0.0279) |
| 22q loss | 89 (18%) | 412 |
1e-05 (0.000105) |
1e-05 (0.000105) |
0.12 (0.204) |
0.502 (0.643) |
1e-05 (0.000105) |
1e-05 (0.000105) |
0.00042 (0.00197) |
4e-05 (0.000288) |
0.00018 (0.000957) |
0.00026 (0.00128) |
| 5p gain | 21 (4%) | 480 |
1e-05 (0.000105) |
0.00187 (0.00685) |
0.529 (0.671) |
0.233 (0.356) |
0.00532 (0.0167) |
7e-05 (0.000459) |
0.0012 (0.00475) |
0.00013 (0.000737) |
0.598 (0.734) |
0.0033 (0.0111) |
| 5q gain | 17 (3%) | 484 |
1e-05 (0.000105) |
3e-05 (0.000242) |
0.849 (0.957) |
0.0791 (0.153) |
0.00017 (0.00092) |
4e-05 (0.000288) |
0.00012 (0.00073) |
2e-05 (0.000182) |
0.839 (0.956) |
0.00419 (0.0137) |
| 7p gain | 18 (4%) | 483 |
1e-05 (0.000105) |
5e-05 (0.000347) |
0.662 (0.789) |
0.0526 (0.11) |
0.00065 (0.00278) |
0.0001 (0.000628) |
0.00038 (0.00181) |
2e-05 (0.000182) |
0.576 (0.712) |
0.00555 (0.0172) |
| 12p gain | 14 (3%) | 487 |
1e-05 (0.000105) |
1e-05 (0.000105) |
0.852 (0.957) |
0.0798 (0.154) |
1e-05 (0.000105) |
1e-05 (0.000105) |
5e-05 (0.000347) |
1e-05 (0.000105) |
0.556 (0.695) |
0.00242 (0.00845) |
| 12q gain | 15 (3%) | 486 |
1e-05 (0.000105) |
1e-05 (0.000105) |
0.85 (0.957) |
0.0813 (0.156) |
1e-05 (0.000105) |
1e-05 (0.000105) |
3e-05 (0.000242) |
2e-05 (0.000182) |
0.386 (0.537) |
0.00098 (0.00399) |
| 14q gain | 11 (2%) | 490 |
1e-05 (0.000105) |
0.00019 (0.001) |
1 (1.00) |
0.159 (0.262) |
0.00057 (0.00257) |
0.00012 (0.00073) |
0.00074 (0.00314) |
0.0001 (0.000628) |
0.851 (0.957) |
0.00217 (0.00771) |
| 16p gain | 15 (3%) | 486 |
1e-05 (0.000105) |
4e-05 (0.000288) |
0.633 (0.763) |
0.0799 (0.154) |
0.00017 (0.00092) |
3e-05 (0.000242) |
0.00011 (0.000683) |
3e-05 (0.000242) |
0.435 (0.584) |
0.00078 (0.00326) |
| 16q gain | 13 (3%) | 488 |
1e-05 (0.000105) |
0.00017 (0.00092) |
0.796 (0.922) |
0.277 (0.404) |
0.00088 (0.00363) |
0.00025 (0.00125) |
0.00163 (0.00601) |
0.00013 (0.000737) |
0.62 (0.755) |
0.00756 (0.0225) |
| 19p gain | 8 (2%) | 493 |
1e-05 (0.000105) |
0.0002 (0.00104) |
0.00043 (0.002) |
0.00023 (0.00116) |
0.00013 (0.000737) |
3e-05 (0.000242) |
0.453 (0.599) |
0.0223 (0.0527) |
||
| 19q gain | 10 (2%) | 491 |
1e-05 (0.000105) |
2e-05 (0.000182) |
0.636 (0.763) |
0.0635 (0.128) |
3e-05 (0.000242) |
4e-05 (0.000288) |
1e-05 (0.000105) |
1e-05 (0.000105) |
0.638 (0.763) |
0.00159 (0.00594) |
| 2p loss | 10 (2%) | 491 |
1e-05 (0.000105) |
4e-05 (0.000288) |
0.775 (0.911) |
0.0915 (0.171) |
7e-05 (0.000459) |
3e-05 (0.000242) |
1e-05 (0.000105) |
2e-05 (0.000182) |
0.234 (0.356) |
0.00044 (0.00203) |
| 2q loss | 10 (2%) | 491 |
1e-05 (0.000105) |
3e-05 (0.000242) |
0.773 (0.911) |
0.0896 (0.17) |
6e-05 (0.000407) |
6e-05 (0.000407) |
1e-05 (0.000105) |
1e-05 (0.000105) |
0.232 (0.356) |
0.00056 (0.00256) |
| 8p loss | 4 (1%) | 497 |
0.0001 (0.000628) |
0.0173 (0.0428) |
0.0364 (0.0814) |
0.0494 (0.105) |
0.00777 (0.0229) |
0.00422 (0.0137) |
0.108 (0.191) |
0.0189 (0.0453) |
||
| 8q loss | 4 (1%) | 497 |
0.00013 (0.000737) |
0.0174 (0.0428) |
0.0362 (0.0812) |
0.0458 (0.0992) |
0.00774 (0.0229) |
0.00421 (0.0137) |
0.111 (0.194) |
0.0187 (0.0451) |
||
| 10p loss | 7 (1%) | 494 |
1e-05 (0.000105) |
0.0116 (0.0322) |
0.496 (0.637) |
0.42 (0.566) |
0.0206 (0.0489) |
0.0154 (0.0401) |
0.00474 (0.0152) |
0.00063 (0.00277) |
1 (1.00) |
0.0112 (0.0313) |
| 11p loss | 7 (1%) | 494 |
1e-05 (0.000105) |
0.011 (0.0309) |
0.495 (0.637) |
0.959 (1.00) |
0.00648 (0.0199) |
0.0175 (0.0429) |
0.00494 (0.0156) |
0.00059 (0.00262) |
0.801 (0.923) |
0.0107 (0.0303) |
| 11q loss | 9 (2%) | 492 |
1e-05 (0.000105) |
0.0173 (0.0428) |
0.321 (0.454) |
0.814 (0.936) |
0.0158 (0.0408) |
0.0184 (0.0447) |
0.00057 (0.00257) |
0.00247 (0.00857) |
0.909 (1.00) |
0.00255 (0.0088) |
| 13q loss | 14 (3%) | 487 |
0.00161 (0.00597) |
0.0148 (0.0387) |
0.0626 (0.127) |
0.138 (0.232) |
0.00534 (0.0167) |
0.00799 (0.0233) |
0.00105 (0.00421) |
0.00041 (0.00194) |
0.102 (0.182) |
0.0127 (0.0346) |
| 15q loss | 8 (2%) | 493 |
0.00014 (0.000787) |
0.0002 (0.00104) |
0.193 (0.306) |
0.09 (0.17) |
0.00059 (0.00262) |
0.00026 (0.00128) |
7e-05 (0.000459) |
2e-05 (0.000182) |
0.411 (0.561) |
0.00195 (0.00706) |
| 19p loss | 5 (1%) | 496 |
0.00932 (0.0269) |
0.00657 (0.02) |
0.324 (0.457) |
0.237 (0.36) |
0.0167 (0.0419) |
0.0129 (0.0349) |
0.00239 (0.00839) |
0.00107 (0.00427) |
0.0874 (0.167) |
0.00145 (0.00548) |
| 4p gain | 5 (1%) | 496 |
1e-05 (0.000105) |
0.00668 (0.0202) |
0.0165 (0.0419) |
0.0125 (0.0341) |
0.0416 (0.0915) |
0.0139 (0.0373) |
1 (1.00) |
0.0754 (0.147) |
||
| 4q gain | 5 (1%) | 496 |
1e-05 (0.000105) |
0.00659 (0.02) |
0.0163 (0.0417) |
0.0125 (0.0341) |
0.0411 (0.0908) |
0.0142 (0.038) |
1 (1.00) |
0.078 (0.152) |
||
| 20p gain | 12 (2%) | 489 |
1e-05 (0.000105) |
0.00029 (0.0014) |
0.794 (0.922) |
0.0944 (0.173) |
0.0026 (0.00892) |
0.00079 (0.00328) |
0.00424 (0.0137) |
0.00075 (0.00316) |
0.915 (1.00) |
0.0955 (0.174) |
| 20q gain | 12 (2%) | 489 |
1e-05 (0.000105) |
0.00031 (0.00149) |
0.795 (0.922) |
0.0926 (0.171) |
0.00277 (0.00945) |
0.00103 (0.00416) |
0.00406 (0.0135) |
0.00065 (0.00278) |
0.912 (1.00) |
0.0947 (0.173) |
| 3q loss | 4 (1%) | 497 |
0.00022 (0.00113) |
0.0174 (0.0428) |
0.496 (0.637) |
0.424 (0.569) |
0.0358 (0.0805) |
0.0459 (0.0992) |
0.00794 (0.0233) |
0.00394 (0.0132) |
1 (1.00) |
0.0675 (0.135) |
| 9p loss | 19 (4%) | 482 |
1e-05 (0.000105) |
0.0379 (0.084) |
0.242 (0.365) |
0.648 (0.774) |
0.031 (0.0701) |
0.0615 (0.125) |
0.0087 (0.0253) |
0.00712 (0.0213) |
0.3 (0.433) |
0.00422 (0.0137) |
| 10q loss | 6 (1%) | 495 |
1e-05 (0.000105) |
0.0292 (0.0663) |
0.0532 (0.11) |
0.0456 (0.0992) |
0.0144 (0.0382) |
0.00445 (0.0143) |
1 (1.00) |
0.028 (0.0642) |
||
| 11p gain | 6 (1%) | 495 |
0.00123 (0.00481) |
0.0279 (0.0642) |
0.781 (0.914) |
0.299 (0.432) |
0.0527 (0.11) |
0.0447 (0.098) |
0.00946 (0.0271) |
0.00645 (0.0199) |
0.469 (0.611) |
0.0965 (0.175) |
| 1q loss | 3 (1%) | 498 |
0.00132 (0.00509) |
0.0478 (0.103) |
0.0997 (0.179) |
0.151 (0.252) |
0.0266 (0.0624) |
0.0167 (0.0419) |
1 (1.00) |
0.192 (0.305) |
||
| 3p loss | 3 (1%) | 498 |
0.00123 (0.00481) |
0.0494 (0.105) |
0.1 (0.179) |
0.153 (0.254) |
0.0267 (0.0624) |
0.0168 (0.042) |
1 (1.00) |
0.193 (0.306) |
||
| 9q loss | 25 (5%) | 476 |
1e-05 (0.000105) |
0.104 (0.184) |
0.172 (0.279) |
0.114 (0.196) |
0.172 (0.279) |
0.0609 (0.124) |
0.00933 (0.0269) |
0.00704 (0.0212) |
0.278 (0.404) |
0.00488 (0.0155) |
| 11q gain | 5 (1%) | 496 |
0.00013 (0.000737) |
0.0602 (0.123) |
0.78 (0.914) |
0.301 (0.433) |
0.111 (0.194) |
0.125 (0.212) |
0.0267 (0.0624) |
0.0214 (0.0506) |
0.31 (0.444) |
0.273 (0.401) |
| 13q gain | 5 (1%) | 496 |
2e-05 (0.000182) |
0.0187 (0.0451) |
0.385 (0.537) |
0.213 (0.329) |
0.0486 (0.104) |
0.103 (0.183) |
0.236 (0.359) |
0.0724 (0.144) |
0.845 (0.957) |
0.196 (0.306) |
| xp gain | 8 (2%) | 493 |
1e-05 (0.000105) |
0.00191 (0.00696) |
0.838 (0.956) |
0.504 (0.644) |
0.045 (0.0984) |
0.0565 (0.116) |
0.178 (0.288) |
0.0737 (0.146) |
0.8 (0.923) |
0.184 (0.295) |
| xq loss | 3 (1%) | 498 |
0.0503 (0.106) |
0.0506 (0.106) |
0.277 (0.404) |
0.196 (0.306) |
0.0994 (0.179) |
0.151 (0.251) |
0.0281 (0.0642) |
0.0166 (0.0419) |
0.11 (0.194) |
0.019 (0.0454) |
| xq gain | 9 (2%) | 492 |
1e-05 (0.000105) |
0.0114 (0.0316) |
0.839 (0.956) |
0.503 (0.644) |
0.0974 (0.176) |
0.14 (0.235) |
0.281 (0.407) |
0.134 (0.226) |
0.909 (1.00) |
0.397 (0.545) |
| 6p loss | 4 (1%) | 497 |
0.00018 (0.000957) |
0.0463 (0.0997) |
0.458 (0.601) |
0.396 (0.545) |
0.113 (0.195) |
0.0641 (0.129) |
0.399 (0.545) |
0.196 (0.306) |
||
| 21q loss | 8 (2%) | 493 |
0.00065 (0.00278) |
0.162 (0.266) |
0.381 (0.533) |
0.214 (0.329) |
0.713 (0.841) |
0.418 (0.564) |
0.0912 (0.171) |
0.0287 (0.0654) |
0.464 (0.607) |
0.0554 (0.114) |
| 1p gain | 5 (1%) | 496 |
0.00146 (0.00549) |
0.842 (0.957) |
0.922 (1.00) |
0.882 (0.989) |
0.108 (0.191) |
0.0513 (0.107) |
0.274 (0.402) |
0.142 (0.238) |
||
| 3p gain | 3 (1%) | 498 |
0.00134 (0.00513) |
0.55 (0.69) |
0.494 (0.637) |
0.589 (0.725) |
0.304 (0.436) |
0.188 (0.3) |
1 (1.00) |
0.194 (0.306) |
||
| 8p gain | 6 (1%) | 495 |
0.00013 (0.000737) |
0.416 (0.564) |
0.454 (0.599) |
0.632 (0.763) |
0.597 (0.734) |
0.534 (0.673) |
0.451 (0.599) |
0.451 (0.599) |
1 (1.00) |
1 (1.00) |
| 8q gain | 7 (1%) | 494 |
4e-05 (0.000288) |
0.201 (0.313) |
0.455 (0.599) |
0.627 (0.763) |
0.826 (0.948) |
0.522 (0.664) |
0.8 (0.923) |
0.63 (0.763) |
0.9 (1.00) |
0.895 (1.00) |
| 9p gain | 3 (1%) | 498 |
0.0013 (0.00505) |
0.553 (0.693) |
0.494 (0.637) |
0.588 (0.725) |
1 (1.00) |
1 (1.00) |
0.418 (0.564) |
0.632 (0.763) |
||
| 9q gain | 4 (1%) | 497 |
0.0107 (0.0303) |
0.79 (0.922) |
0.902 (1.00) |
0.364 (0.51) |
0.838 (0.956) |
1 (1.00) |
0.166 (0.273) |
0.571 (0.711) |
||
| 18p gain | 6 (1%) | 495 |
1e-05 (0.000105) |
0.0926 (0.171) |
0.121 (0.206) |
0.268 (0.397) |
0.255 (0.381) |
0.113 (0.195) |
0.454 (0.599) |
0.396 (0.545) |
||
| 18q gain | 6 (1%) | 495 |
4e-05 (0.000288) |
0.0918 (0.171) |
0.119 (0.204) |
0.265 (0.394) |
0.256 (0.381) |
0.111 (0.194) |
0.454 (0.599) |
0.399 (0.545) |
||
| 21q gain | 5 (1%) | 496 |
0.00023 (0.00116) |
0.168 (0.275) |
0.241 (0.365) |
0.491 (0.637) |
0.453 (0.599) |
0.311 (0.444) |
0.692 (0.818) |
0.683 (0.81) |
||
| 1p loss | 4 (1%) | 497 |
0.0001 (0.000628) |
0.184 (0.295) |
0.244 (0.367) |
0.316 (0.45) |
0.114 (0.196) |
0.0646 (0.129) |
0.846 (0.957) |
0.195 (0.306) |
||
| 17p loss | 8 (2%) | 493 |
0.0144 (0.0382) |
0.414 (0.563) |
1 (1.00) |
0.389 (0.538) |
0.636 (0.763) |
0.601 (0.736) |
0.0922 (0.171) |
0.16 (0.264) |
0.126 (0.213) |
0.205 (0.318) |
| 18p loss | 4 (1%) | 497 |
0.002 (0.0072) |
0.183 (0.295) |
1 (1.00) |
0.389 (0.538) |
0.245 (0.367) |
0.317 (0.451) |
0.0741 (0.146) |
0.0922 (0.171) |
0.533 (0.673) |
0.273 (0.401) |
| 18q loss | 4 (1%) | 497 |
0.00214 (0.00765) |
0.185 (0.295) |
1 (1.00) |
0.387 (0.537) |
0.245 (0.367) |
0.32 (0.453) |
0.0743 (0.146) |
0.0902 (0.17) |
0.532 (0.673) |
0.273 (0.401) |
| xp loss | 4 (1%) | 497 |
0.113 (0.195) |
0.184 (0.295) |
0.279 (0.405) |
0.196 (0.306) |
0.458 (0.601) |
0.209 (0.323) |
0.0747 (0.146) |
0.0921 (0.171) |
0.0375 (0.0835) |
0.0952 (0.174) |
| 17q loss | 4 (1%) | 497 |
0.114 (0.196) |
0.792 (0.922) |
0.9 (1.00) |
0.665 (0.791) |
0.467 (0.609) |
0.568 (0.709) |
0.332 (0.467) |
0.684 (0.81) |
P value = 0.00146 (Fisher's exact test), Q value = 0.0055
Table S1. Gene #1: '1p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 1P GAIN MUTATED | 4 | 1 | 0 |
| 1P GAIN WILD-TYPE | 52 | 355 | 89 |
Figure S1. Get High-res Image Gene #1: '1p gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S2. Gene #2: '1q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 1Q GAIN MUTATED | 18 | 7 | 1 |
| 1Q GAIN WILD-TYPE | 38 | 349 | 88 |
Figure S2. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #1: 'CN_CNMF'
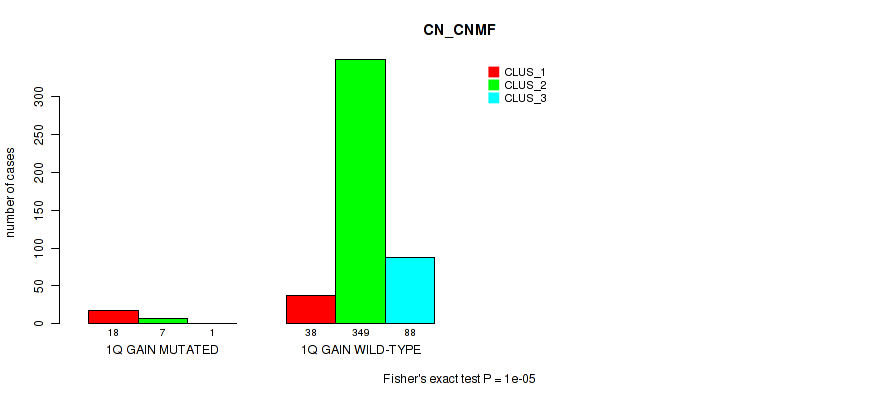
P value = 0.0161 (Fisher's exact test), Q value = 0.041
Table S3. Gene #2: '1q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 1Q GAIN MUTATED | 19 | 2 | 5 |
| 1Q GAIN WILD-TYPE | 258 | 152 | 65 |
Figure S3. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
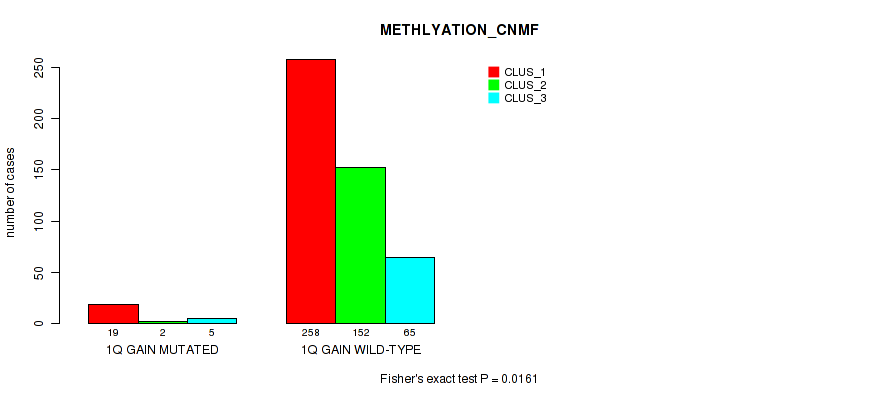
P value = 0.00302 (Fisher's exact test), Q value = 0.01
Table S4. Gene #2: '1q gain' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 79 | 81 |
| 1Q GAIN MUTATED | 0 | 9 | 2 |
| 1Q GAIN WILD-TYPE | 61 | 70 | 79 |
Figure S4. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #3: 'RPPA_CNMF'

P value = 0.00144 (Fisher's exact test), Q value = 0.0055
Table S5. Gene #2: '1q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 1Q GAIN MUTATED | 17 | 2 | 4 | 3 |
| 1Q GAIN WILD-TYPE | 151 | 153 | 55 | 114 |
Figure S5. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
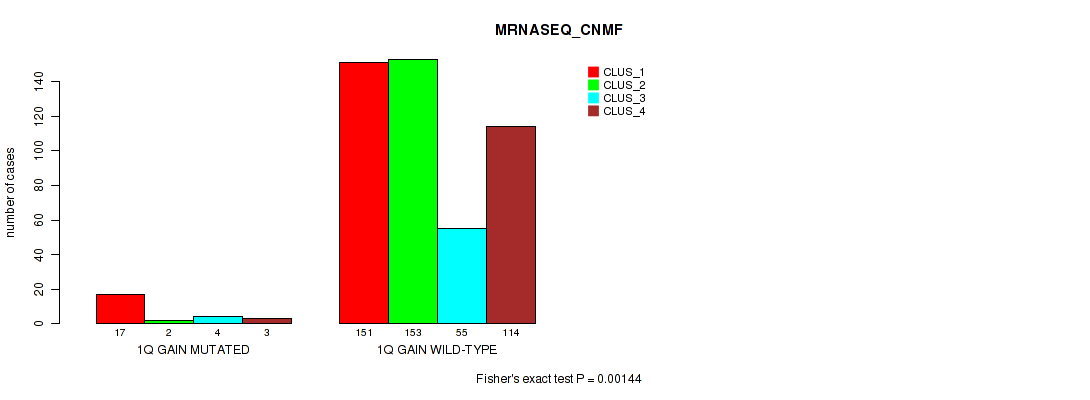
P value = 4e-05 (Fisher's exact test), Q value = 0.00029
Table S6. Gene #2: '1q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 1Q GAIN MUTATED | 16 | 2 | 1 | 3 | 4 |
| 1Q GAIN WILD-TYPE | 89 | 132 | 85 | 93 | 74 |
Figure S6. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
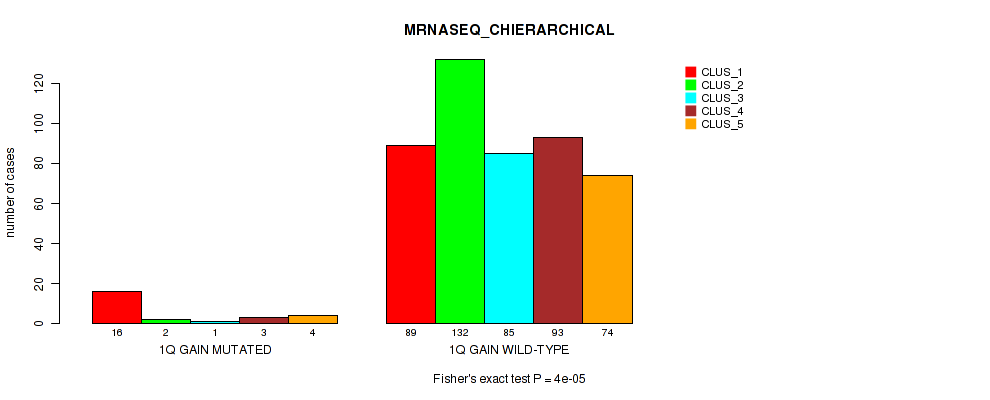
P value = 0.0493 (Fisher's exact test), Q value = 0.1
Table S7. Gene #2: '1q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 1Q GAIN MUTATED | 15 | 3 | 8 |
| 1Q GAIN WILD-TYPE | 164 | 125 | 185 |
Figure S7. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
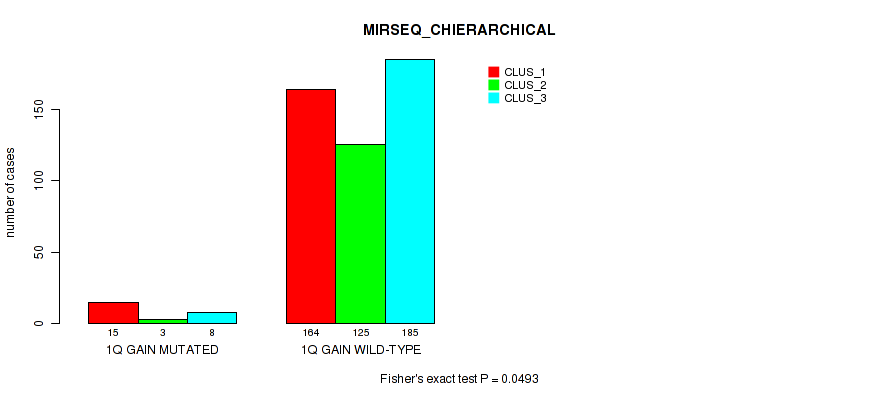
P value = 0.0146 (Fisher's exact test), Q value = 0.038
Table S8. Gene #2: '1q gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 160 | 164 | 146 |
| 1Q GAIN MUTATED | 14 | 5 | 3 |
| 1Q GAIN WILD-TYPE | 146 | 159 | 143 |
Figure S8. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
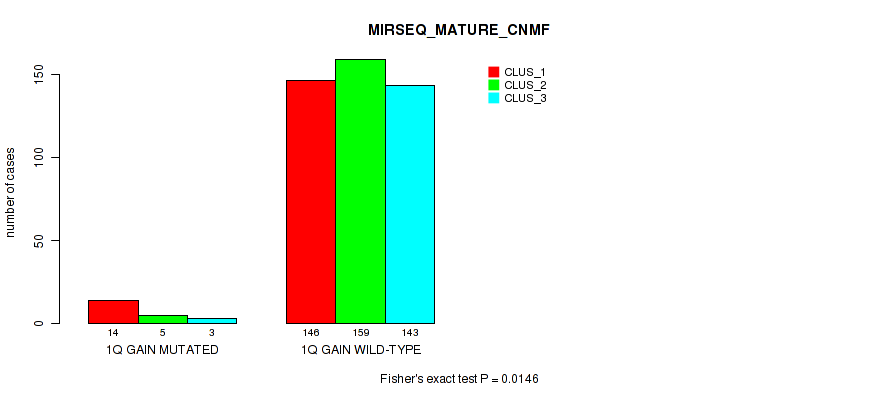
P value = 0.0154 (Fisher's exact test), Q value = 0.04
Table S9. Gene #2: '1q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 1Q GAIN MUTATED | 14 | 2 | 6 |
| 1Q GAIN WILD-TYPE | 151 | 124 | 173 |
Figure S9. Get High-res Image Gene #2: '1q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
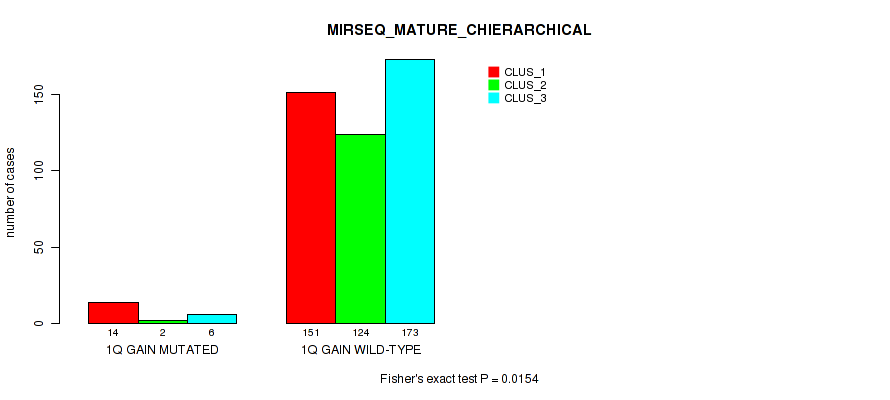
P value = 0.00134 (Fisher's exact test), Q value = 0.0051
Table S10. Gene #3: '3p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 3P GAIN MUTATED | 3 | 0 | 0 |
| 3P GAIN WILD-TYPE | 53 | 356 | 89 |
Figure S10. Get High-res Image Gene #3: '3p gain' versus Molecular Subtype #1: 'CN_CNMF'
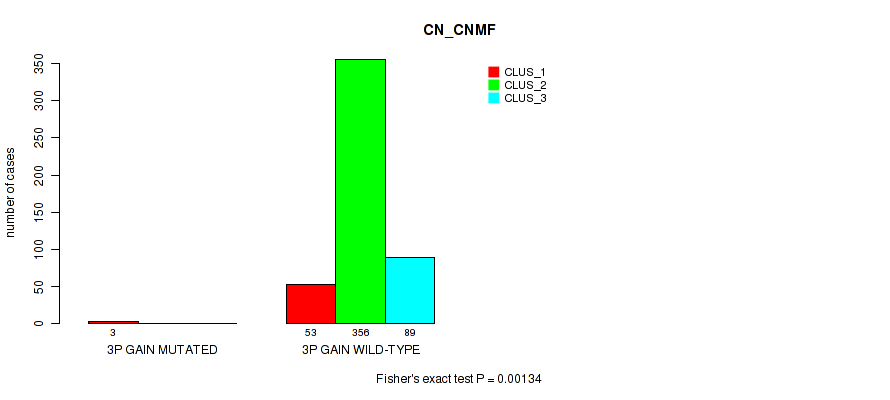
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S11. Gene #4: '4p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 4P GAIN MUTATED | 5 | 0 | 0 |
| 4P GAIN WILD-TYPE | 51 | 356 | 89 |
Figure S11. Get High-res Image Gene #4: '4p gain' versus Molecular Subtype #1: 'CN_CNMF'
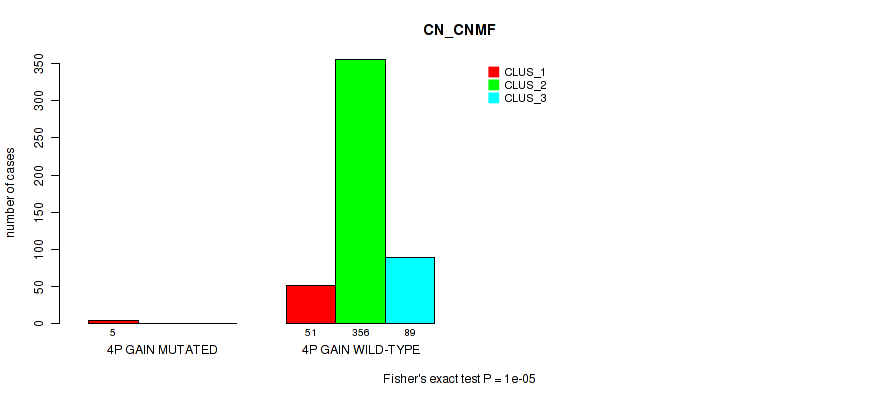
P value = 0.00668 (Fisher's exact test), Q value = 0.02
Table S12. Gene #4: '4p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 4P GAIN MUTATED | 0 | 5 | 0 |
| 4P GAIN WILD-TYPE | 277 | 149 | 70 |
Figure S12. Get High-res Image Gene #4: '4p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.0165 (Fisher's exact test), Q value = 0.042
Table S13. Gene #4: '4p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 4P GAIN MUTATED | 0 | 5 | 0 | 0 |
| 4P GAIN WILD-TYPE | 168 | 150 | 59 | 117 |
Figure S13. Get High-res Image Gene #4: '4p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
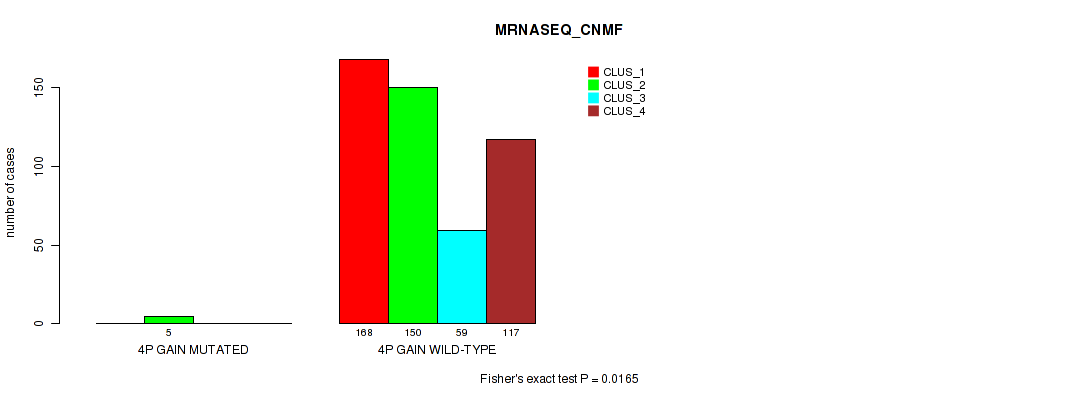
P value = 0.0125 (Fisher's exact test), Q value = 0.034
Table S14. Gene #4: '4p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 4P GAIN MUTATED | 0 | 5 | 0 | 0 | 0 |
| 4P GAIN WILD-TYPE | 105 | 129 | 86 | 96 | 78 |
Figure S14. Get High-res Image Gene #4: '4p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
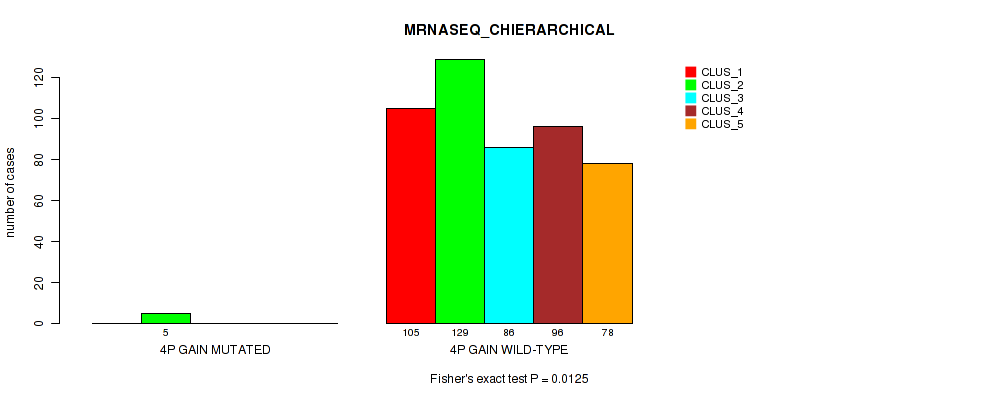
P value = 0.0416 (Fisher's exact test), Q value = 0.091
Table S15. Gene #4: '4p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 4P GAIN MUTATED | 1 | 4 | 0 |
| 4P GAIN WILD-TYPE | 181 | 147 | 167 |
Figure S15. Get High-res Image Gene #4: '4p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
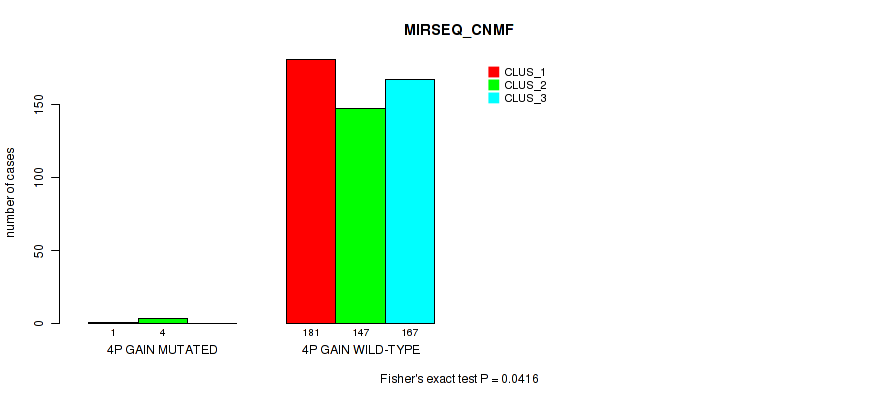
P value = 0.0139 (Fisher's exact test), Q value = 0.037
Table S16. Gene #4: '4p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 4P GAIN MUTATED | 1 | 4 | 0 |
| 4P GAIN WILD-TYPE | 178 | 124 | 193 |
Figure S16. Get High-res Image Gene #4: '4p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S17. Gene #5: '4q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 4Q GAIN MUTATED | 5 | 0 | 0 |
| 4Q GAIN WILD-TYPE | 51 | 356 | 89 |
Figure S17. Get High-res Image Gene #5: '4q gain' versus Molecular Subtype #1: 'CN_CNMF'
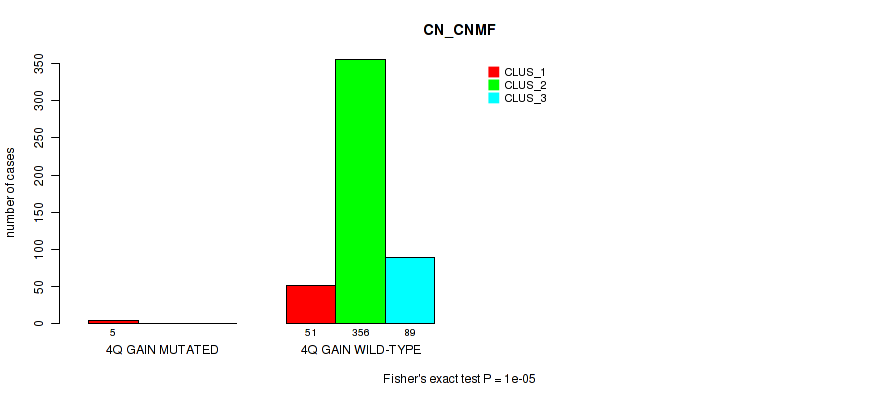
P value = 0.00659 (Fisher's exact test), Q value = 0.02
Table S18. Gene #5: '4q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 4Q GAIN MUTATED | 0 | 5 | 0 |
| 4Q GAIN WILD-TYPE | 277 | 149 | 70 |
Figure S18. Get High-res Image Gene #5: '4q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
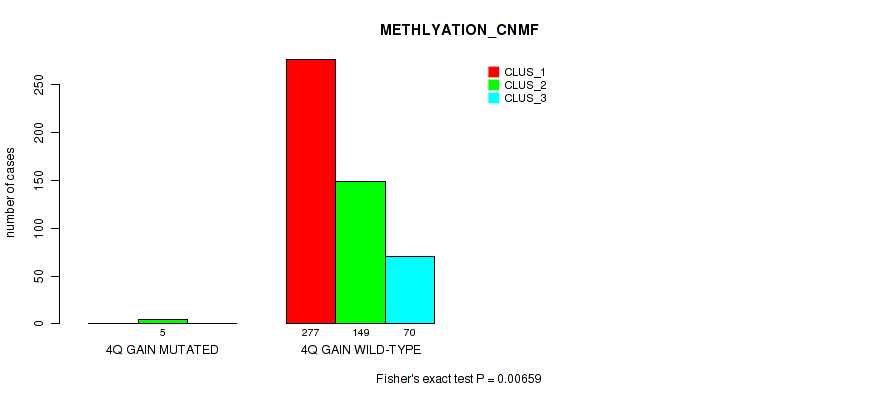
P value = 0.0163 (Fisher's exact test), Q value = 0.042
Table S19. Gene #5: '4q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 4Q GAIN MUTATED | 0 | 5 | 0 | 0 |
| 4Q GAIN WILD-TYPE | 168 | 150 | 59 | 117 |
Figure S19. Get High-res Image Gene #5: '4q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
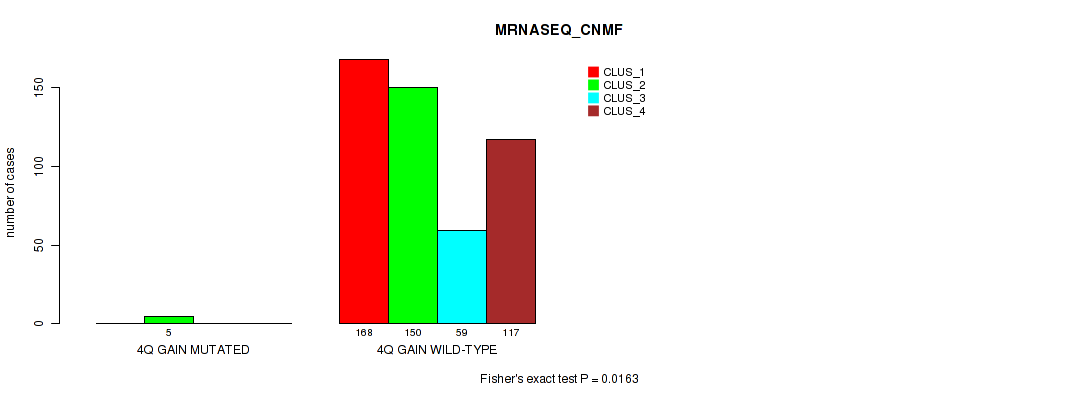
P value = 0.0125 (Fisher's exact test), Q value = 0.034
Table S20. Gene #5: '4q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 4Q GAIN MUTATED | 0 | 5 | 0 | 0 | 0 |
| 4Q GAIN WILD-TYPE | 105 | 129 | 86 | 96 | 78 |
Figure S20. Get High-res Image Gene #5: '4q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
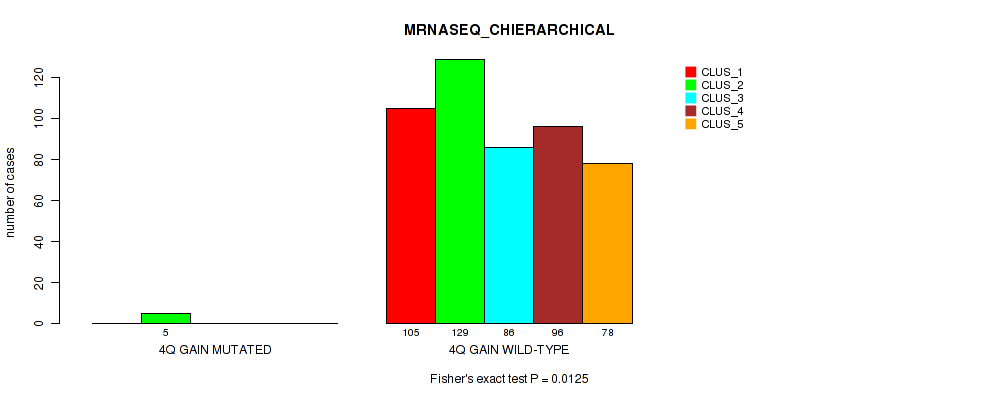
P value = 0.0411 (Fisher's exact test), Q value = 0.091
Table S21. Gene #5: '4q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 4Q GAIN MUTATED | 1 | 4 | 0 |
| 4Q GAIN WILD-TYPE | 181 | 147 | 167 |
Figure S21. Get High-res Image Gene #5: '4q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
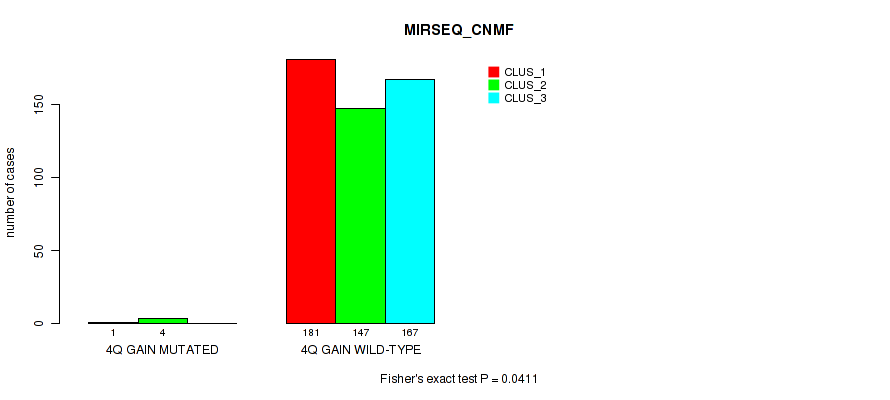
P value = 0.0142 (Fisher's exact test), Q value = 0.038
Table S22. Gene #5: '4q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 4Q GAIN MUTATED | 1 | 4 | 0 |
| 4Q GAIN WILD-TYPE | 178 | 124 | 193 |
Figure S22. Get High-res Image Gene #5: '4q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
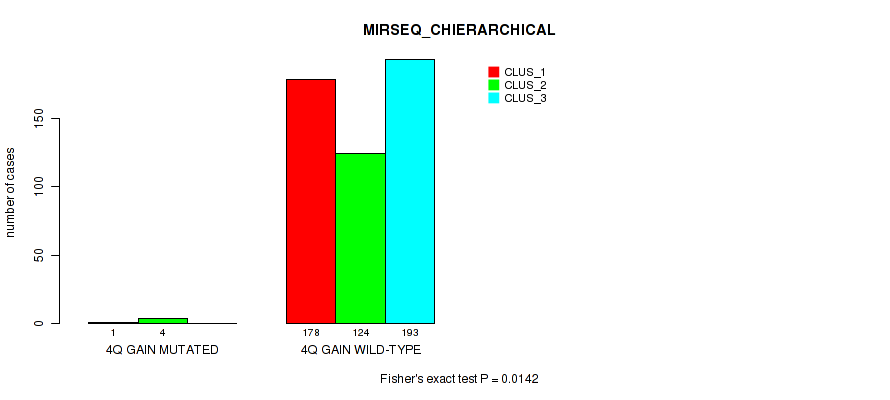
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S23. Gene #6: '5p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 5P GAIN MUTATED | 21 | 0 | 0 |
| 5P GAIN WILD-TYPE | 35 | 356 | 89 |
Figure S23. Get High-res Image Gene #6: '5p gain' versus Molecular Subtype #1: 'CN_CNMF'
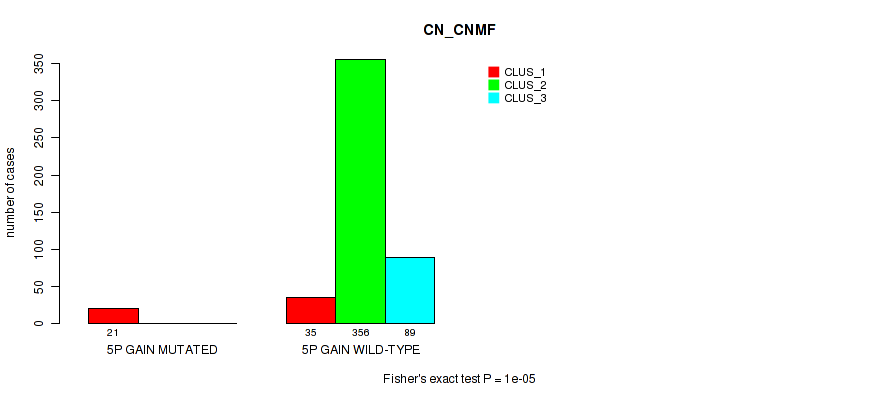
P value = 0.00187 (Fisher's exact test), Q value = 0.0069
Table S24. Gene #6: '5p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 5P GAIN MUTATED | 5 | 14 | 2 |
| 5P GAIN WILD-TYPE | 272 | 140 | 68 |
Figure S24. Get High-res Image Gene #6: '5p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00532 (Fisher's exact test), Q value = 0.017
Table S25. Gene #6: '5p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 5P GAIN MUTATED | 5 | 14 | 1 | 1 |
| 5P GAIN WILD-TYPE | 163 | 141 | 58 | 116 |
Figure S25. Get High-res Image Gene #6: '5p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
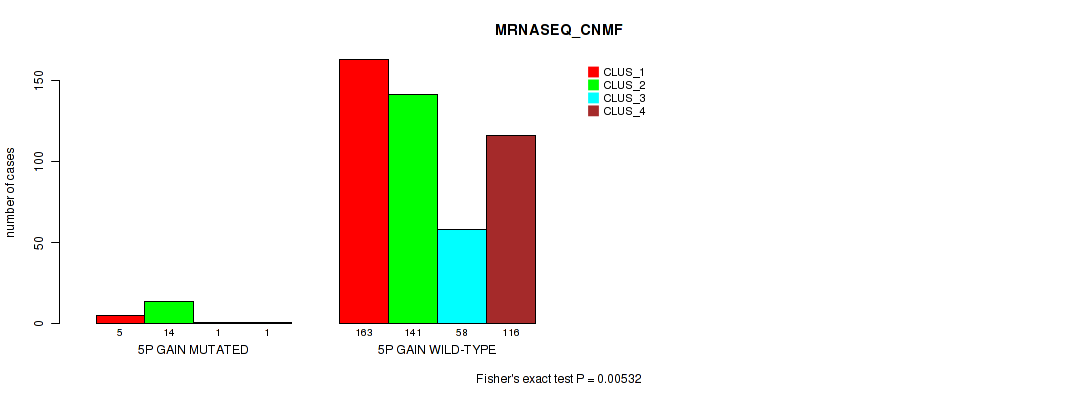
P value = 7e-05 (Fisher's exact test), Q value = 0.00046
Table S26. Gene #6: '5p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 5P GAIN MUTATED | 6 | 14 | 0 | 1 | 0 |
| 5P GAIN WILD-TYPE | 99 | 120 | 86 | 95 | 78 |
Figure S26. Get High-res Image Gene #6: '5p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
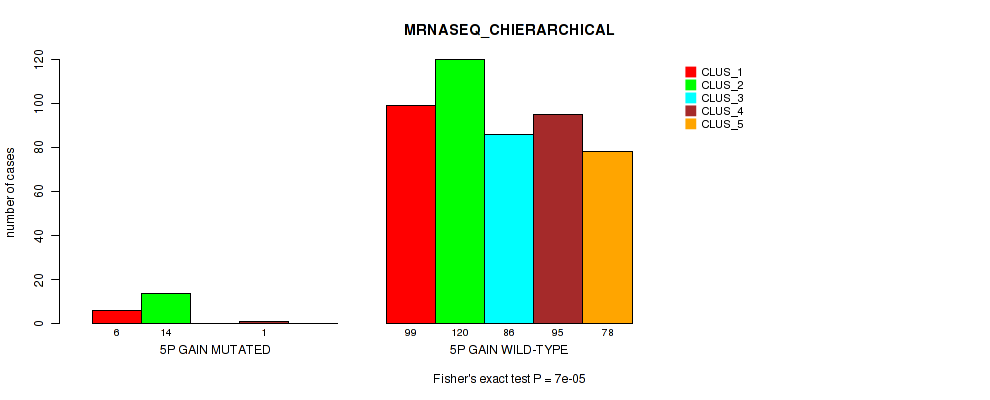
P value = 0.0012 (Fisher's exact test), Q value = 0.0048
Table S27. Gene #6: '5p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 5P GAIN MUTATED | 7 | 13 | 1 |
| 5P GAIN WILD-TYPE | 175 | 138 | 166 |
Figure S27. Get High-res Image Gene #6: '5p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
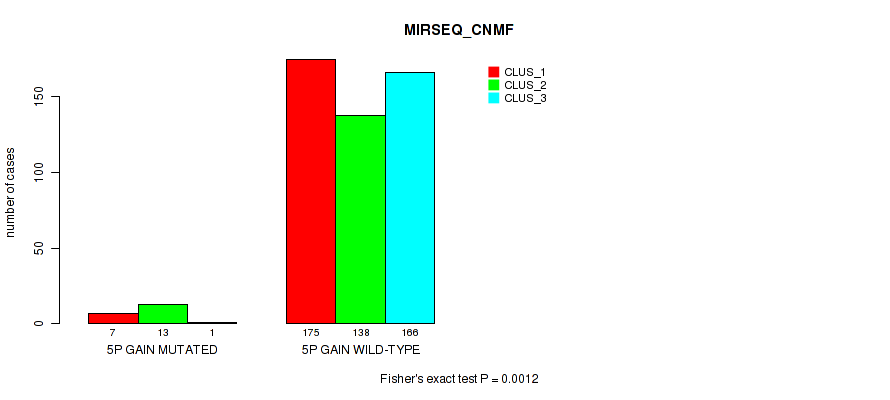
P value = 0.00013 (Fisher's exact test), Q value = 0.00074
Table S28. Gene #6: '5p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 5P GAIN MUTATED | 7 | 13 | 1 |
| 5P GAIN WILD-TYPE | 172 | 115 | 192 |
Figure S28. Get High-res Image Gene #6: '5p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
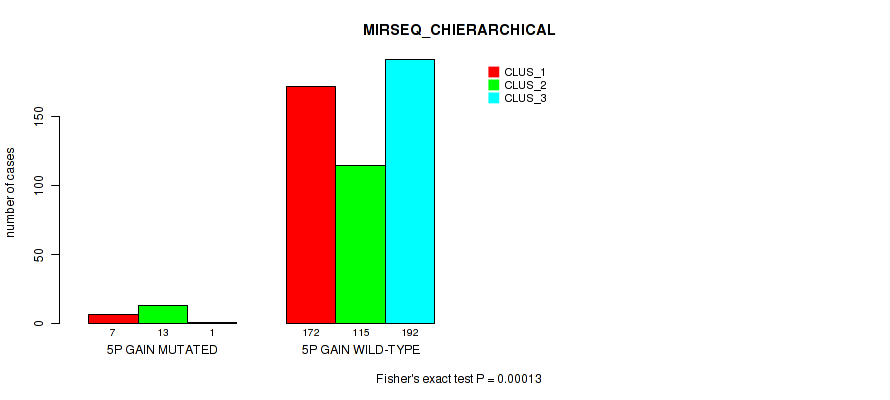
P value = 0.0033 (Fisher's exact test), Q value = 0.011
Table S29. Gene #6: '5p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 5P GAIN MUTATED | 8 | 9 | 1 |
| 5P GAIN WILD-TYPE | 157 | 117 | 178 |
Figure S29. Get High-res Image Gene #6: '5p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S30. Gene #7: '5q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 5Q GAIN MUTATED | 17 | 0 | 0 |
| 5Q GAIN WILD-TYPE | 39 | 356 | 89 |
Figure S30. Get High-res Image Gene #7: '5q gain' versus Molecular Subtype #1: 'CN_CNMF'
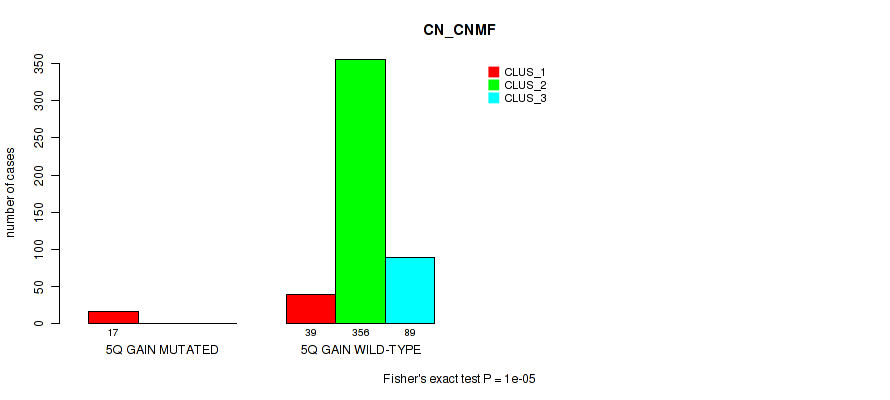
P value = 3e-05 (Fisher's exact test), Q value = 0.00024
Table S31. Gene #7: '5q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 5Q GAIN MUTATED | 2 | 14 | 1 |
| 5Q GAIN WILD-TYPE | 275 | 140 | 69 |
Figure S31. Get High-res Image Gene #7: '5q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00017 (Fisher's exact test), Q value = 0.00092
Table S32. Gene #7: '5q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 5Q GAIN MUTATED | 2 | 14 | 0 | 1 |
| 5Q GAIN WILD-TYPE | 166 | 141 | 59 | 116 |
Figure S32. Get High-res Image Gene #7: '5q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'

P value = 4e-05 (Fisher's exact test), Q value = 0.00029
Table S33. Gene #7: '5q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 5Q GAIN MUTATED | 2 | 14 | 0 | 1 | 0 |
| 5Q GAIN WILD-TYPE | 103 | 120 | 86 | 95 | 78 |
Figure S33. Get High-res Image Gene #7: '5q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
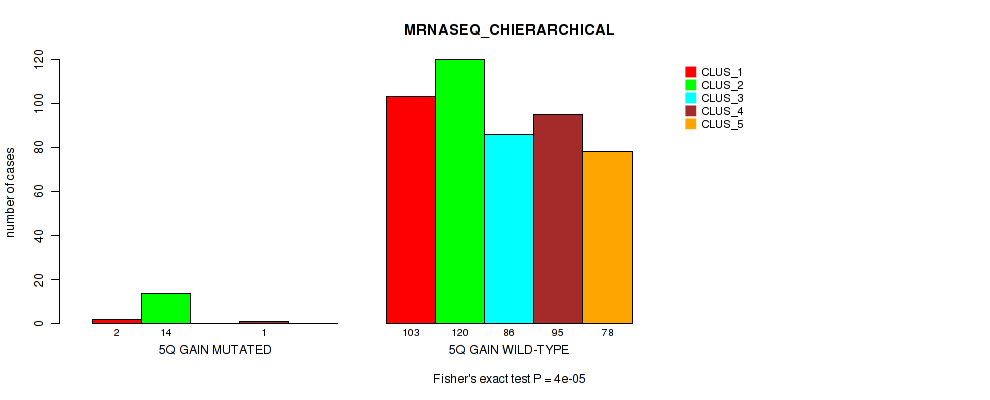
P value = 0.00012 (Fisher's exact test), Q value = 0.00073
Table S34. Gene #7: '5q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 5Q GAIN MUTATED | 3 | 13 | 1 |
| 5Q GAIN WILD-TYPE | 179 | 138 | 166 |
Figure S34. Get High-res Image Gene #7: '5q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'

P value = 2e-05 (Fisher's exact test), Q value = 0.00018
Table S35. Gene #7: '5q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 5Q GAIN MUTATED | 3 | 13 | 1 |
| 5Q GAIN WILD-TYPE | 176 | 115 | 192 |
Figure S35. Get High-res Image Gene #7: '5q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
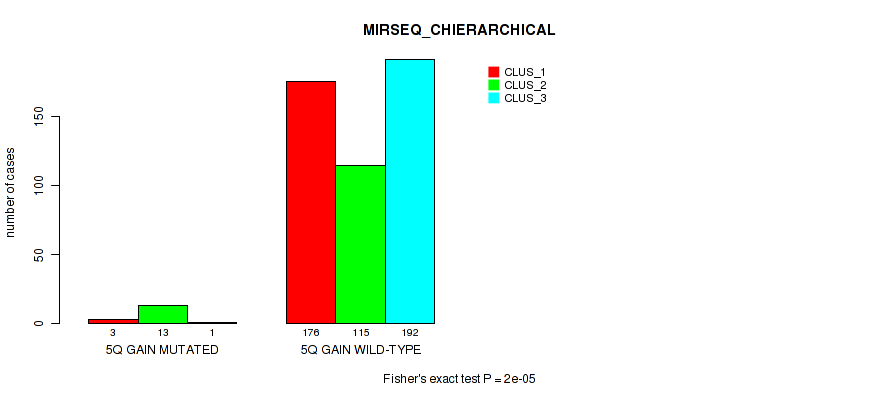
P value = 0.00419 (Fisher's exact test), Q value = 0.014
Table S36. Gene #7: '5q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 5Q GAIN MUTATED | 4 | 9 | 1 |
| 5Q GAIN WILD-TYPE | 161 | 117 | 178 |
Figure S36. Get High-res Image Gene #7: '5q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
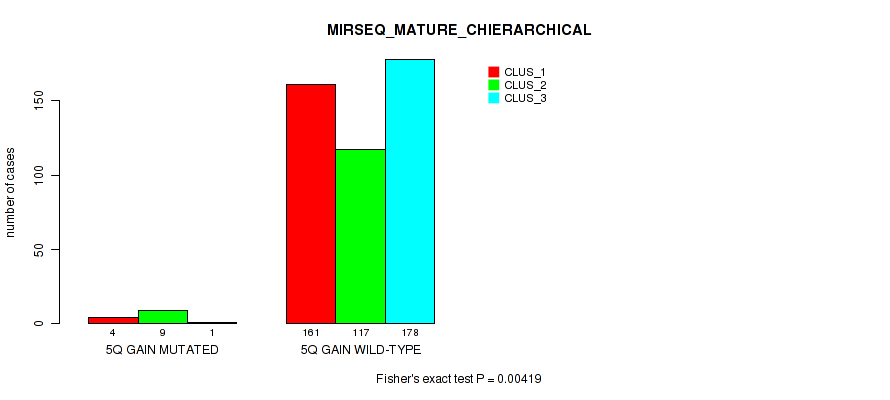
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S37. Gene #8: '7p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 7P GAIN MUTATED | 18 | 0 | 0 |
| 7P GAIN WILD-TYPE | 38 | 356 | 89 |
Figure S37. Get High-res Image Gene #8: '7p gain' versus Molecular Subtype #1: 'CN_CNMF'
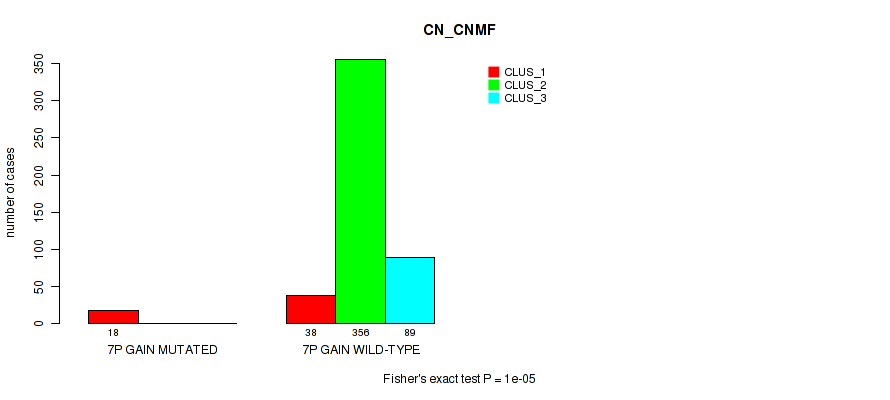
P value = 5e-05 (Fisher's exact test), Q value = 0.00035
Table S38. Gene #8: '7p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 7P GAIN MUTATED | 2 | 14 | 2 |
| 7P GAIN WILD-TYPE | 275 | 140 | 68 |
Figure S38. Get High-res Image Gene #8: '7p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
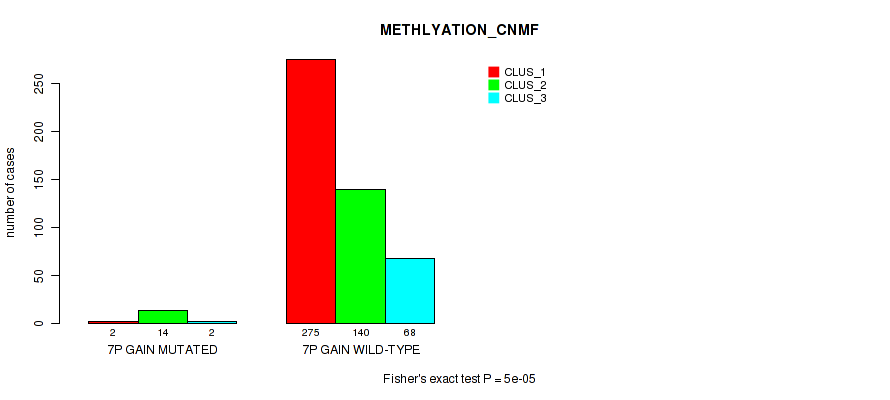
P value = 0.00065 (Fisher's exact test), Q value = 0.0028
Table S39. Gene #8: '7p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 7P GAIN MUTATED | 2 | 14 | 1 | 1 |
| 7P GAIN WILD-TYPE | 166 | 141 | 58 | 116 |
Figure S39. Get High-res Image Gene #8: '7p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
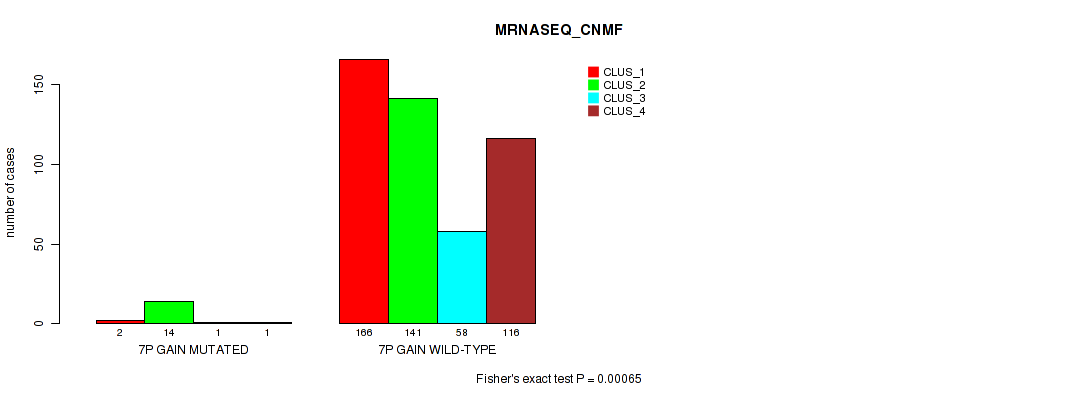
P value = 1e-04 (Fisher's exact test), Q value = 0.00063
Table S40. Gene #8: '7p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 7P GAIN MUTATED | 2 | 14 | 1 | 1 | 0 |
| 7P GAIN WILD-TYPE | 103 | 120 | 85 | 95 | 78 |
Figure S40. Get High-res Image Gene #8: '7p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
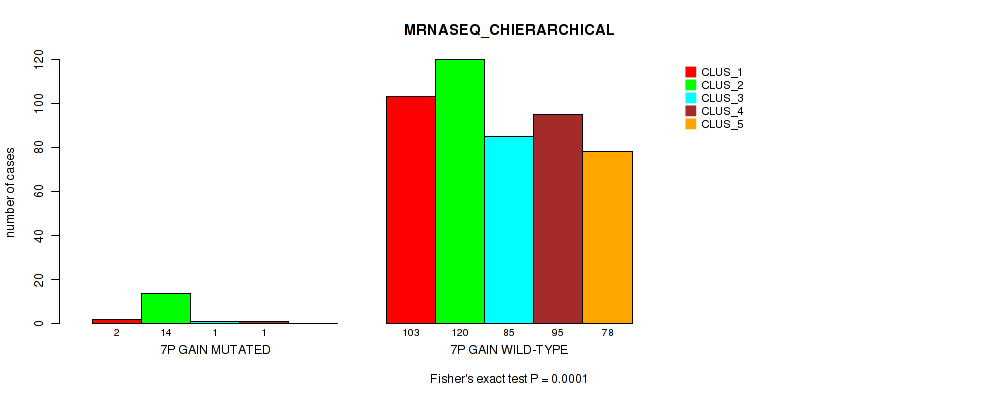
P value = 0.00038 (Fisher's exact test), Q value = 0.0018
Table S41. Gene #8: '7p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 7P GAIN MUTATED | 4 | 13 | 1 |
| 7P GAIN WILD-TYPE | 178 | 138 | 166 |
Figure S41. Get High-res Image Gene #8: '7p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
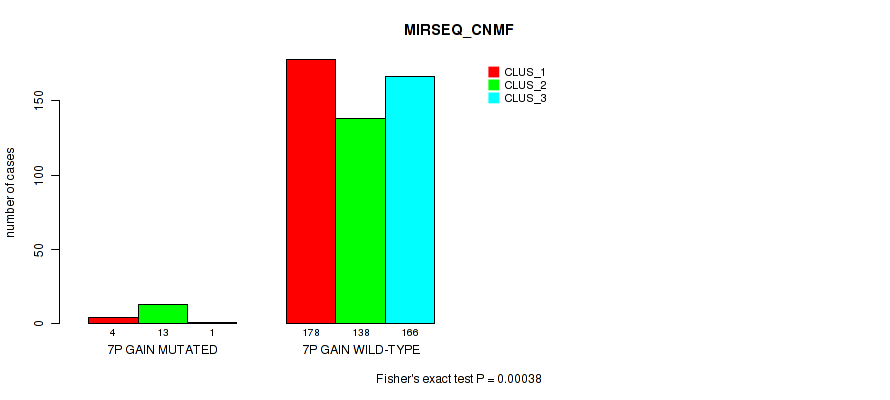
P value = 2e-05 (Fisher's exact test), Q value = 0.00018
Table S42. Gene #8: '7p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 7P GAIN MUTATED | 4 | 13 | 1 |
| 7P GAIN WILD-TYPE | 175 | 115 | 192 |
Figure S42. Get High-res Image Gene #8: '7p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
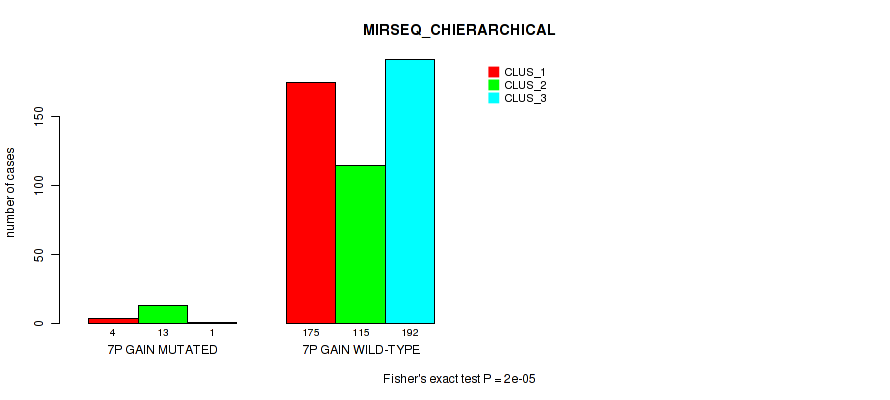
P value = 0.00555 (Fisher's exact test), Q value = 0.017
Table S43. Gene #8: '7p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 7P GAIN MUTATED | 5 | 9 | 1 |
| 7P GAIN WILD-TYPE | 160 | 117 | 178 |
Figure S43. Get High-res Image Gene #8: '7p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
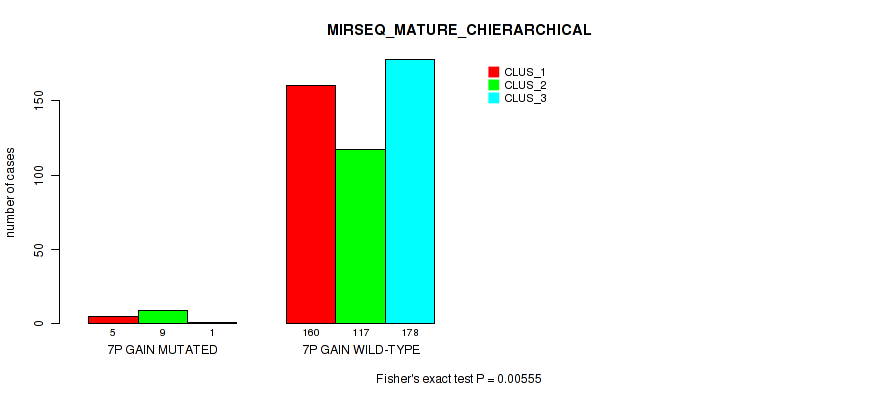
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S44. Gene #9: '7q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 7Q GAIN MUTATED | 19 | 0 | 1 |
| 7Q GAIN WILD-TYPE | 37 | 356 | 88 |
Figure S44. Get High-res Image Gene #9: '7q gain' versus Molecular Subtype #1: 'CN_CNMF'
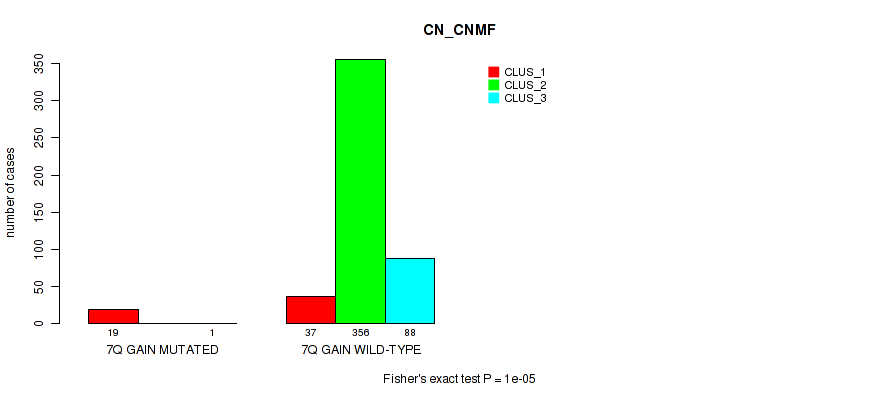
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S45. Gene #9: '7q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 7Q GAIN MUTATED | 2 | 16 | 2 |
| 7Q GAIN WILD-TYPE | 275 | 138 | 68 |
Figure S45. Get High-res Image Gene #9: '7q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
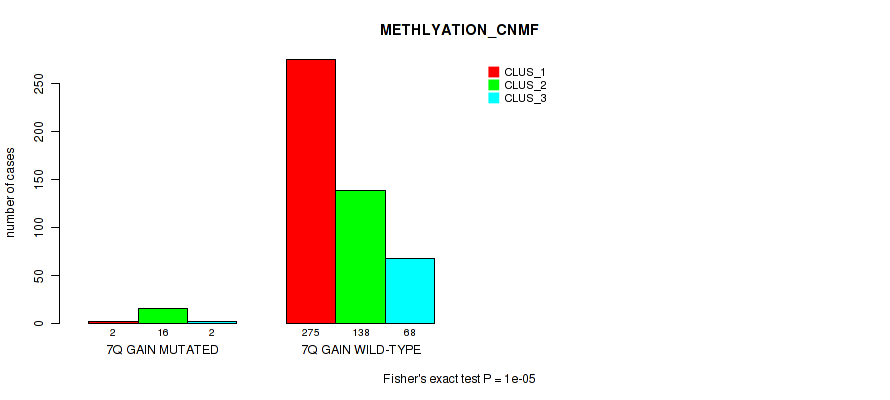
P value = 0.0132 (Fisher's exact test), Q value = 0.036
Table S46. Gene #9: '7q gain' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 46 | 28 | 33 | 29 | 49 | 27 | 9 |
| 7Q GAIN MUTATED | 4 | 1 | 0 | 2 | 0 | 5 | 0 |
| 7Q GAIN WILD-TYPE | 42 | 27 | 33 | 27 | 49 | 22 | 9 |
Figure S46. Get High-res Image Gene #9: '7q gain' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
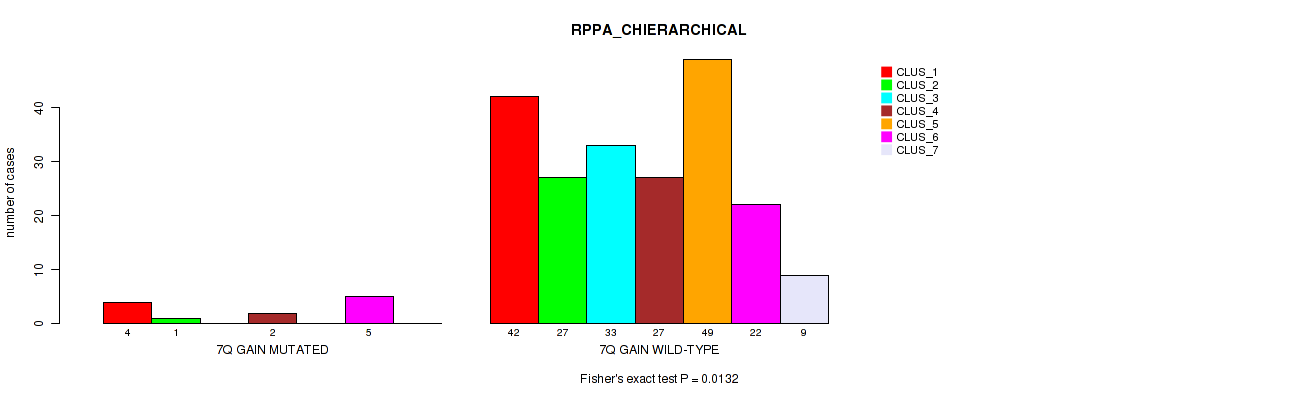
P value = 0.00013 (Fisher's exact test), Q value = 0.00074
Table S47. Gene #9: '7q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 7Q GAIN MUTATED | 2 | 16 | 1 | 1 |
| 7Q GAIN WILD-TYPE | 166 | 139 | 58 | 116 |
Figure S47. Get High-res Image Gene #9: '7q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
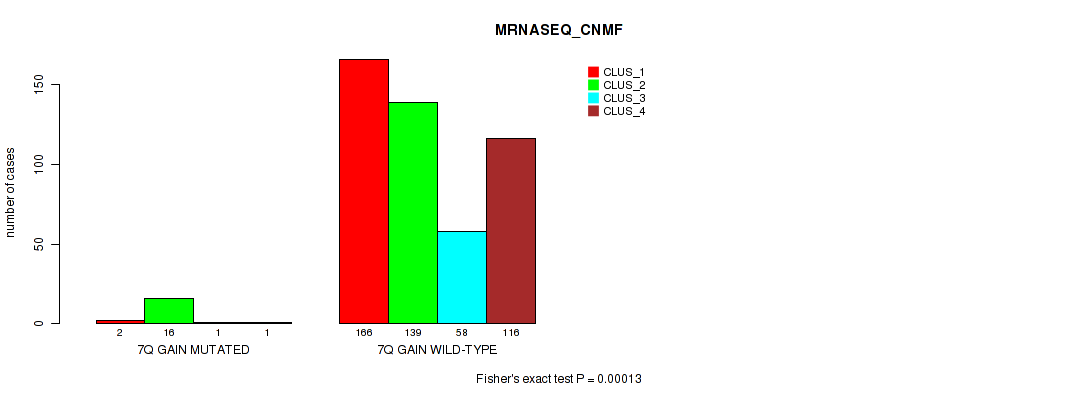
P value = 2e-05 (Fisher's exact test), Q value = 0.00018
Table S48. Gene #9: '7q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 7Q GAIN MUTATED | 2 | 16 | 1 | 1 | 0 |
| 7Q GAIN WILD-TYPE | 103 | 118 | 85 | 95 | 78 |
Figure S48. Get High-res Image Gene #9: '7q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
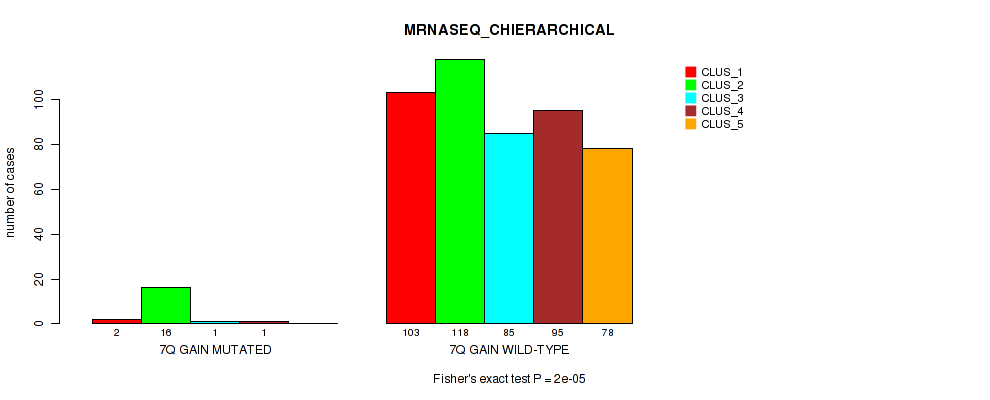
P value = 5e-05 (Fisher's exact test), Q value = 0.00035
Table S49. Gene #9: '7q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 7Q GAIN MUTATED | 4 | 15 | 1 |
| 7Q GAIN WILD-TYPE | 178 | 136 | 166 |
Figure S49. Get High-res Image Gene #9: '7q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
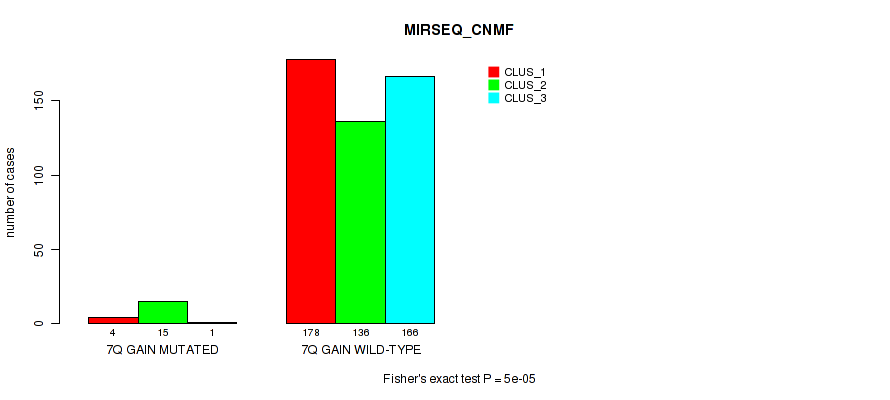
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S50. Gene #9: '7q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 7Q GAIN MUTATED | 4 | 15 | 1 |
| 7Q GAIN WILD-TYPE | 175 | 113 | 192 |
Figure S50. Get High-res Image Gene #9: '7q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
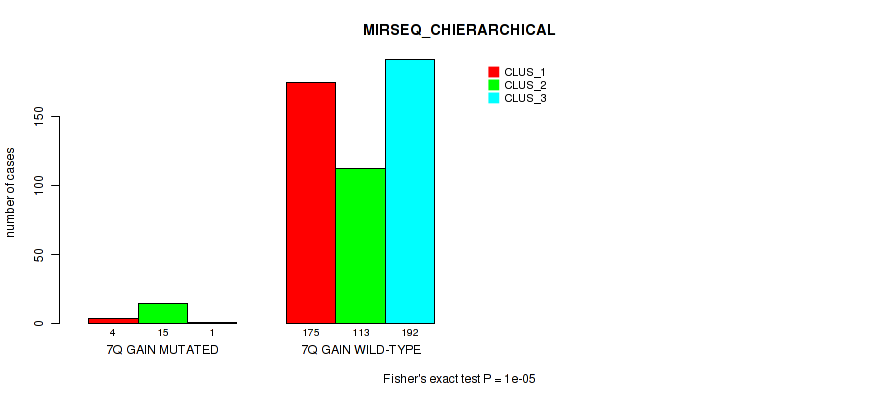
P value = 0.00089 (Fisher's exact test), Q value = 0.0036
Table S51. Gene #9: '7q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 7Q GAIN MUTATED | 5 | 11 | 1 |
| 7Q GAIN WILD-TYPE | 160 | 115 | 178 |
Figure S51. Get High-res Image Gene #9: '7q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
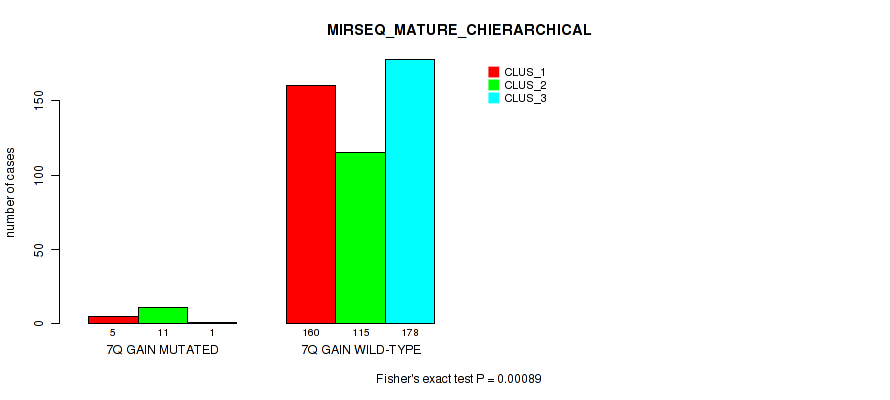
P value = 0.00013 (Fisher's exact test), Q value = 0.00074
Table S52. Gene #10: '8p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 8P GAIN MUTATED | 5 | 1 | 0 |
| 8P GAIN WILD-TYPE | 51 | 355 | 89 |
Figure S52. Get High-res Image Gene #10: '8p gain' versus Molecular Subtype #1: 'CN_CNMF'
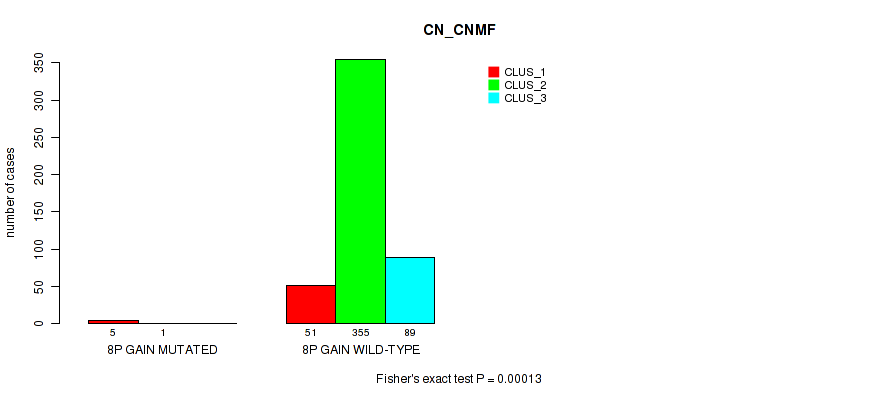
P value = 4e-05 (Fisher's exact test), Q value = 0.00029
Table S53. Gene #11: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 8Q GAIN MUTATED | 6 | 1 | 0 |
| 8Q GAIN WILD-TYPE | 50 | 355 | 89 |
Figure S53. Get High-res Image Gene #11: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'
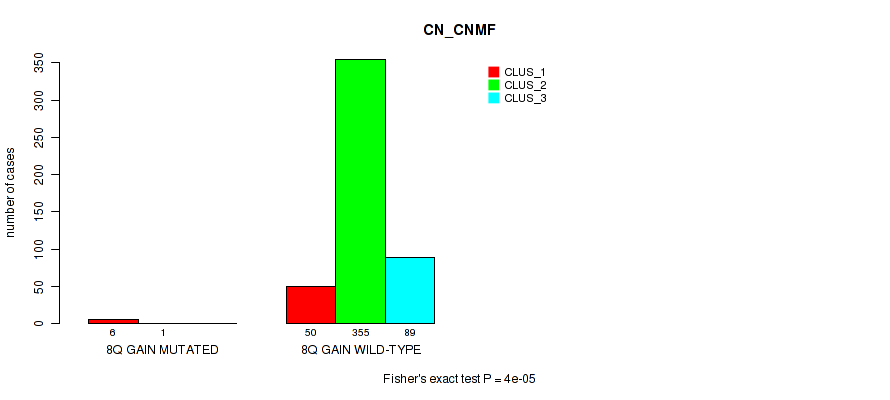
P value = 0.0013 (Fisher's exact test), Q value = 0.005
Table S54. Gene #12: '9p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 9P GAIN MUTATED | 3 | 0 | 0 |
| 9P GAIN WILD-TYPE | 53 | 356 | 89 |
Figure S54. Get High-res Image Gene #12: '9p gain' versus Molecular Subtype #1: 'CN_CNMF'
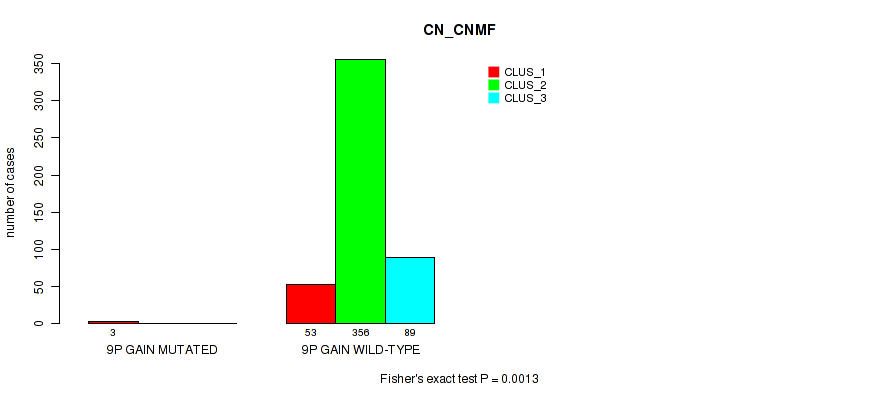
P value = 0.0107 (Fisher's exact test), Q value = 0.03
Table S55. Gene #13: '9q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 9Q GAIN MUTATED | 3 | 1 | 0 |
| 9Q GAIN WILD-TYPE | 53 | 355 | 89 |
Figure S55. Get High-res Image Gene #13: '9q gain' versus Molecular Subtype #1: 'CN_CNMF'
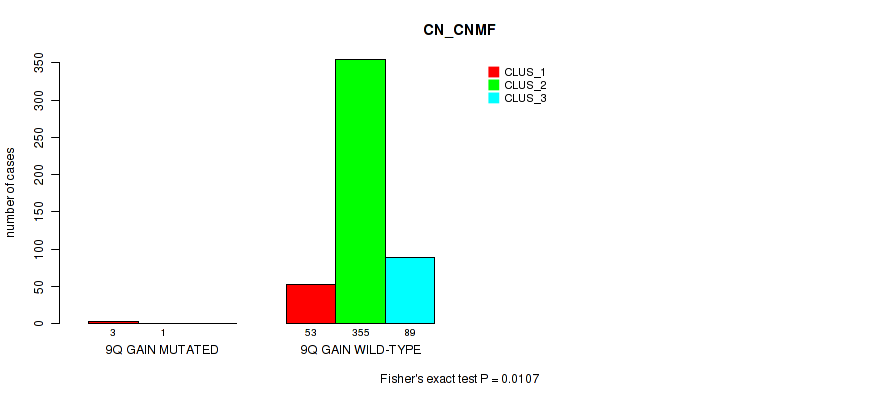
P value = 0.00123 (Fisher's exact test), Q value = 0.0048
Table S56. Gene #14: '11p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 11P GAIN MUTATED | 4 | 1 | 1 |
| 11P GAIN WILD-TYPE | 52 | 355 | 88 |
Figure S56. Get High-res Image Gene #14: '11p gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.0279 (Fisher's exact test), Q value = 0.064
Table S57. Gene #14: '11p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 11P GAIN MUTATED | 1 | 5 | 0 |
| 11P GAIN WILD-TYPE | 276 | 149 | 70 |
Figure S57. Get High-res Image Gene #14: '11p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
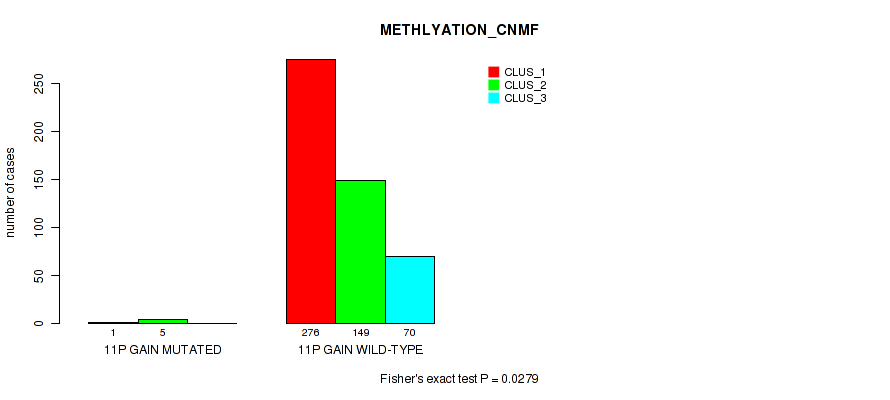
P value = 0.0447 (Fisher's exact test), Q value = 0.098
Table S58. Gene #14: '11p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 11P GAIN MUTATED | 0 | 5 | 0 | 1 | 0 |
| 11P GAIN WILD-TYPE | 105 | 129 | 86 | 95 | 78 |
Figure S58. Get High-res Image Gene #14: '11p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'

P value = 0.00946 (Fisher's exact test), Q value = 0.027
Table S59. Gene #14: '11p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 11P GAIN MUTATED | 0 | 5 | 1 |
| 11P GAIN WILD-TYPE | 182 | 146 | 166 |
Figure S59. Get High-res Image Gene #14: '11p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
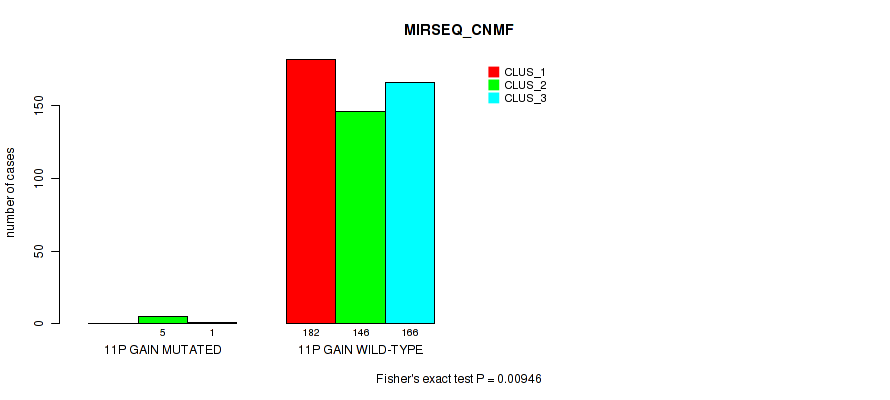
P value = 0.00645 (Fisher's exact test), Q value = 0.02
Table S60. Gene #14: '11p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 11P GAIN MUTATED | 0 | 5 | 1 |
| 11P GAIN WILD-TYPE | 179 | 123 | 192 |
Figure S60. Get High-res Image Gene #14: '11p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'

P value = 0.00013 (Fisher's exact test), Q value = 0.00074
Table S61. Gene #15: '11q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 11Q GAIN MUTATED | 4 | 0 | 1 |
| 11Q GAIN WILD-TYPE | 52 | 356 | 88 |
Figure S61. Get High-res Image Gene #15: '11q gain' versus Molecular Subtype #1: 'CN_CNMF'
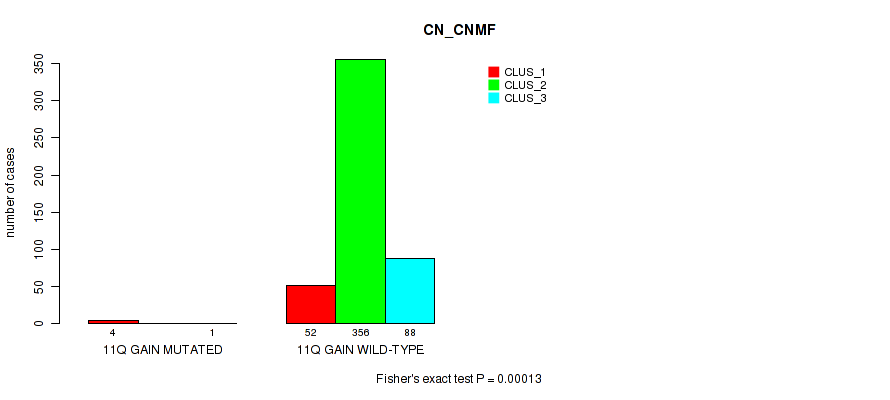
P value = 0.0267 (Fisher's exact test), Q value = 0.062
Table S62. Gene #15: '11q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 11Q GAIN MUTATED | 0 | 4 | 1 |
| 11Q GAIN WILD-TYPE | 182 | 147 | 166 |
Figure S62. Get High-res Image Gene #15: '11q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
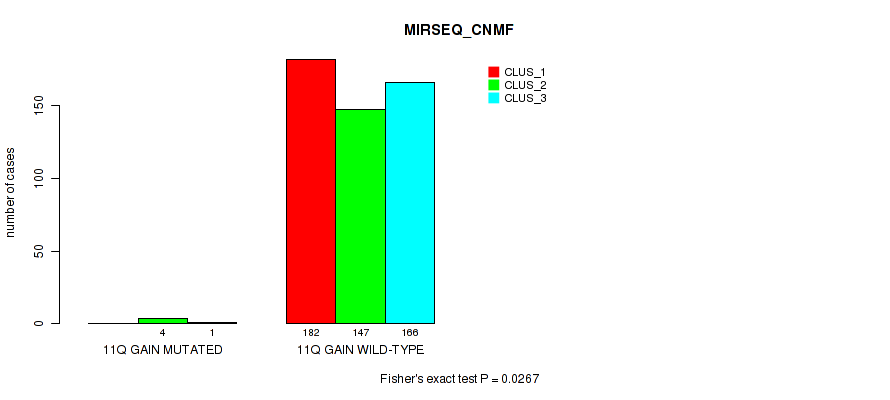
P value = 0.0214 (Fisher's exact test), Q value = 0.051
Table S63. Gene #15: '11q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 11Q GAIN MUTATED | 0 | 4 | 1 |
| 11Q GAIN WILD-TYPE | 179 | 124 | 192 |
Figure S63. Get High-res Image Gene #15: '11q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
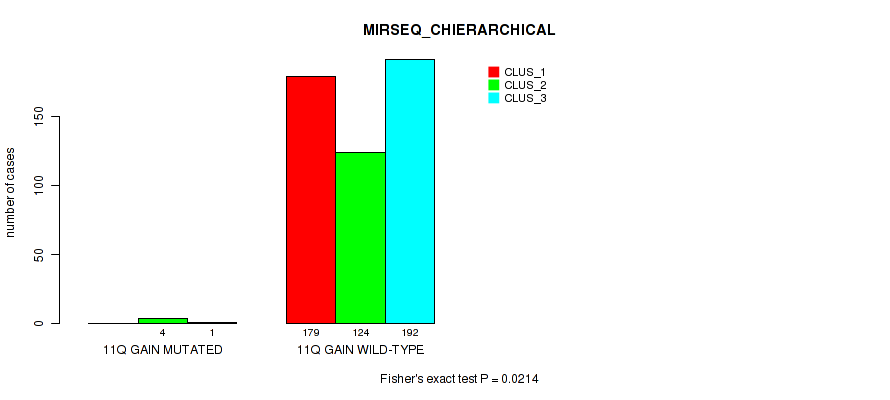
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S64. Gene #16: '12p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 12P GAIN MUTATED | 13 | 0 | 1 |
| 12P GAIN WILD-TYPE | 43 | 356 | 88 |
Figure S64. Get High-res Image Gene #16: '12p gain' versus Molecular Subtype #1: 'CN_CNMF'
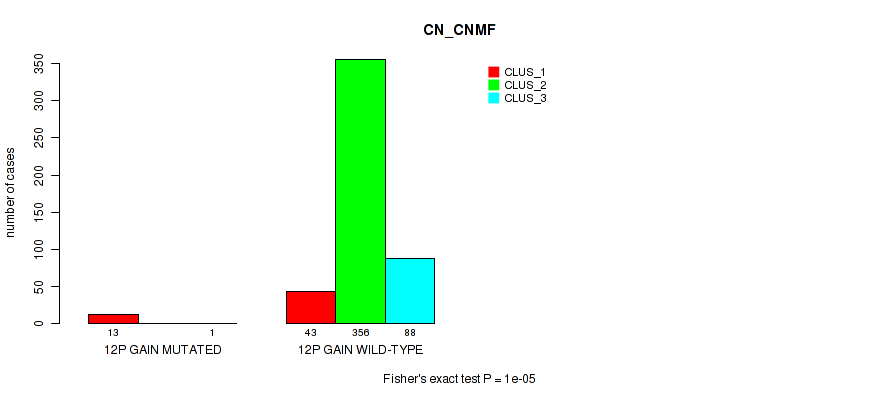
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S65. Gene #16: '12p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 12P GAIN MUTATED | 1 | 13 | 0 |
| 12P GAIN WILD-TYPE | 276 | 141 | 70 |
Figure S65. Get High-res Image Gene #16: '12p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
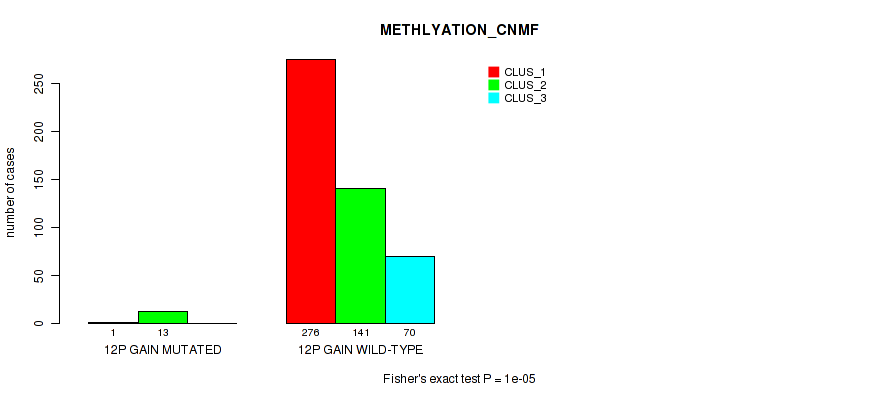
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S66. Gene #16: '12p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 12P GAIN MUTATED | 0 | 13 | 0 | 1 |
| 12P GAIN WILD-TYPE | 168 | 142 | 59 | 116 |
Figure S66. Get High-res Image Gene #16: '12p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
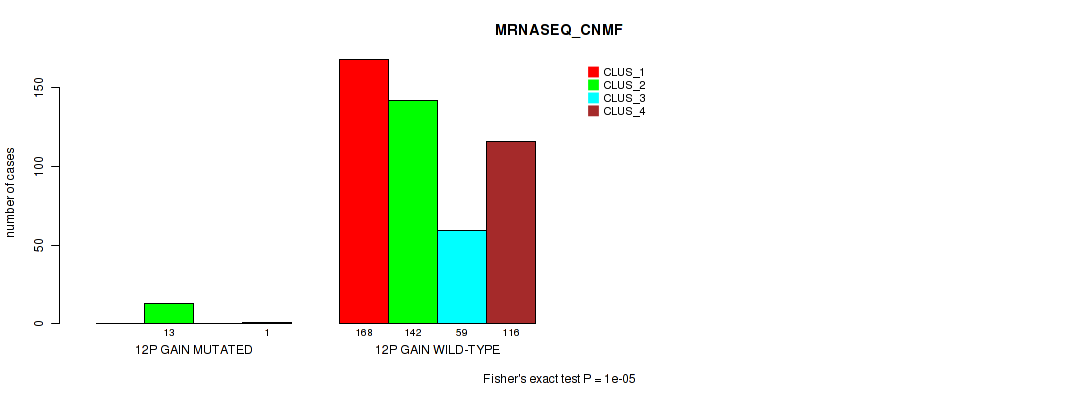
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S67. Gene #16: '12p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 12P GAIN MUTATED | 0 | 13 | 0 | 1 | 0 |
| 12P GAIN WILD-TYPE | 105 | 121 | 86 | 95 | 78 |
Figure S67. Get High-res Image Gene #16: '12p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'

P value = 5e-05 (Fisher's exact test), Q value = 0.00035
Table S68. Gene #16: '12p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 12P GAIN MUTATED | 1 | 12 | 1 |
| 12P GAIN WILD-TYPE | 181 | 139 | 166 |
Figure S68. Get High-res Image Gene #16: '12p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S69. Gene #16: '12p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 12P GAIN MUTATED | 1 | 12 | 1 |
| 12P GAIN WILD-TYPE | 178 | 116 | 192 |
Figure S69. Get High-res Image Gene #16: '12p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
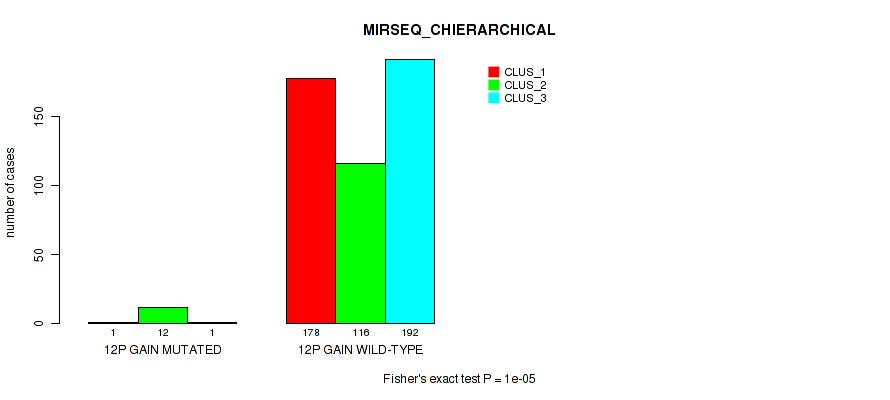
P value = 0.00242 (Fisher's exact test), Q value = 0.0084
Table S70. Gene #16: '12p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 12P GAIN MUTATED | 2 | 8 | 1 |
| 12P GAIN WILD-TYPE | 163 | 118 | 178 |
Figure S70. Get High-res Image Gene #16: '12p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
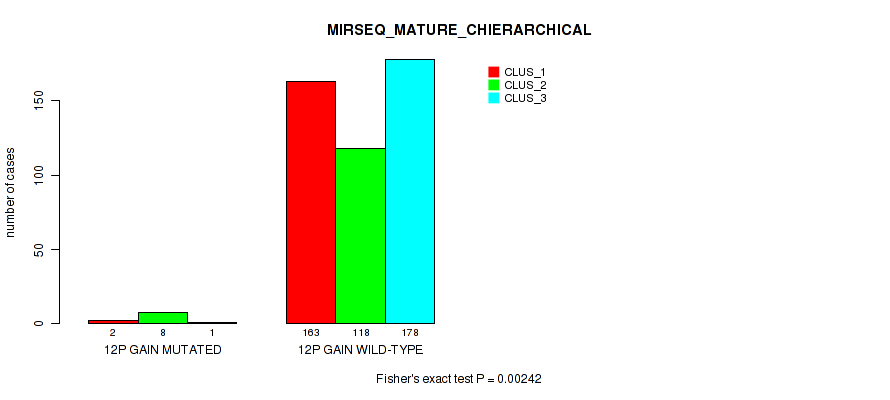
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S71. Gene #17: '12q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 12Q GAIN MUTATED | 14 | 0 | 1 |
| 12Q GAIN WILD-TYPE | 42 | 356 | 88 |
Figure S71. Get High-res Image Gene #17: '12q gain' versus Molecular Subtype #1: 'CN_CNMF'
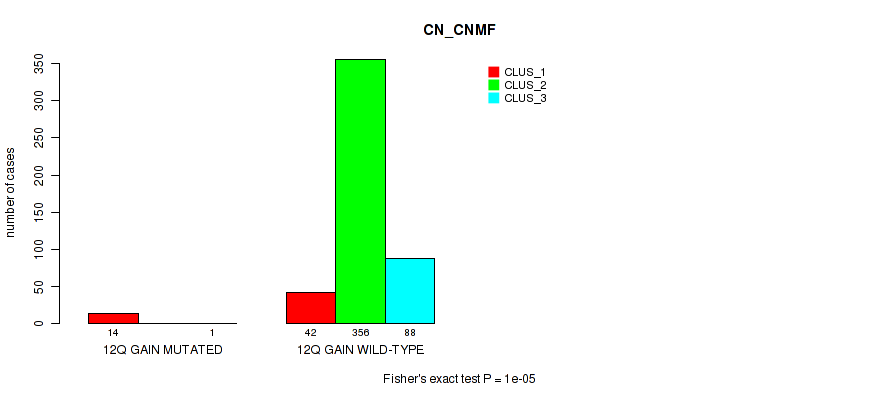
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S72. Gene #17: '12q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 12Q GAIN MUTATED | 1 | 14 | 0 |
| 12Q GAIN WILD-TYPE | 276 | 140 | 70 |
Figure S72. Get High-res Image Gene #17: '12q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
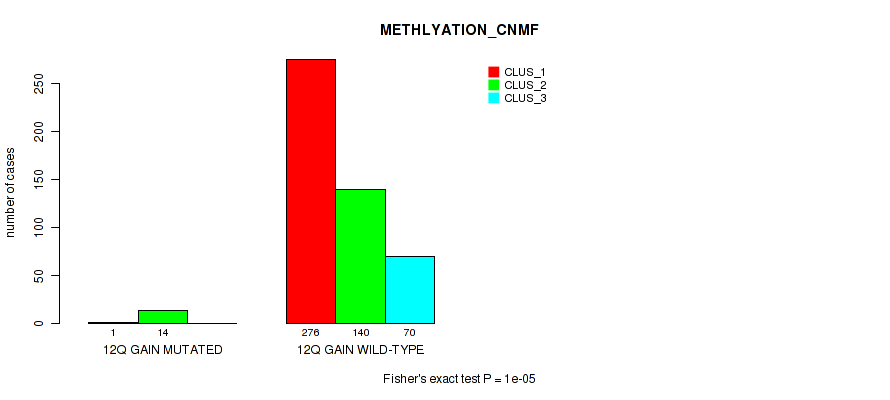
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S73. Gene #17: '12q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 12Q GAIN MUTATED | 0 | 14 | 0 | 1 |
| 12Q GAIN WILD-TYPE | 168 | 141 | 59 | 116 |
Figure S73. Get High-res Image Gene #17: '12q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
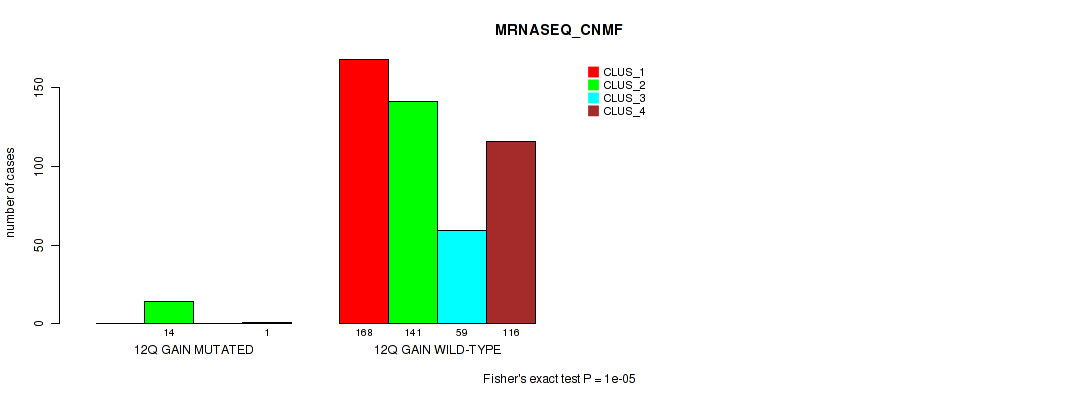
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S74. Gene #17: '12q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 12Q GAIN MUTATED | 0 | 14 | 0 | 1 | 0 |
| 12Q GAIN WILD-TYPE | 105 | 120 | 86 | 95 | 78 |
Figure S74. Get High-res Image Gene #17: '12q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
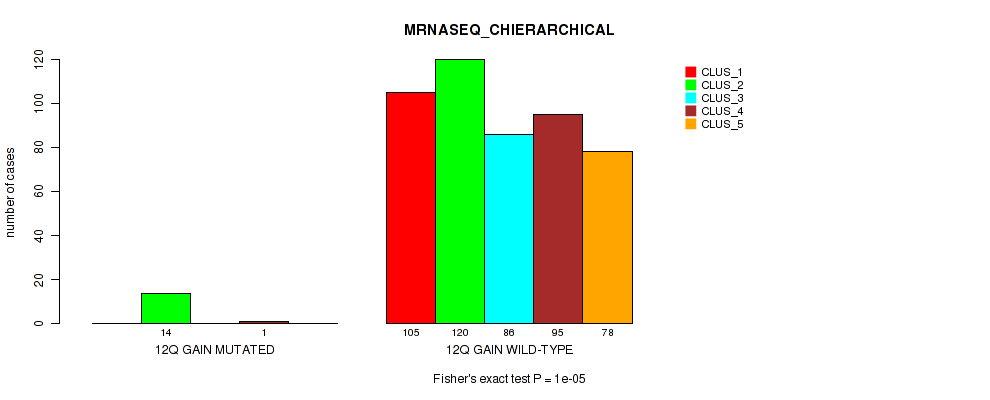
P value = 3e-05 (Fisher's exact test), Q value = 0.00024
Table S75. Gene #17: '12q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 12Q GAIN MUTATED | 1 | 13 | 1 |
| 12Q GAIN WILD-TYPE | 181 | 138 | 166 |
Figure S75. Get High-res Image Gene #17: '12q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
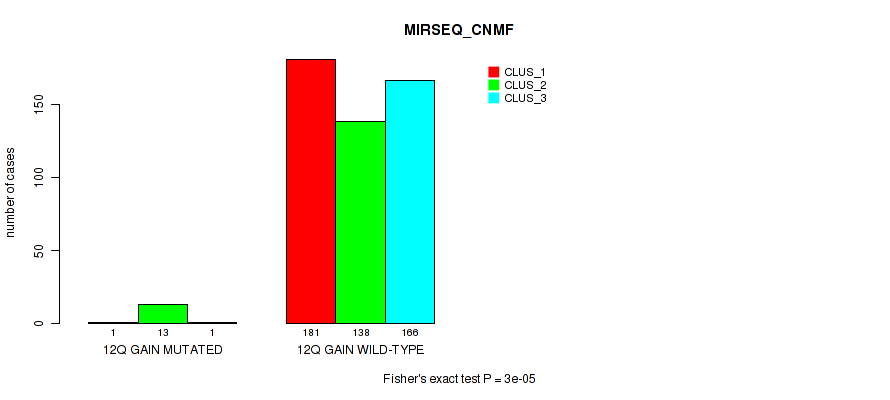
P value = 2e-05 (Fisher's exact test), Q value = 0.00018
Table S76. Gene #17: '12q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 12Q GAIN MUTATED | 1 | 13 | 1 |
| 12Q GAIN WILD-TYPE | 178 | 115 | 192 |
Figure S76. Get High-res Image Gene #17: '12q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
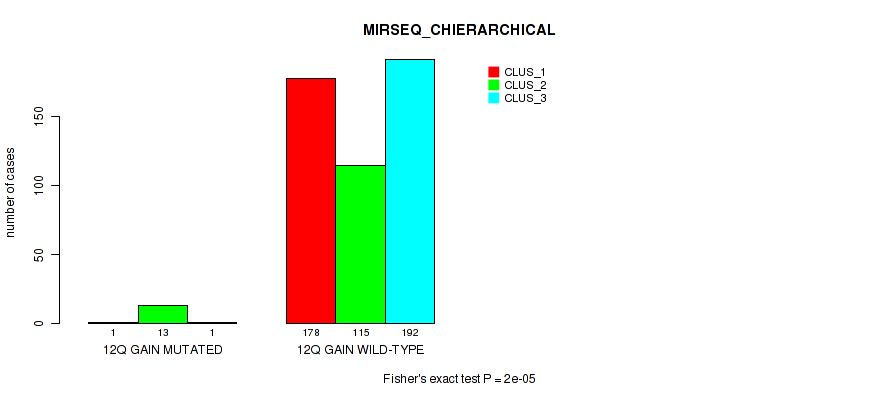
P value = 0.00098 (Fisher's exact test), Q value = 0.004
Table S77. Gene #17: '12q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 12Q GAIN MUTATED | 2 | 9 | 1 |
| 12Q GAIN WILD-TYPE | 163 | 117 | 178 |
Figure S77. Get High-res Image Gene #17: '12q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 2e-05 (Fisher's exact test), Q value = 0.00018
Table S78. Gene #18: '13q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 13Q GAIN MUTATED | 5 | 0 | 0 |
| 13Q GAIN WILD-TYPE | 51 | 356 | 89 |
Figure S78. Get High-res Image Gene #18: '13q gain' versus Molecular Subtype #1: 'CN_CNMF'
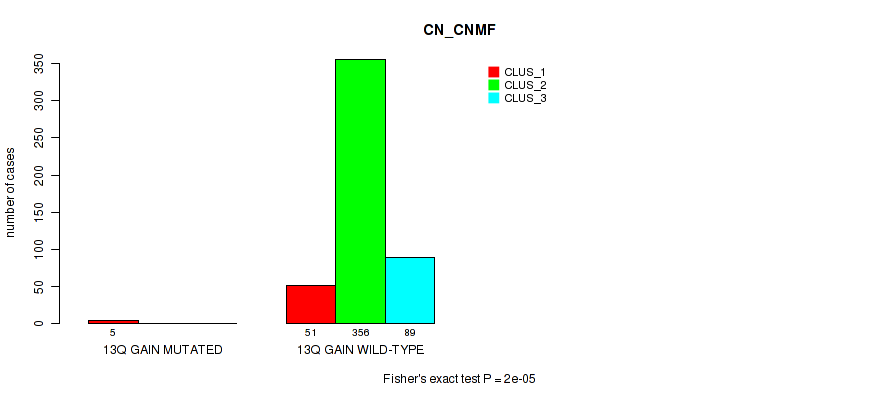
P value = 0.0187 (Fisher's exact test), Q value = 0.045
Table S79. Gene #18: '13q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 13Q GAIN MUTATED | 0 | 4 | 1 |
| 13Q GAIN WILD-TYPE | 277 | 150 | 69 |
Figure S79. Get High-res Image Gene #18: '13q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
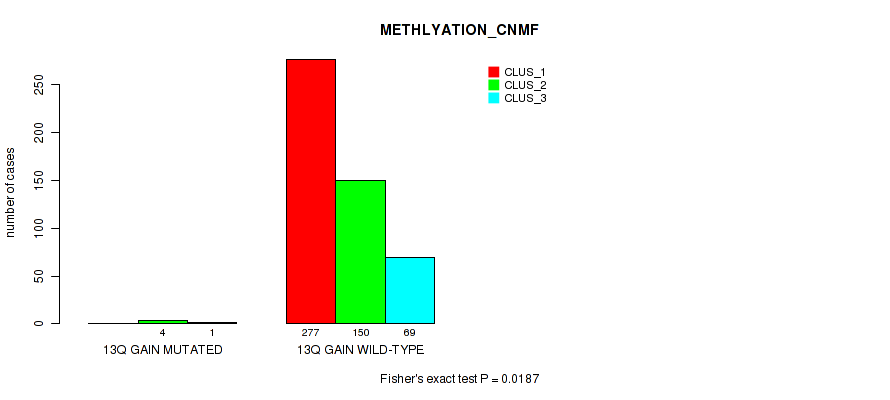
P value = 0.0486 (Fisher's exact test), Q value = 0.1
Table S80. Gene #18: '13q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 13Q GAIN MUTATED | 0 | 4 | 1 | 0 |
| 13Q GAIN WILD-TYPE | 168 | 151 | 58 | 117 |
Figure S80. Get High-res Image Gene #18: '13q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
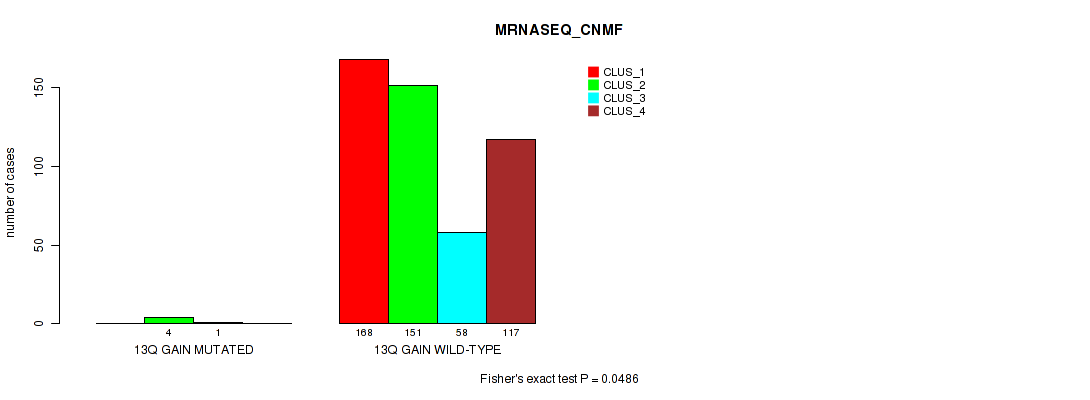
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S81. Gene #19: '14q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 14Q GAIN MUTATED | 10 | 0 | 1 |
| 14Q GAIN WILD-TYPE | 46 | 356 | 88 |
Figure S81. Get High-res Image Gene #19: '14q gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00019 (Fisher's exact test), Q value = 0.001
Table S82. Gene #19: '14q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 14Q GAIN MUTATED | 1 | 10 | 0 |
| 14Q GAIN WILD-TYPE | 276 | 144 | 70 |
Figure S82. Get High-res Image Gene #19: '14q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00057 (Fisher's exact test), Q value = 0.0026
Table S83. Gene #19: '14q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 14Q GAIN MUTATED | 1 | 10 | 0 | 0 |
| 14Q GAIN WILD-TYPE | 167 | 145 | 59 | 117 |
Figure S83. Get High-res Image Gene #19: '14q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
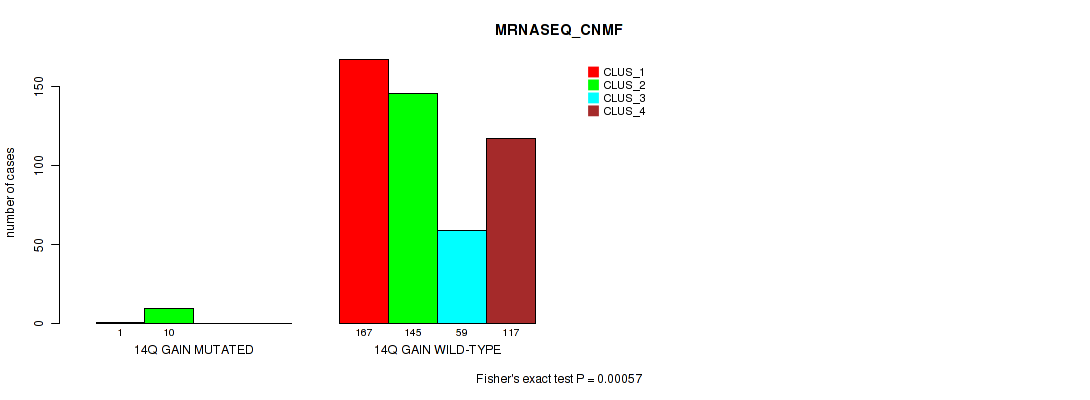
P value = 0.00012 (Fisher's exact test), Q value = 0.00073
Table S84. Gene #19: '14q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 14Q GAIN MUTATED | 0 | 10 | 0 | 0 | 1 |
| 14Q GAIN WILD-TYPE | 105 | 124 | 86 | 96 | 77 |
Figure S84. Get High-res Image Gene #19: '14q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'

P value = 0.00074 (Fisher's exact test), Q value = 0.0031
Table S85. Gene #19: '14q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 14Q GAIN MUTATED | 2 | 9 | 0 |
| 14Q GAIN WILD-TYPE | 180 | 142 | 167 |
Figure S85. Get High-res Image Gene #19: '14q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
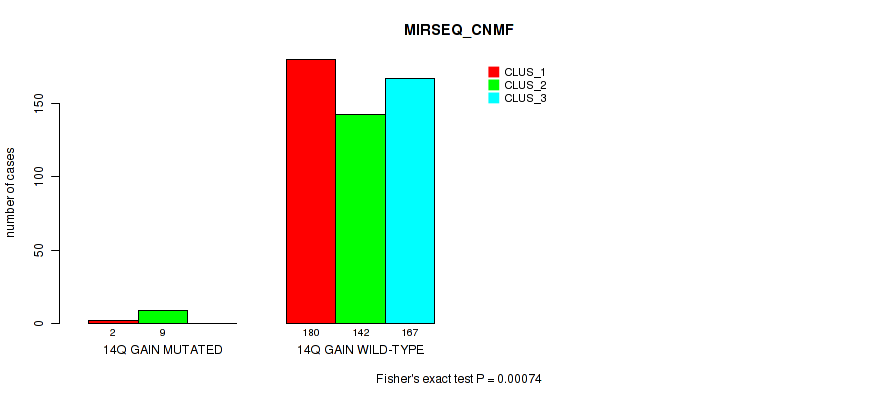
P value = 1e-04 (Fisher's exact test), Q value = 0.00063
Table S86. Gene #19: '14q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 14Q GAIN MUTATED | 2 | 9 | 0 |
| 14Q GAIN WILD-TYPE | 177 | 119 | 193 |
Figure S86. Get High-res Image Gene #19: '14q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
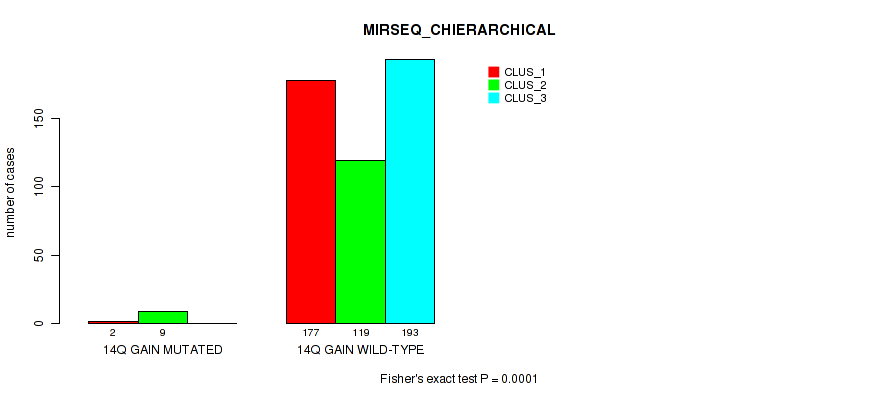
P value = 0.00217 (Fisher's exact test), Q value = 0.0077
Table S87. Gene #19: '14q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 14Q GAIN MUTATED | 3 | 7 | 0 |
| 14Q GAIN WILD-TYPE | 162 | 119 | 179 |
Figure S87. Get High-res Image Gene #19: '14q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
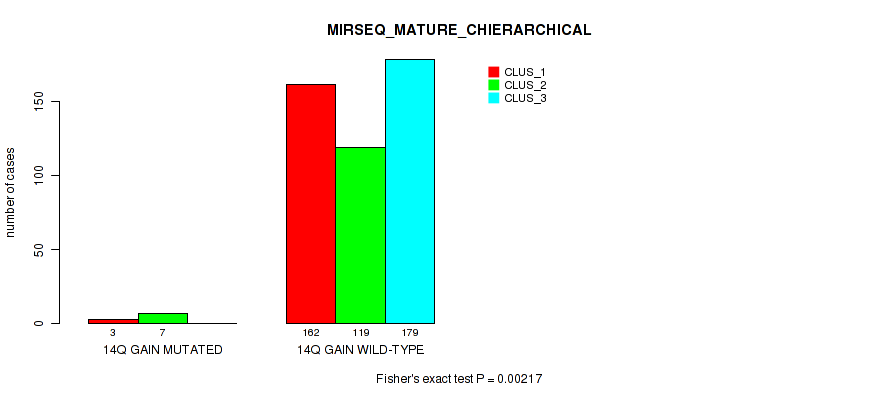
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S88. Gene #20: '16p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 16P GAIN MUTATED | 14 | 0 | 1 |
| 16P GAIN WILD-TYPE | 42 | 356 | 88 |
Figure S88. Get High-res Image Gene #20: '16p gain' versus Molecular Subtype #1: 'CN_CNMF'
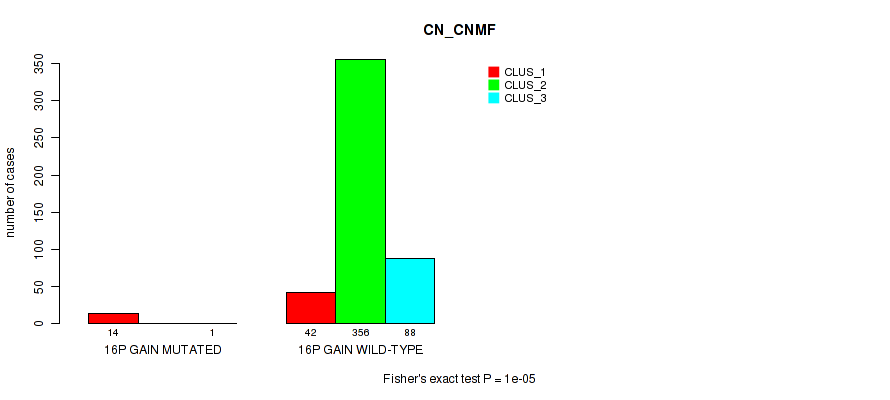
P value = 4e-05 (Fisher's exact test), Q value = 0.00029
Table S89. Gene #20: '16p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 16P GAIN MUTATED | 1 | 13 | 1 |
| 16P GAIN WILD-TYPE | 276 | 141 | 69 |
Figure S89. Get High-res Image Gene #20: '16p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
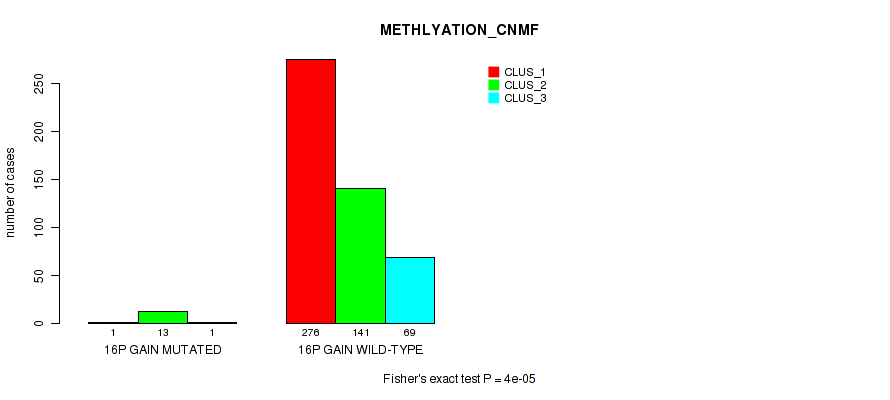
P value = 0.00017 (Fisher's exact test), Q value = 0.00092
Table S90. Gene #20: '16p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 16P GAIN MUTATED | 1 | 13 | 0 | 1 |
| 16P GAIN WILD-TYPE | 167 | 142 | 59 | 116 |
Figure S90. Get High-res Image Gene #20: '16p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
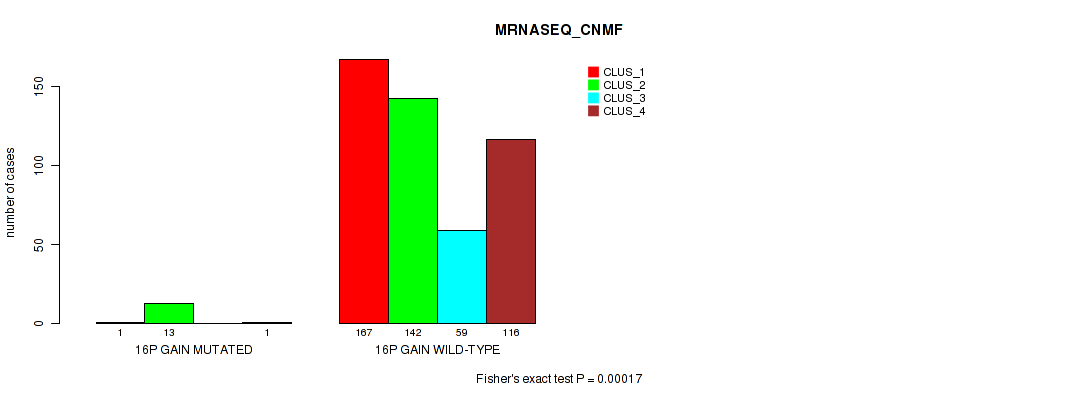
P value = 3e-05 (Fisher's exact test), Q value = 0.00024
Table S91. Gene #20: '16p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 16P GAIN MUTATED | 1 | 13 | 0 | 1 | 0 |
| 16P GAIN WILD-TYPE | 104 | 121 | 86 | 95 | 78 |
Figure S91. Get High-res Image Gene #20: '16p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
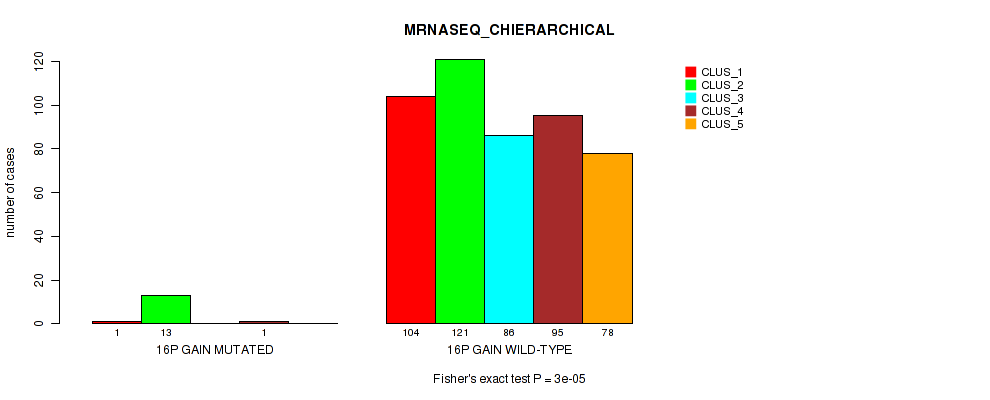
P value = 0.00011 (Fisher's exact test), Q value = 0.00068
Table S92. Gene #20: '16p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 16P GAIN MUTATED | 2 | 12 | 1 |
| 16P GAIN WILD-TYPE | 180 | 139 | 166 |
Figure S92. Get High-res Image Gene #20: '16p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
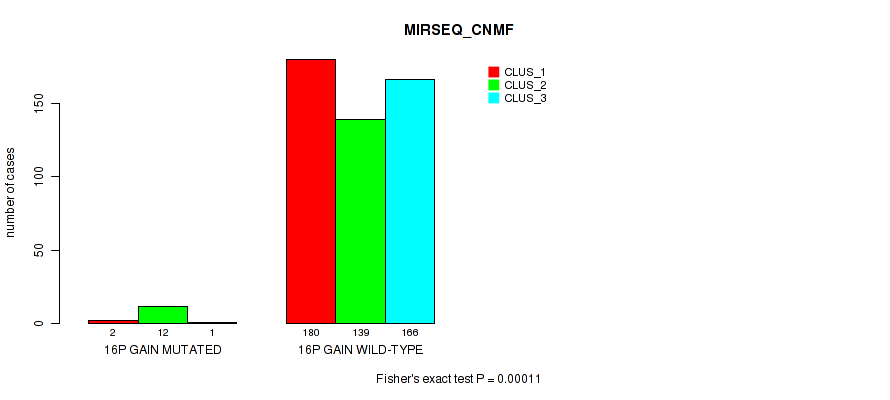
P value = 3e-05 (Fisher's exact test), Q value = 0.00024
Table S93. Gene #20: '16p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 16P GAIN MUTATED | 2 | 12 | 1 |
| 16P GAIN WILD-TYPE | 177 | 116 | 192 |
Figure S93. Get High-res Image Gene #20: '16p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
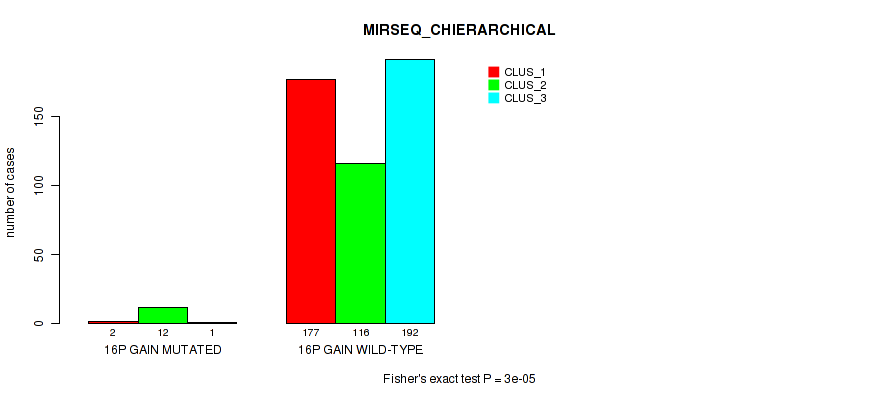
P value = 0.00078 (Fisher's exact test), Q value = 0.0033
Table S94. Gene #20: '16p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 16P GAIN MUTATED | 3 | 10 | 1 |
| 16P GAIN WILD-TYPE | 162 | 116 | 178 |
Figure S94. Get High-res Image Gene #20: '16p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
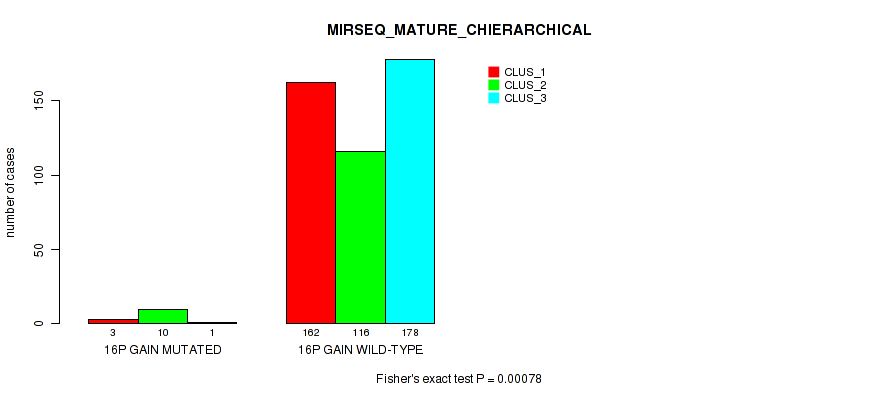
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S95. Gene #21: '16q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 16Q GAIN MUTATED | 13 | 0 | 0 |
| 16Q GAIN WILD-TYPE | 43 | 356 | 89 |
Figure S95. Get High-res Image Gene #21: '16q gain' versus Molecular Subtype #1: 'CN_CNMF'
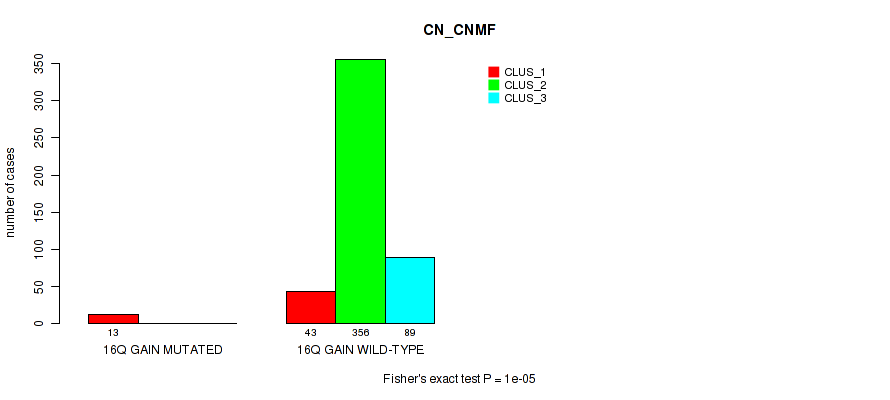
P value = 0.00017 (Fisher's exact test), Q value = 0.00092
Table S96. Gene #21: '16q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 16Q GAIN MUTATED | 1 | 11 | 1 |
| 16Q GAIN WILD-TYPE | 276 | 143 | 69 |
Figure S96. Get High-res Image Gene #21: '16q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
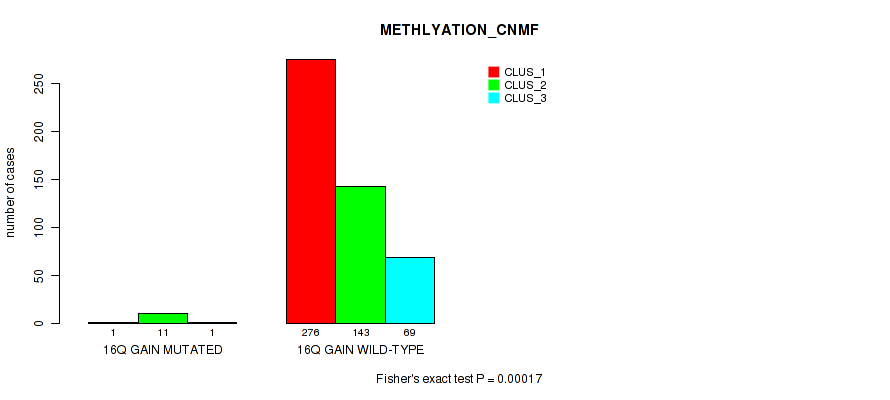
P value = 0.00088 (Fisher's exact test), Q value = 0.0036
Table S97. Gene #21: '16q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 16Q GAIN MUTATED | 1 | 11 | 0 | 1 |
| 16Q GAIN WILD-TYPE | 167 | 144 | 59 | 116 |
Figure S97. Get High-res Image Gene #21: '16q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
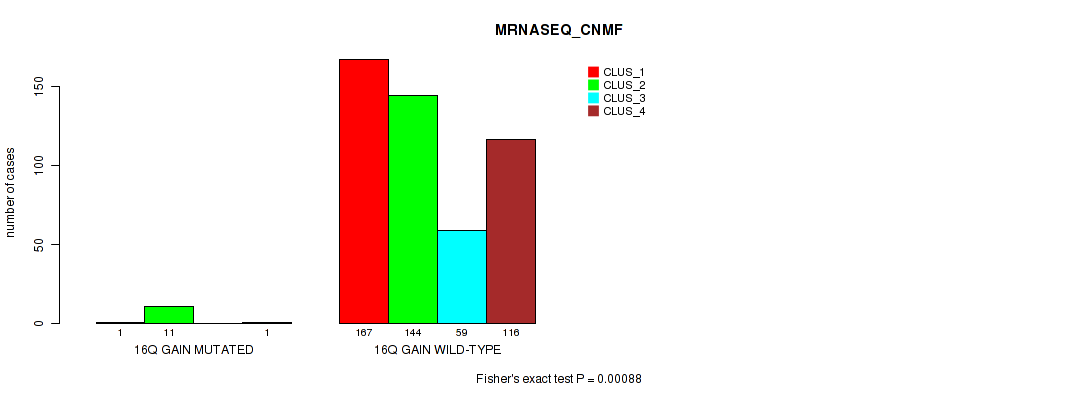
P value = 0.00025 (Fisher's exact test), Q value = 0.0012
Table S98. Gene #21: '16q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 16Q GAIN MUTATED | 1 | 11 | 0 | 1 | 0 |
| 16Q GAIN WILD-TYPE | 104 | 123 | 86 | 95 | 78 |
Figure S98. Get High-res Image Gene #21: '16q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
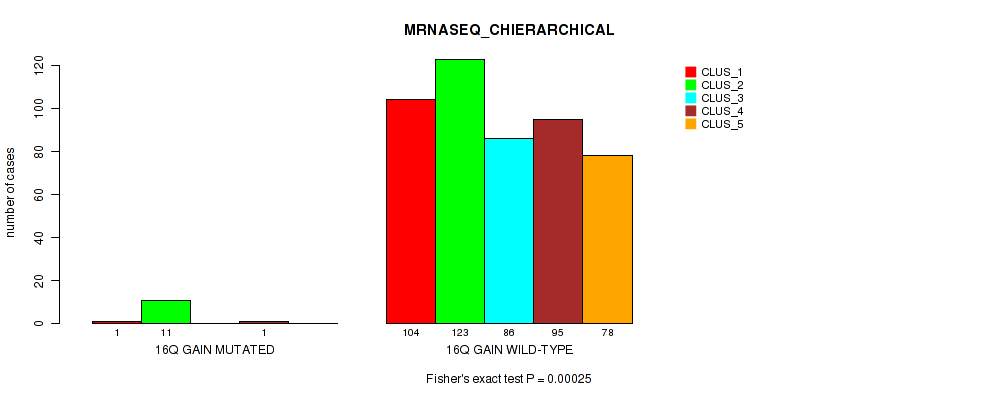
P value = 0.00163 (Fisher's exact test), Q value = 0.006
Table S99. Gene #21: '16q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 16Q GAIN MUTATED | 2 | 10 | 1 |
| 16Q GAIN WILD-TYPE | 180 | 141 | 166 |
Figure S99. Get High-res Image Gene #21: '16q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
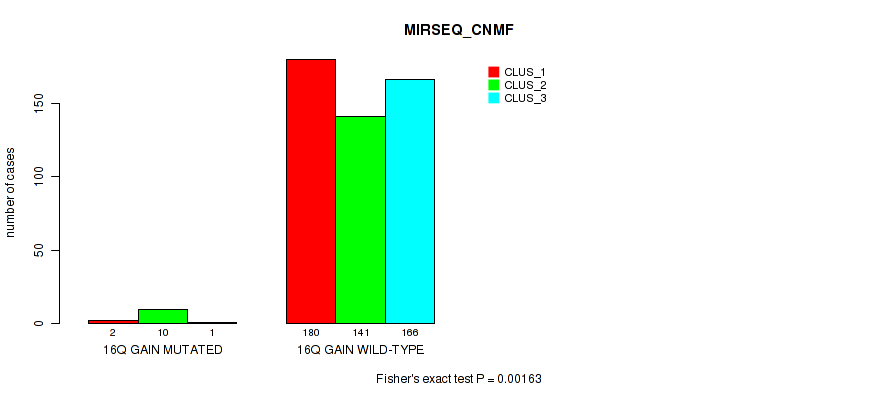
P value = 0.00013 (Fisher's exact test), Q value = 0.00074
Table S100. Gene #21: '16q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 16Q GAIN MUTATED | 2 | 10 | 1 |
| 16Q GAIN WILD-TYPE | 177 | 118 | 192 |
Figure S100. Get High-res Image Gene #21: '16q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
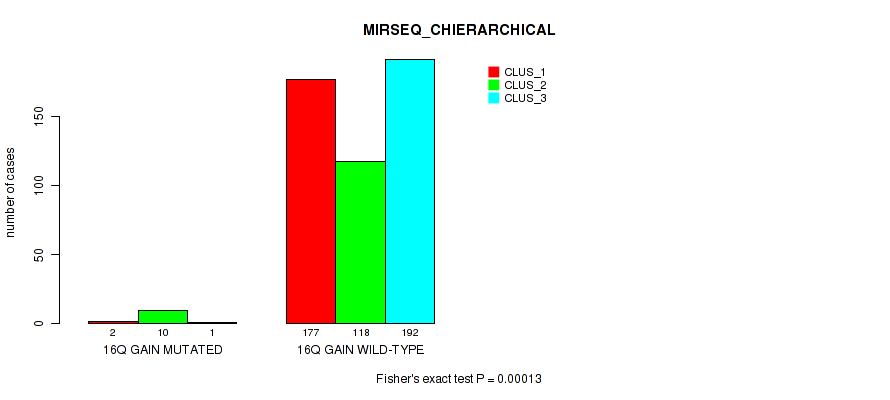
P value = 0.00756 (Fisher's exact test), Q value = 0.023
Table S101. Gene #21: '16q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 16Q GAIN MUTATED | 3 | 8 | 1 |
| 16Q GAIN WILD-TYPE | 162 | 118 | 178 |
Figure S101. Get High-res Image Gene #21: '16q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
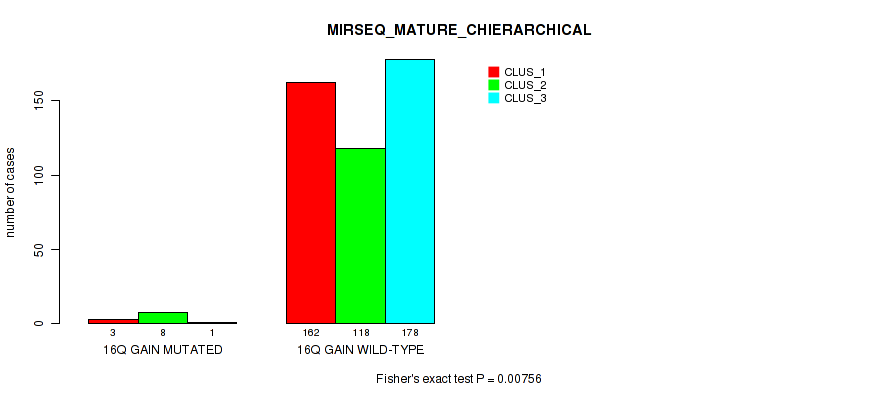
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S102. Gene #22: '17p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 17P GAIN MUTATED | 12 | 2 | 2 |
| 17P GAIN WILD-TYPE | 44 | 354 | 87 |
Figure S102. Get High-res Image Gene #22: '17p gain' versus Molecular Subtype #1: 'CN_CNMF'
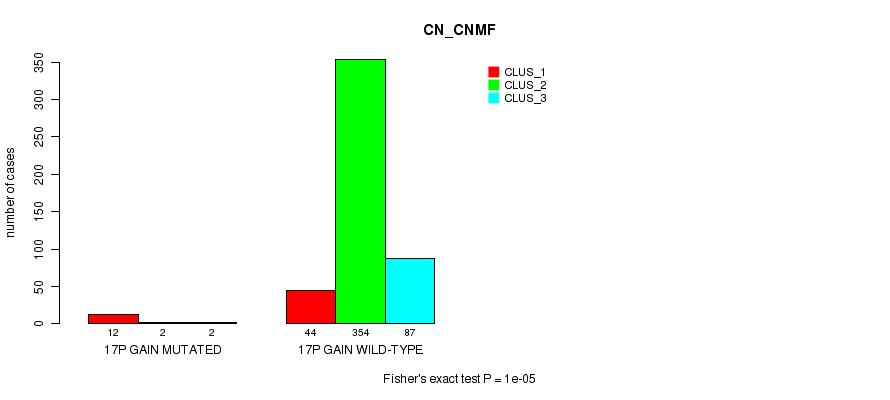
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S103. Gene #22: '17p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 17P GAIN MUTATED | 1 | 15 | 0 |
| 17P GAIN WILD-TYPE | 276 | 139 | 70 |
Figure S103. Get High-res Image Gene #22: '17p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
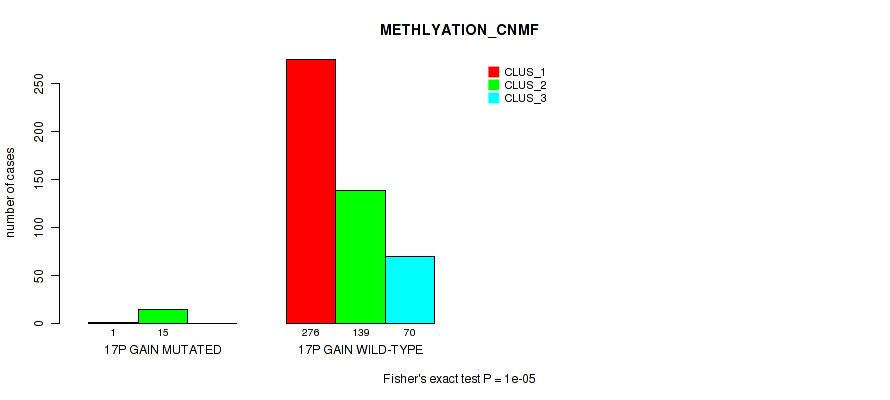
P value = 0.0278 (Fisher's exact test), Q value = 0.064
Table S104. Gene #22: '17p gain' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 46 | 28 | 33 | 29 | 49 | 27 | 9 |
| 17P GAIN MUTATED | 1 | 1 | 0 | 2 | 0 | 4 | 0 |
| 17P GAIN WILD-TYPE | 45 | 27 | 33 | 27 | 49 | 23 | 9 |
Figure S104. Get High-res Image Gene #22: '17p gain' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S105. Gene #22: '17p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 17P GAIN MUTATED | 0 | 15 | 0 | 1 |
| 17P GAIN WILD-TYPE | 168 | 140 | 59 | 116 |
Figure S105. Get High-res Image Gene #22: '17p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
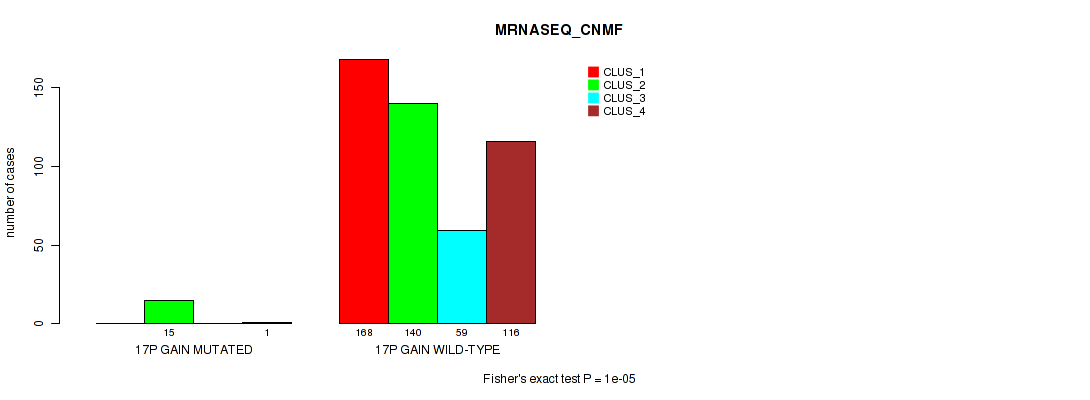
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S106. Gene #22: '17p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 17P GAIN MUTATED | 0 | 15 | 0 | 1 | 0 |
| 17P GAIN WILD-TYPE | 105 | 119 | 86 | 95 | 78 |
Figure S106. Get High-res Image Gene #22: '17p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
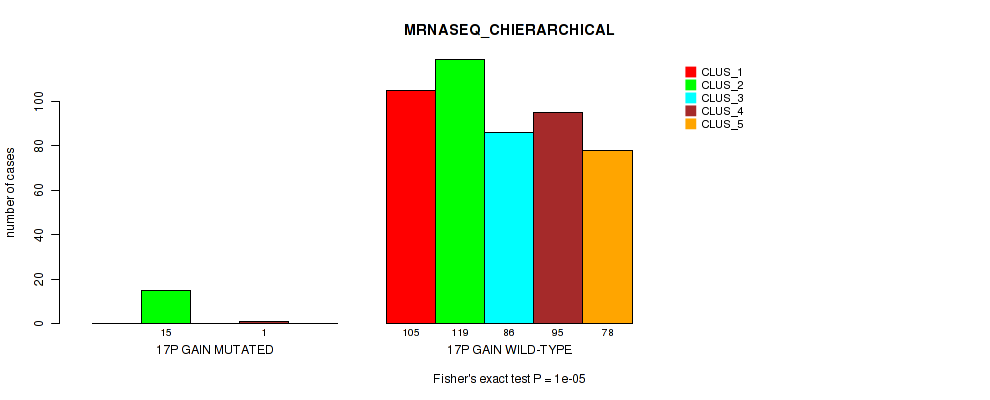
P value = 2e-05 (Fisher's exact test), Q value = 0.00018
Table S107. Gene #22: '17p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 17P GAIN MUTATED | 1 | 14 | 1 |
| 17P GAIN WILD-TYPE | 181 | 137 | 166 |
Figure S107. Get High-res Image Gene #22: '17p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
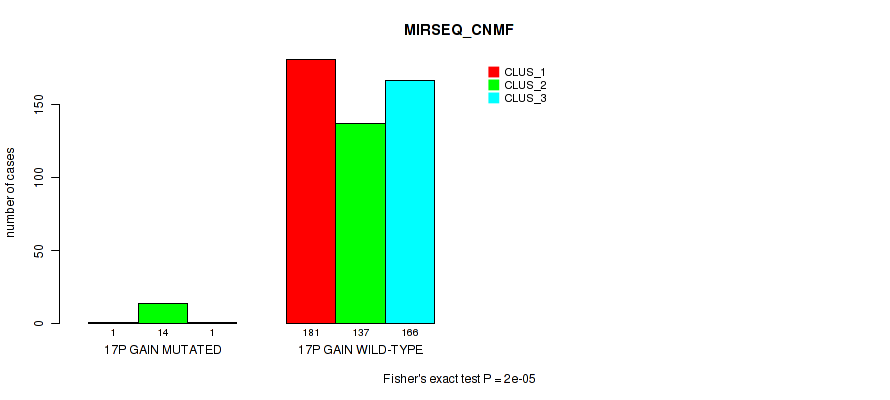
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S108. Gene #22: '17p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 17P GAIN MUTATED | 1 | 14 | 1 |
| 17P GAIN WILD-TYPE | 178 | 114 | 192 |
Figure S108. Get High-res Image Gene #22: '17p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
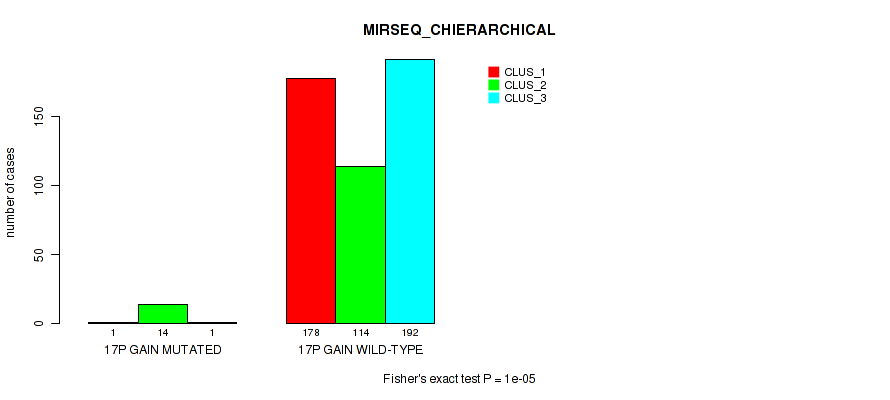
P value = 0.00027 (Fisher's exact test), Q value = 0.0013
Table S109. Gene #22: '17p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 17P GAIN MUTATED | 2 | 10 | 1 |
| 17P GAIN WILD-TYPE | 163 | 116 | 178 |
Figure S109. Get High-res Image Gene #22: '17p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
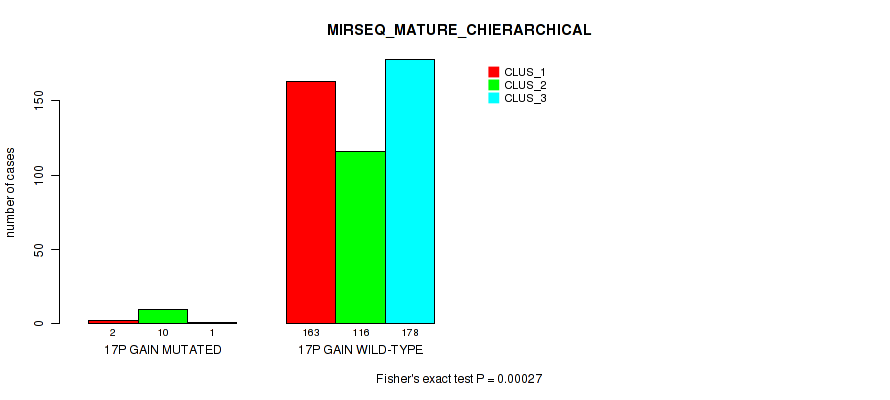
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S110. Gene #23: '17q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 17Q GAIN MUTATED | 13 | 2 | 3 |
| 17Q GAIN WILD-TYPE | 43 | 354 | 86 |
Figure S110. Get High-res Image Gene #23: '17q gain' versus Molecular Subtype #1: 'CN_CNMF'
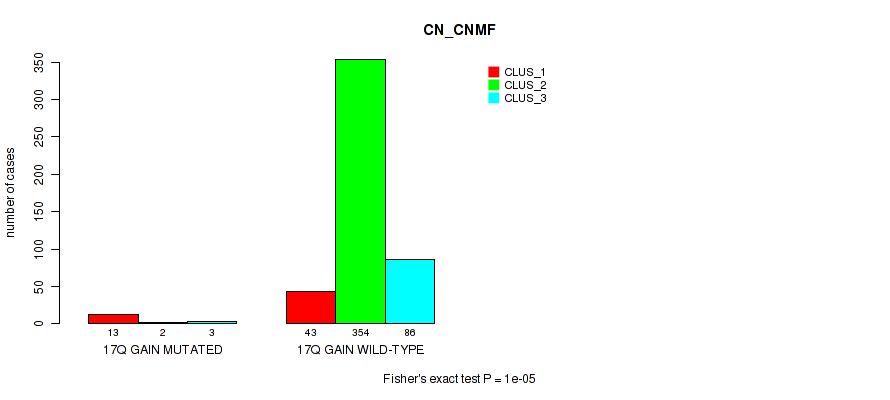
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S111. Gene #23: '17q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 17Q GAIN MUTATED | 2 | 16 | 0 |
| 17Q GAIN WILD-TYPE | 275 | 138 | 70 |
Figure S111. Get High-res Image Gene #23: '17q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.016 (Fisher's exact test), Q value = 0.041
Table S112. Gene #23: '17q gain' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 | CLUS_7 |
|---|---|---|---|---|---|---|---|
| ALL | 46 | 28 | 33 | 29 | 49 | 27 | 9 |
| 17Q GAIN MUTATED | 1 | 1 | 0 | 3 | 0 | 4 | 0 |
| 17Q GAIN WILD-TYPE | 45 | 27 | 33 | 26 | 49 | 23 | 9 |
Figure S112. Get High-res Image Gene #23: '17q gain' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
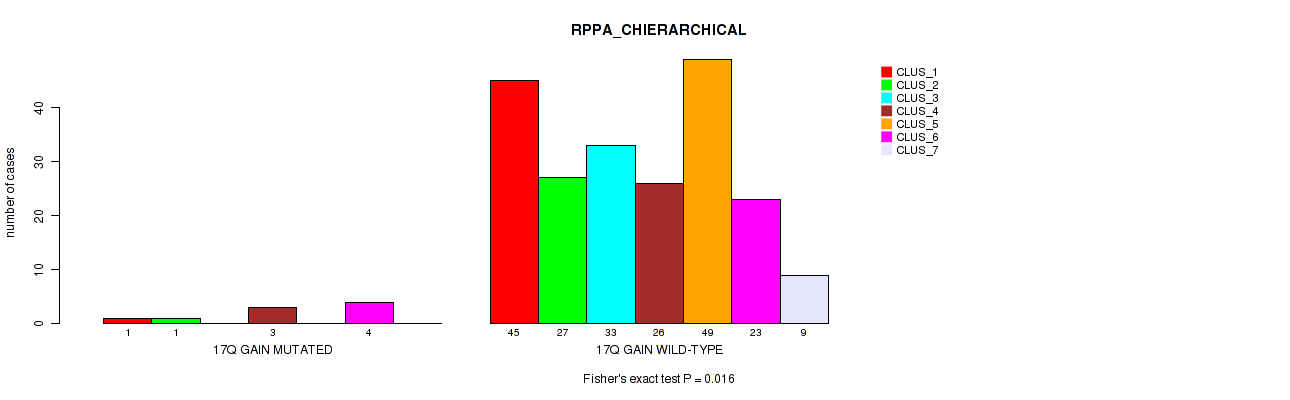
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S113. Gene #23: '17q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 17Q GAIN MUTATED | 1 | 16 | 0 | 1 |
| 17Q GAIN WILD-TYPE | 167 | 139 | 59 | 116 |
Figure S113. Get High-res Image Gene #23: '17q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
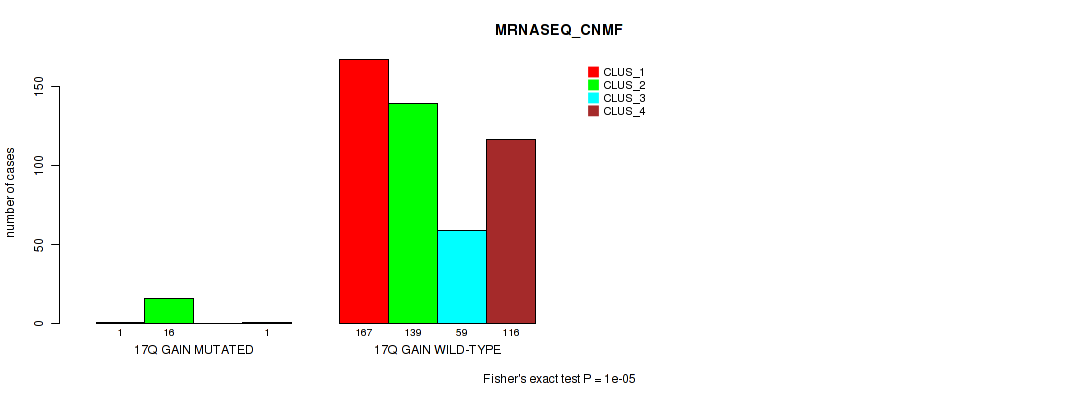
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S114. Gene #23: '17q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 17Q GAIN MUTATED | 1 | 16 | 0 | 1 | 0 |
| 17Q GAIN WILD-TYPE | 104 | 118 | 86 | 95 | 78 |
Figure S114. Get High-res Image Gene #23: '17q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
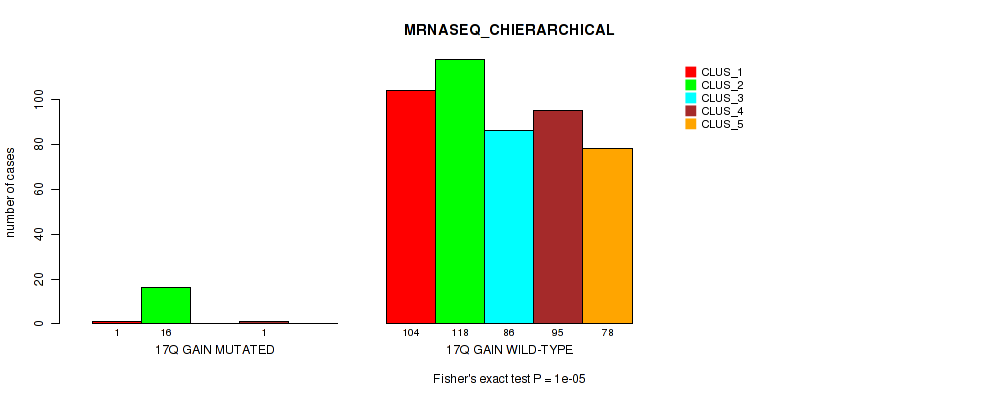
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S115. Gene #23: '17q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 17Q GAIN MUTATED | 2 | 15 | 1 |
| 17Q GAIN WILD-TYPE | 180 | 136 | 166 |
Figure S115. Get High-res Image Gene #23: '17q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
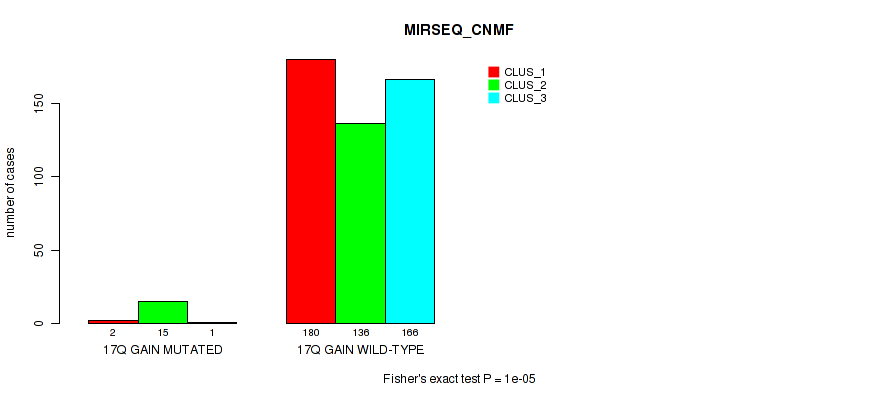
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S116. Gene #23: '17q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 17Q GAIN MUTATED | 2 | 15 | 1 |
| 17Q GAIN WILD-TYPE | 177 | 113 | 192 |
Figure S116. Get High-res Image Gene #23: '17q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
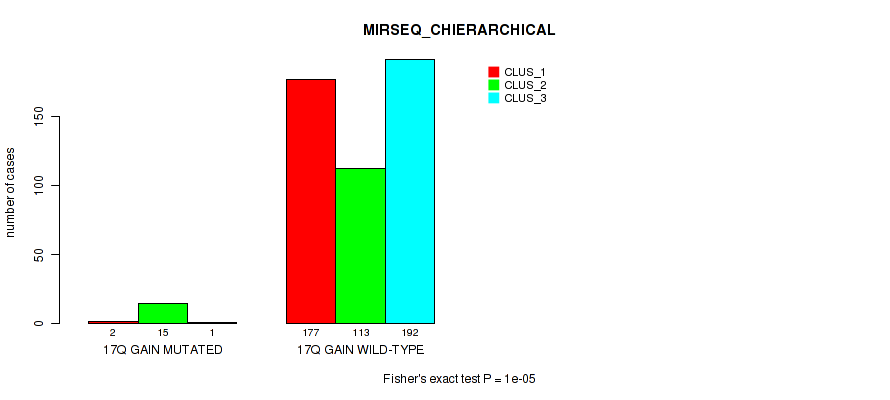
P value = 0.00017 (Fisher's exact test), Q value = 0.00092
Table S117. Gene #23: '17q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 17Q GAIN MUTATED | 3 | 11 | 1 |
| 17Q GAIN WILD-TYPE | 162 | 115 | 178 |
Figure S117. Get High-res Image Gene #23: '17q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
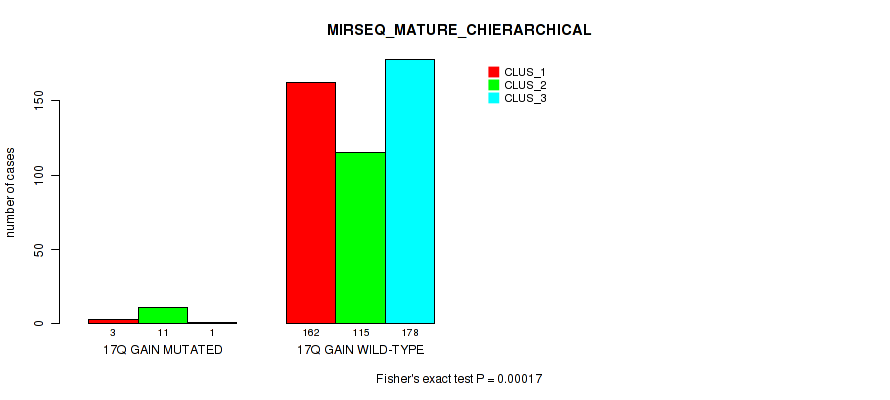
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S118. Gene #24: '18p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 18P GAIN MUTATED | 5 | 0 | 1 |
| 18P GAIN WILD-TYPE | 51 | 356 | 88 |
Figure S118. Get High-res Image Gene #24: '18p gain' versus Molecular Subtype #1: 'CN_CNMF'
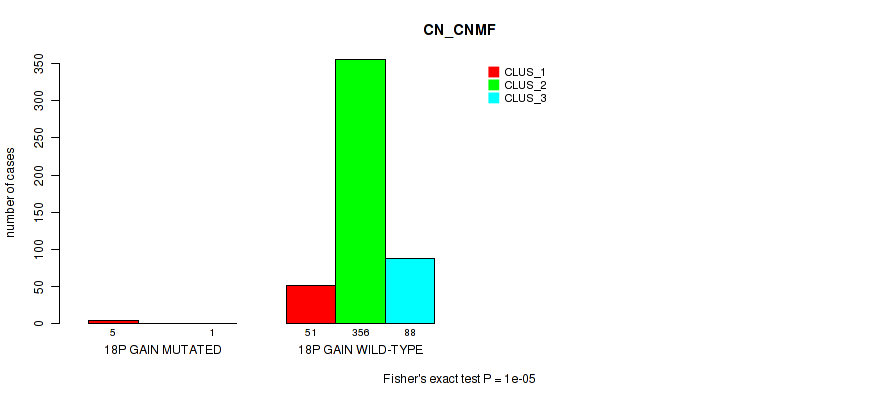
P value = 4e-05 (Fisher's exact test), Q value = 0.00029
Table S119. Gene #25: '18q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 18Q GAIN MUTATED | 5 | 0 | 1 |
| 18Q GAIN WILD-TYPE | 51 | 356 | 88 |
Figure S119. Get High-res Image Gene #25: '18q gain' versus Molecular Subtype #1: 'CN_CNMF'
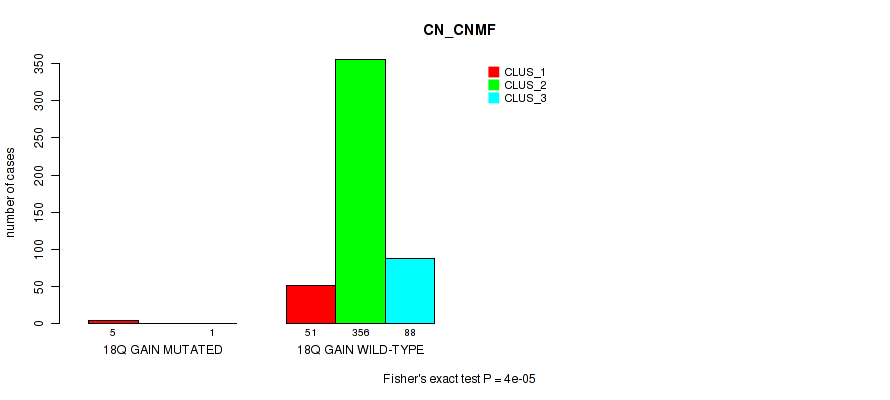
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S120. Gene #26: '19p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 19P GAIN MUTATED | 8 | 0 | 0 |
| 19P GAIN WILD-TYPE | 48 | 356 | 89 |
Figure S120. Get High-res Image Gene #26: '19p gain' versus Molecular Subtype #1: 'CN_CNMF'
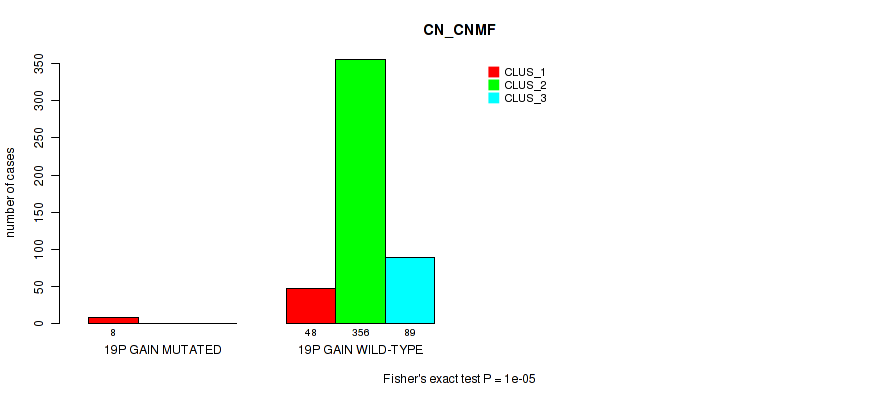
P value = 2e-04 (Fisher's exact test), Q value = 0.001
Table S121. Gene #26: '19p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 19P GAIN MUTATED | 0 | 8 | 0 |
| 19P GAIN WILD-TYPE | 277 | 146 | 70 |
Figure S121. Get High-res Image Gene #26: '19p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
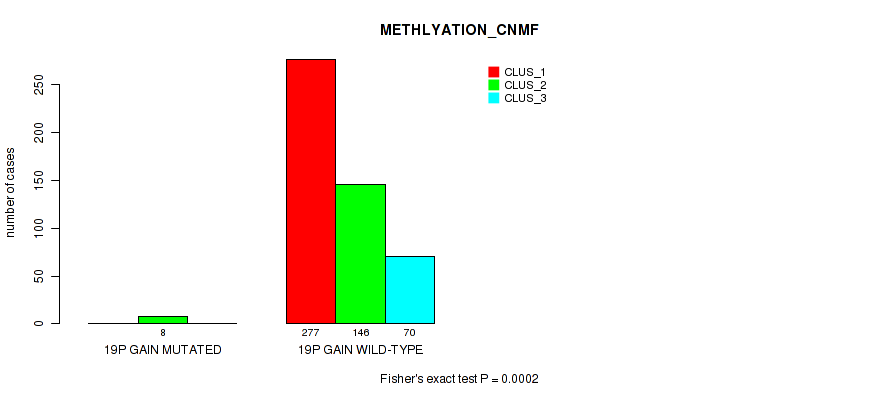
P value = 0.00043 (Fisher's exact test), Q value = 0.002
Table S122. Gene #26: '19p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 19P GAIN MUTATED | 0 | 8 | 0 | 0 |
| 19P GAIN WILD-TYPE | 168 | 147 | 59 | 117 |
Figure S122. Get High-res Image Gene #26: '19p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
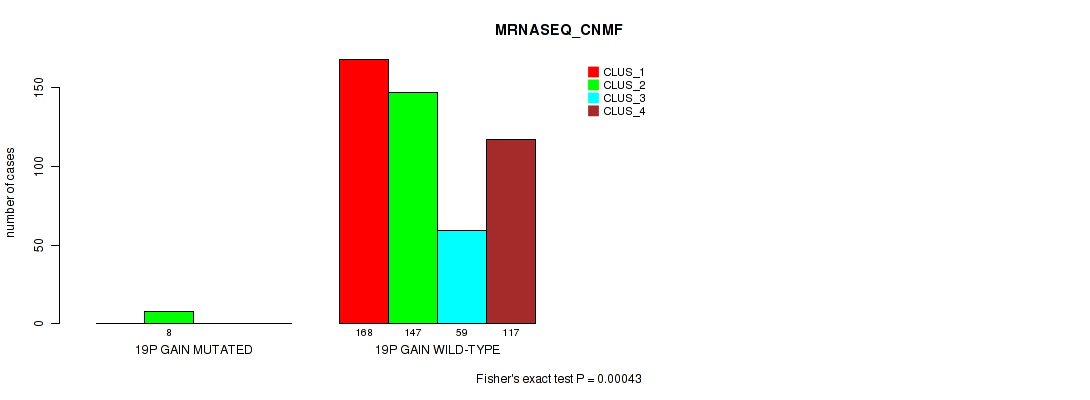
P value = 0.00023 (Fisher's exact test), Q value = 0.0012
Table S123. Gene #26: '19p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 19P GAIN MUTATED | 0 | 8 | 0 | 0 | 0 |
| 19P GAIN WILD-TYPE | 105 | 126 | 86 | 96 | 78 |
Figure S123. Get High-res Image Gene #26: '19p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
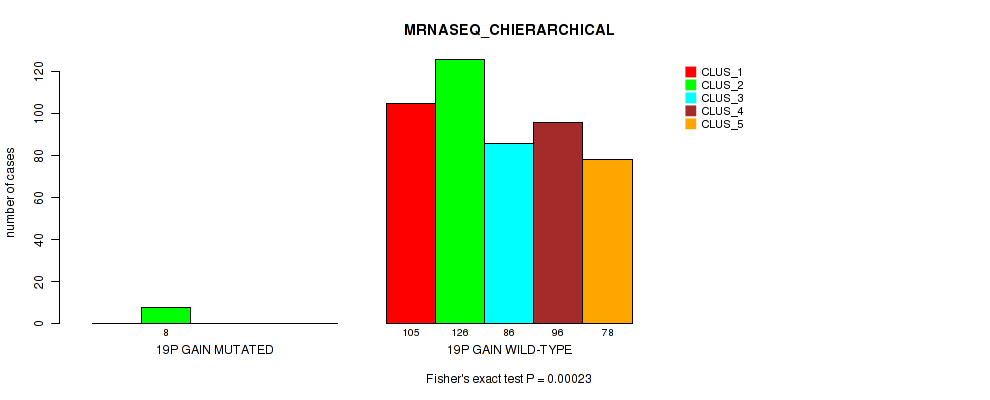
P value = 0.00013 (Fisher's exact test), Q value = 0.00074
Table S124. Gene #26: '19p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 19P GAIN MUTATED | 0 | 8 | 0 |
| 19P GAIN WILD-TYPE | 182 | 143 | 167 |
Figure S124. Get High-res Image Gene #26: '19p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
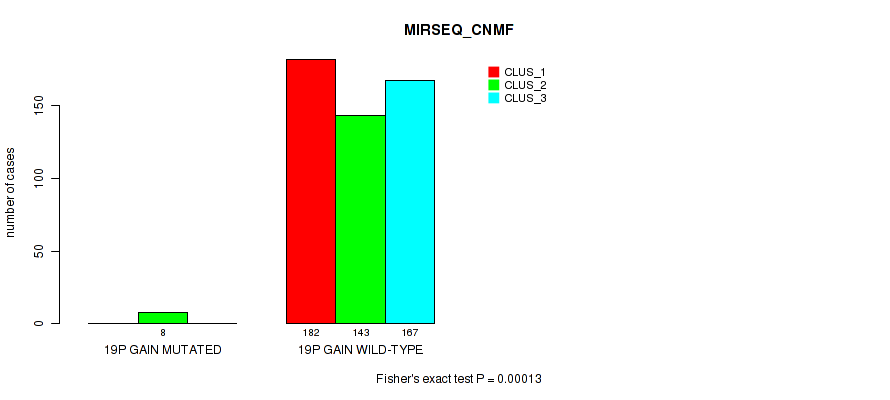
P value = 3e-05 (Fisher's exact test), Q value = 0.00024
Table S125. Gene #26: '19p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 19P GAIN MUTATED | 0 | 8 | 0 |
| 19P GAIN WILD-TYPE | 179 | 120 | 193 |
Figure S125. Get High-res Image Gene #26: '19p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
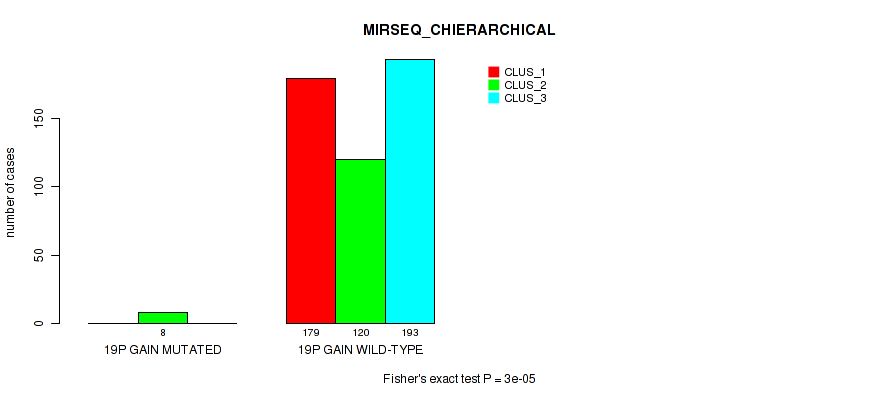
P value = 0.0223 (Fisher's exact test), Q value = 0.053
Table S126. Gene #26: '19p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 19P GAIN MUTATED | 1 | 4 | 0 |
| 19P GAIN WILD-TYPE | 164 | 122 | 179 |
Figure S126. Get High-res Image Gene #26: '19p gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
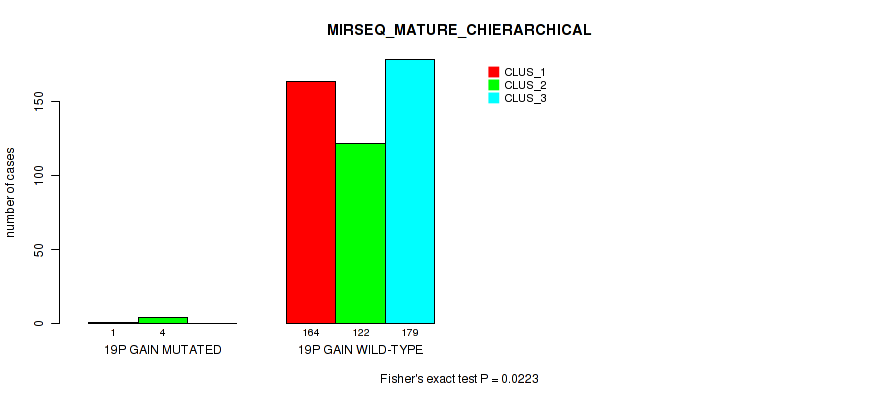
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S127. Gene #27: '19q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 19Q GAIN MUTATED | 10 | 0 | 0 |
| 19Q GAIN WILD-TYPE | 46 | 356 | 89 |
Figure S127. Get High-res Image Gene #27: '19q gain' versus Molecular Subtype #1: 'CN_CNMF'
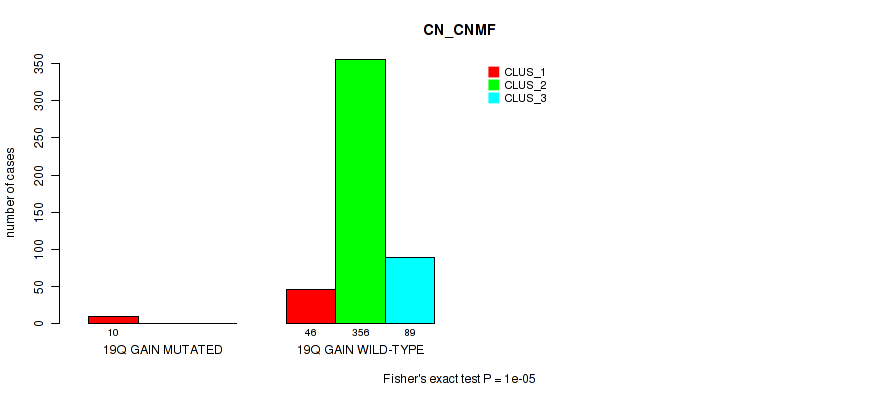
P value = 2e-05 (Fisher's exact test), Q value = 0.00018
Table S128. Gene #27: '19q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 19Q GAIN MUTATED | 0 | 10 | 0 |
| 19Q GAIN WILD-TYPE | 277 | 144 | 70 |
Figure S128. Get High-res Image Gene #27: '19q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
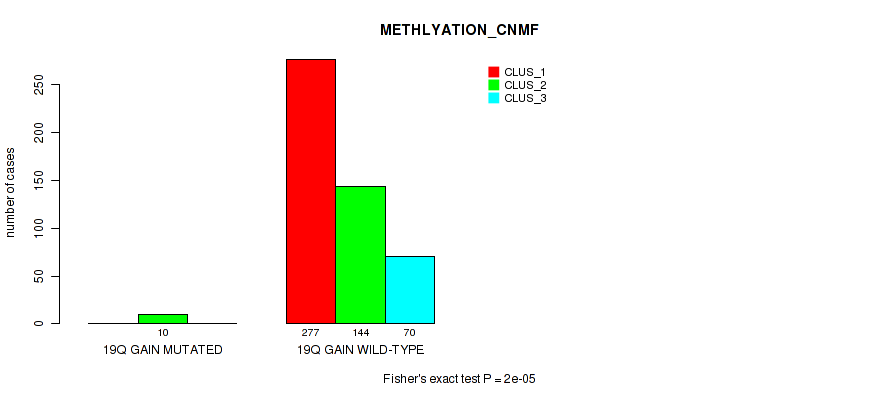
P value = 3e-05 (Fisher's exact test), Q value = 0.00024
Table S129. Gene #27: '19q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 19Q GAIN MUTATED | 0 | 10 | 0 | 0 |
| 19Q GAIN WILD-TYPE | 168 | 145 | 59 | 117 |
Figure S129. Get High-res Image Gene #27: '19q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
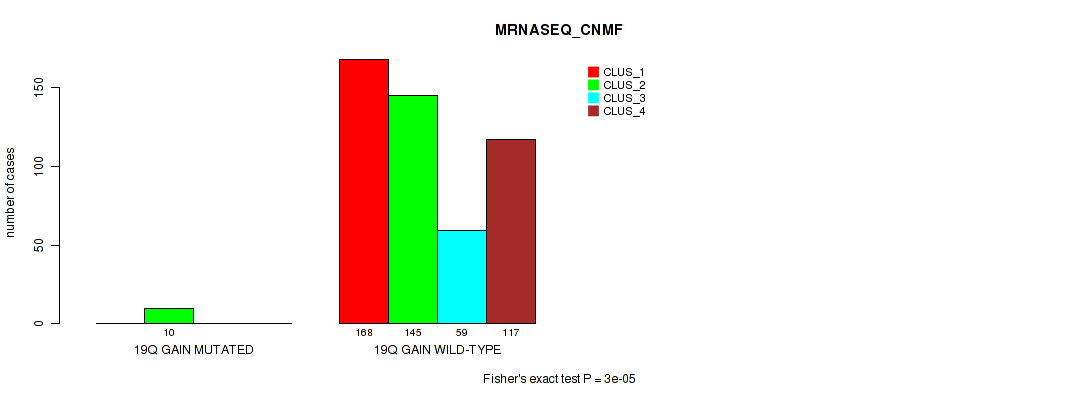
P value = 4e-05 (Fisher's exact test), Q value = 0.00029
Table S130. Gene #27: '19q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 19Q GAIN MUTATED | 0 | 10 | 0 | 0 | 0 |
| 19Q GAIN WILD-TYPE | 105 | 124 | 86 | 96 | 78 |
Figure S130. Get High-res Image Gene #27: '19q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
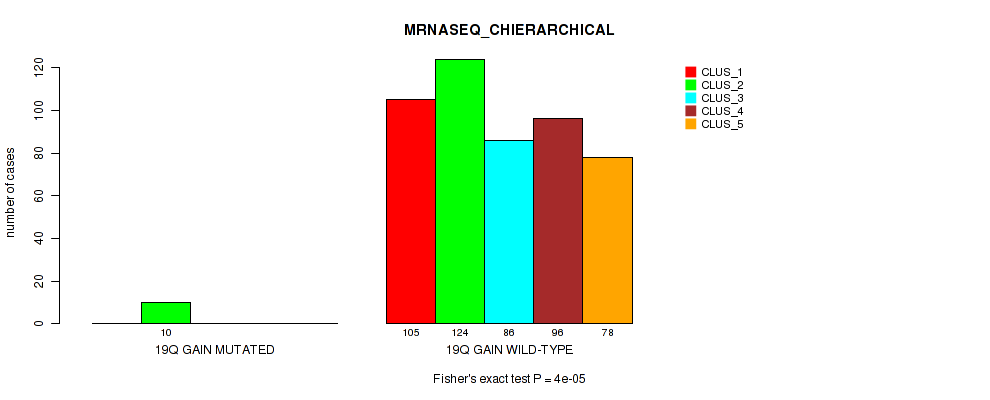
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S131. Gene #27: '19q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 19Q GAIN MUTATED | 0 | 10 | 0 |
| 19Q GAIN WILD-TYPE | 182 | 141 | 167 |
Figure S131. Get High-res Image Gene #27: '19q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S132. Gene #27: '19q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 19Q GAIN MUTATED | 0 | 10 | 0 |
| 19Q GAIN WILD-TYPE | 179 | 118 | 193 |
Figure S132. Get High-res Image Gene #27: '19q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'

P value = 0.00159 (Fisher's exact test), Q value = 0.0059
Table S133. Gene #27: '19q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 19Q GAIN MUTATED | 1 | 6 | 0 |
| 19Q GAIN WILD-TYPE | 164 | 120 | 179 |
Figure S133. Get High-res Image Gene #27: '19q gain' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
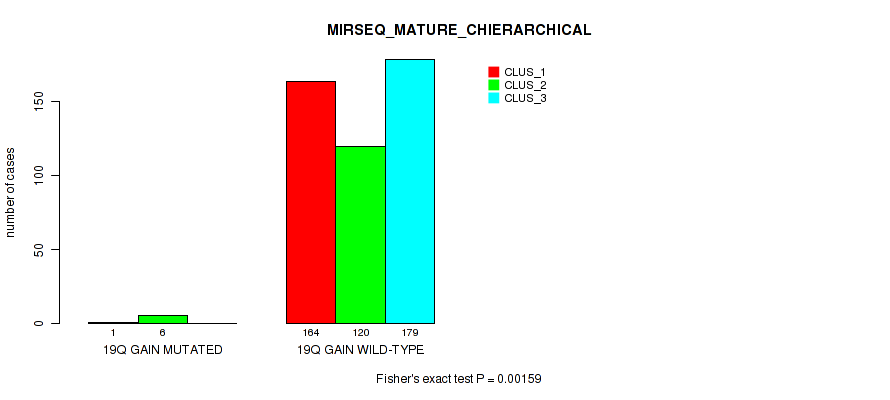
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S134. Gene #28: '20p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 20P GAIN MUTATED | 12 | 0 | 0 |
| 20P GAIN WILD-TYPE | 44 | 356 | 89 |
Figure S134. Get High-res Image Gene #28: '20p gain' versus Molecular Subtype #1: 'CN_CNMF'
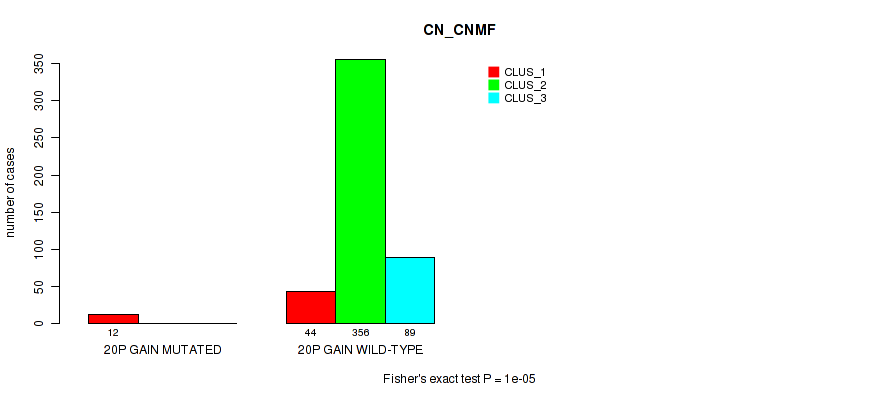
P value = 0.00029 (Fisher's exact test), Q value = 0.0014
Table S135. Gene #28: '20p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 20P GAIN MUTATED | 1 | 10 | 1 |
| 20P GAIN WILD-TYPE | 276 | 144 | 69 |
Figure S135. Get High-res Image Gene #28: '20p gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
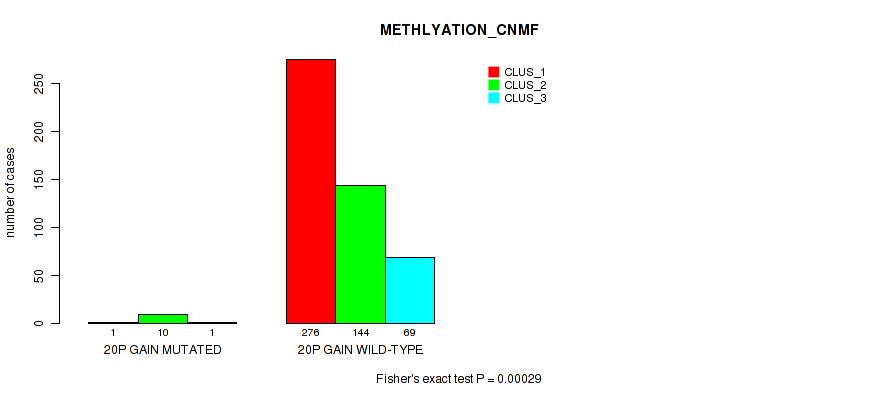
P value = 0.0026 (Fisher's exact test), Q value = 0.0089
Table S136. Gene #28: '20p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 20P GAIN MUTATED | 1 | 10 | 0 | 1 |
| 20P GAIN WILD-TYPE | 167 | 145 | 59 | 116 |
Figure S136. Get High-res Image Gene #28: '20p gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
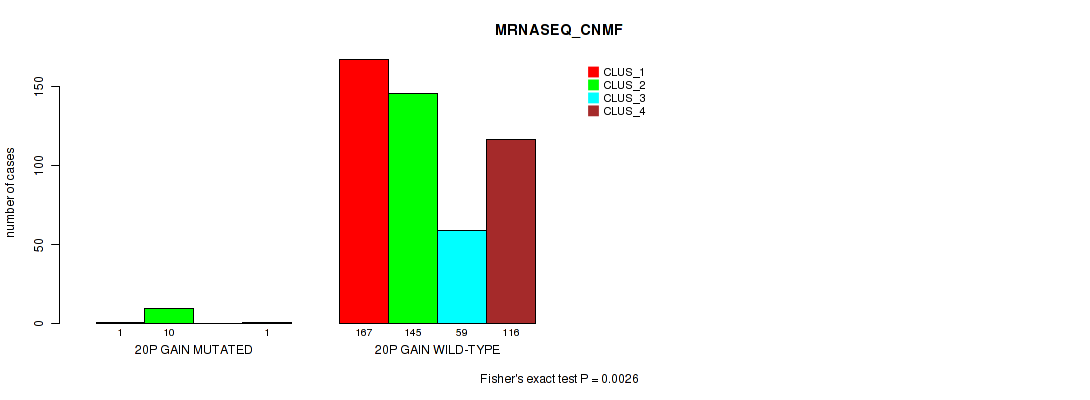
P value = 0.00079 (Fisher's exact test), Q value = 0.0033
Table S137. Gene #28: '20p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 20P GAIN MUTATED | 1 | 10 | 0 | 1 | 0 |
| 20P GAIN WILD-TYPE | 104 | 124 | 86 | 95 | 78 |
Figure S137. Get High-res Image Gene #28: '20p gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
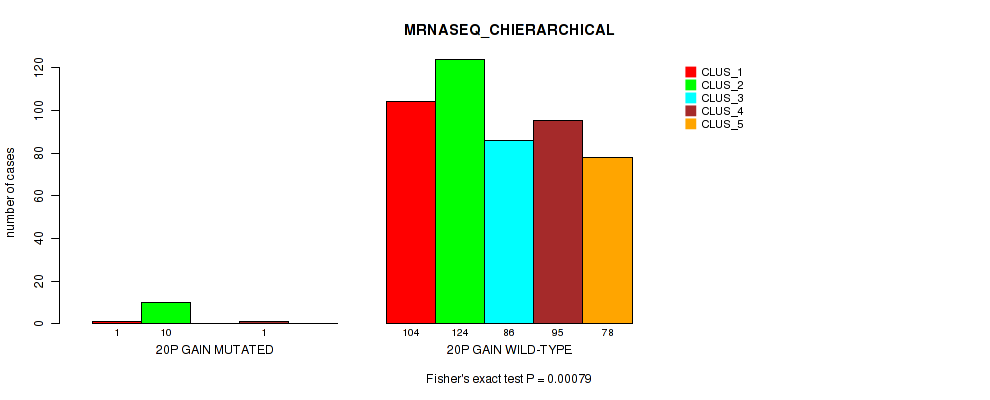
P value = 0.00424 (Fisher's exact test), Q value = 0.014
Table S138. Gene #28: '20p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 20P GAIN MUTATED | 2 | 9 | 1 |
| 20P GAIN WILD-TYPE | 180 | 142 | 166 |
Figure S138. Get High-res Image Gene #28: '20p gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
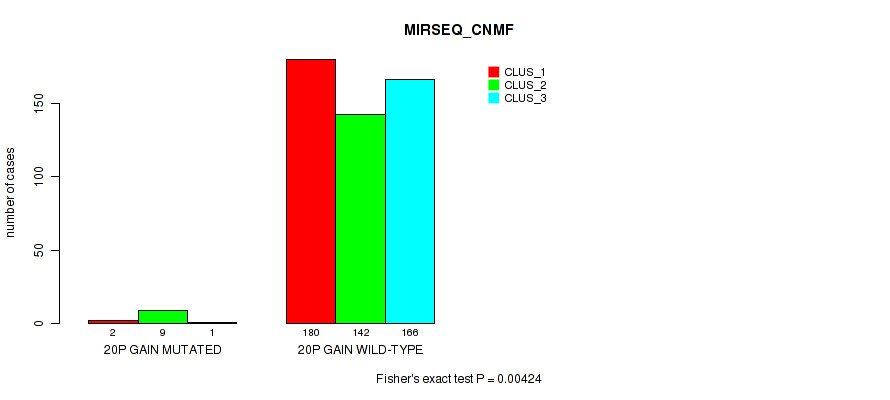
P value = 0.00075 (Fisher's exact test), Q value = 0.0032
Table S139. Gene #28: '20p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 20P GAIN MUTATED | 2 | 9 | 1 |
| 20P GAIN WILD-TYPE | 177 | 119 | 192 |
Figure S139. Get High-res Image Gene #28: '20p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
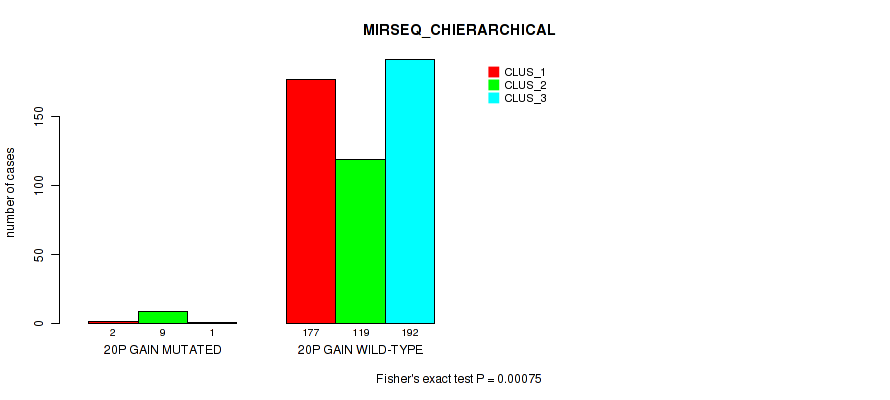
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S140. Gene #29: '20q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 20Q GAIN MUTATED | 12 | 0 | 0 |
| 20Q GAIN WILD-TYPE | 44 | 356 | 89 |
Figure S140. Get High-res Image Gene #29: '20q gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00031 (Fisher's exact test), Q value = 0.0015
Table S141. Gene #29: '20q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 20Q GAIN MUTATED | 1 | 10 | 1 |
| 20Q GAIN WILD-TYPE | 276 | 144 | 69 |
Figure S141. Get High-res Image Gene #29: '20q gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
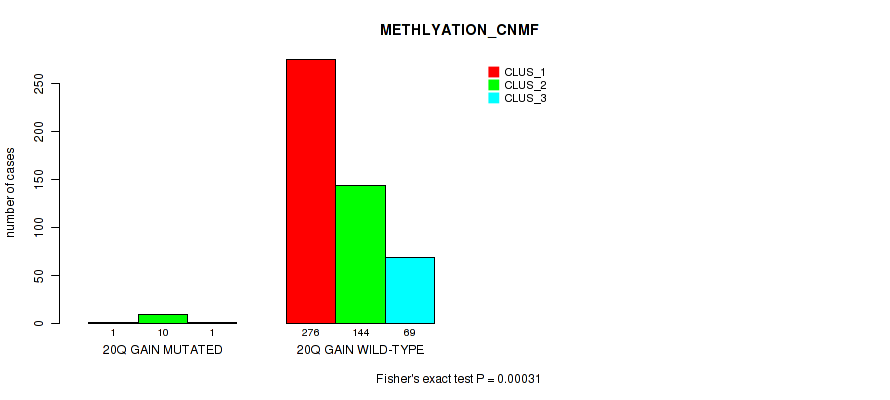
P value = 0.00277 (Fisher's exact test), Q value = 0.0094
Table S142. Gene #29: '20q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 20Q GAIN MUTATED | 1 | 10 | 0 | 1 |
| 20Q GAIN WILD-TYPE | 167 | 145 | 59 | 116 |
Figure S142. Get High-res Image Gene #29: '20q gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
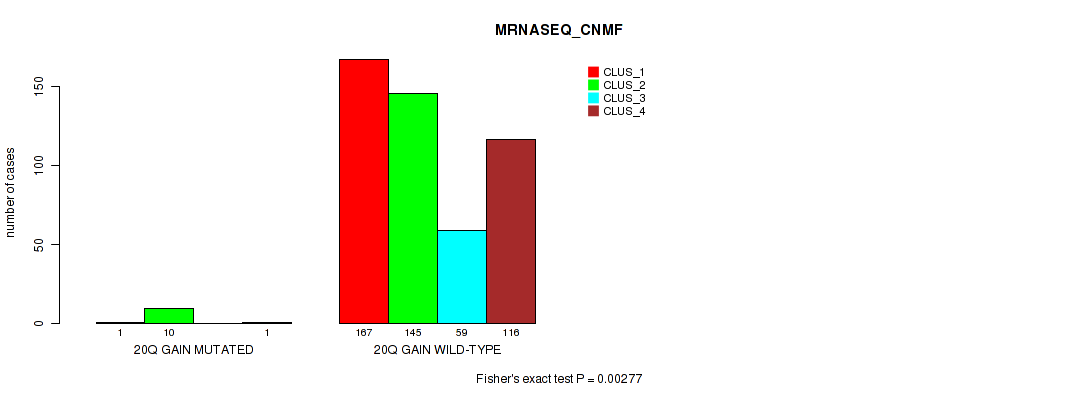
P value = 0.00103 (Fisher's exact test), Q value = 0.0042
Table S143. Gene #29: '20q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 20Q GAIN MUTATED | 1 | 10 | 0 | 1 | 0 |
| 20Q GAIN WILD-TYPE | 104 | 124 | 86 | 95 | 78 |
Figure S143. Get High-res Image Gene #29: '20q gain' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
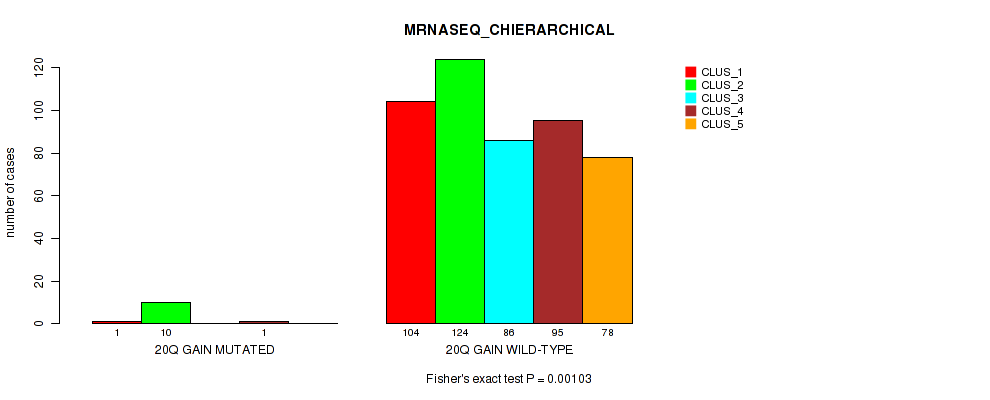
P value = 0.00406 (Fisher's exact test), Q value = 0.014
Table S144. Gene #29: '20q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 20Q GAIN MUTATED | 2 | 9 | 1 |
| 20Q GAIN WILD-TYPE | 180 | 142 | 166 |
Figure S144. Get High-res Image Gene #29: '20q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
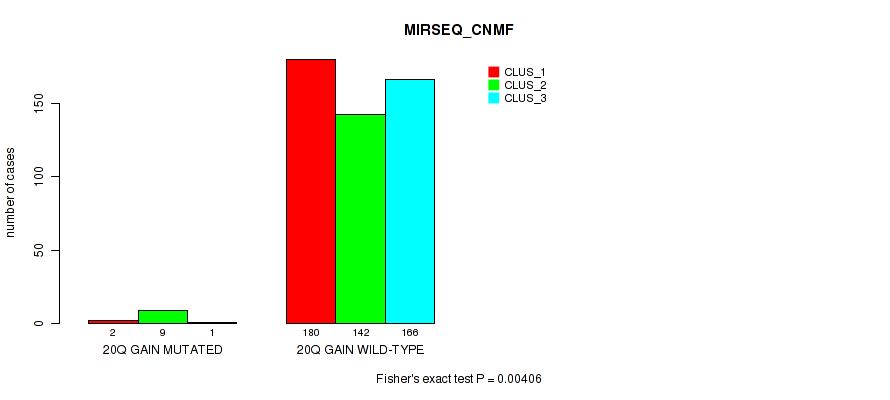
P value = 0.00065 (Fisher's exact test), Q value = 0.0028
Table S145. Gene #29: '20q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 20Q GAIN MUTATED | 2 | 9 | 1 |
| 20Q GAIN WILD-TYPE | 177 | 119 | 192 |
Figure S145. Get High-res Image Gene #29: '20q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
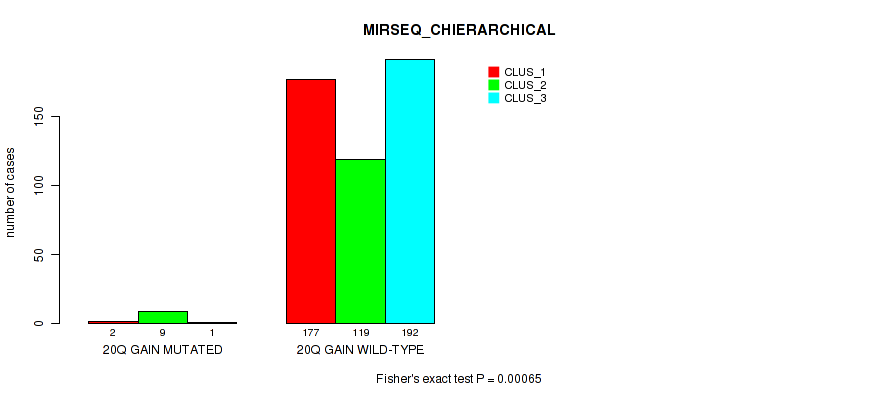
P value = 0.00023 (Fisher's exact test), Q value = 0.0012
Table S146. Gene #30: '21q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 21Q GAIN MUTATED | 4 | 0 | 1 |
| 21Q GAIN WILD-TYPE | 52 | 356 | 88 |
Figure S146. Get High-res Image Gene #30: '21q gain' versus Molecular Subtype #1: 'CN_CNMF'
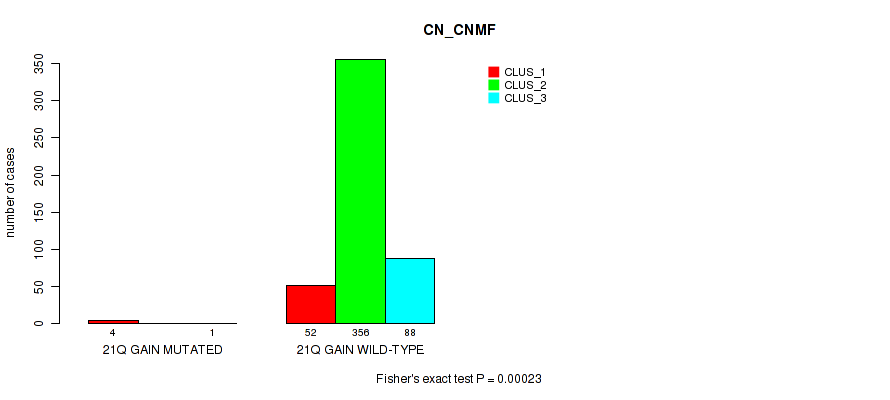
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S147. Gene #31: 'xp gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| XP GAIN MUTATED | 8 | 0 | 0 |
| XP GAIN WILD-TYPE | 48 | 356 | 89 |
Figure S147. Get High-res Image Gene #31: 'xp gain' versus Molecular Subtype #1: 'CN_CNMF'
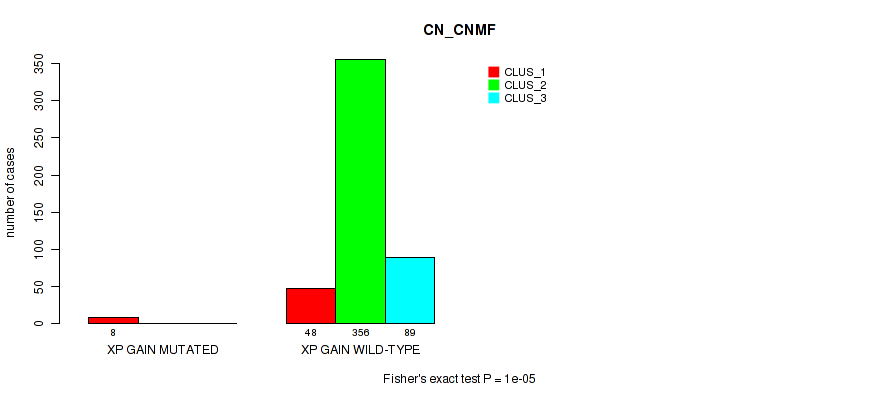
P value = 0.00191 (Fisher's exact test), Q value = 0.007
Table S148. Gene #31: 'xp gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| XP GAIN MUTATED | 0 | 6 | 2 |
| XP GAIN WILD-TYPE | 277 | 148 | 68 |
Figure S148. Get High-res Image Gene #31: 'xp gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
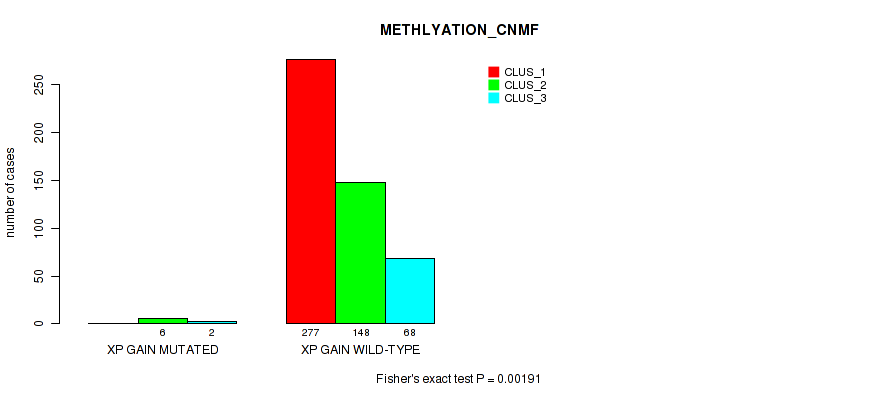
P value = 0.045 (Fisher's exact test), Q value = 0.098
Table S149. Gene #31: 'xp gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| XP GAIN MUTATED | 1 | 6 | 1 | 0 |
| XP GAIN WILD-TYPE | 167 | 149 | 58 | 117 |
Figure S149. Get High-res Image Gene #31: 'xp gain' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
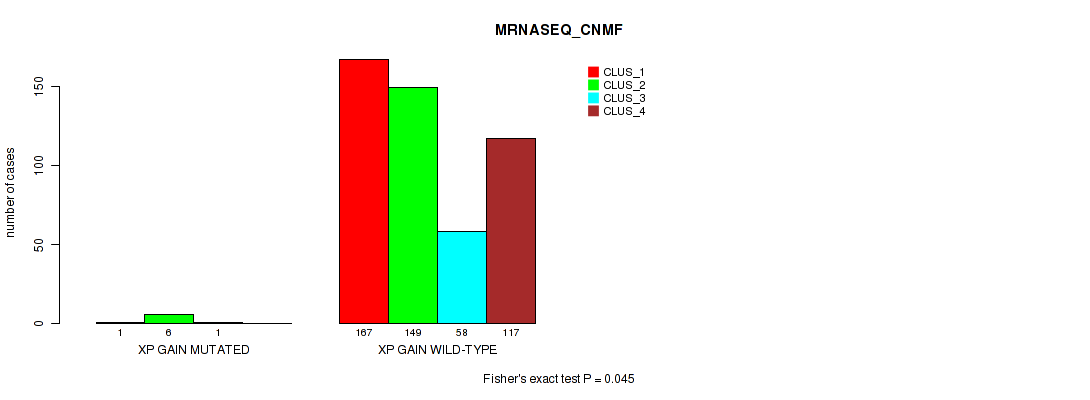
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S150. Gene #32: 'xq gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| XQ GAIN MUTATED | 8 | 1 | 0 |
| XQ GAIN WILD-TYPE | 48 | 355 | 89 |
Figure S150. Get High-res Image Gene #32: 'xq gain' versus Molecular Subtype #1: 'CN_CNMF'
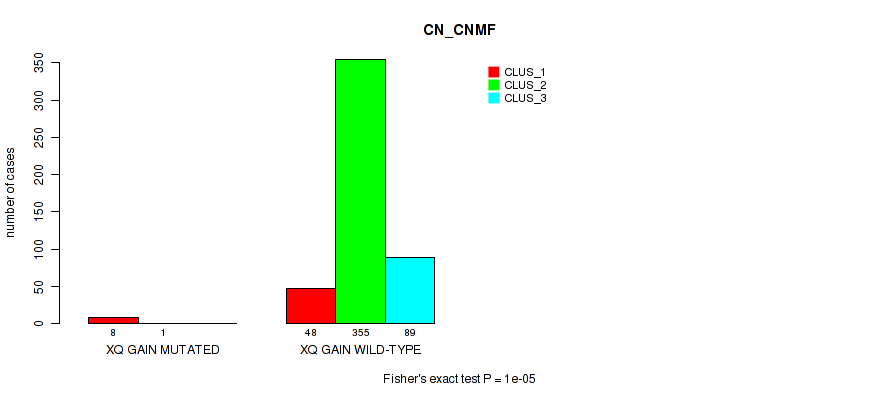
P value = 0.0114 (Fisher's exact test), Q value = 0.032
Table S151. Gene #32: 'xq gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| XQ GAIN MUTATED | 1 | 6 | 2 |
| XQ GAIN WILD-TYPE | 276 | 148 | 68 |
Figure S151. Get High-res Image Gene #32: 'xq gain' versus Molecular Subtype #2: 'METHLYATION_CNMF'
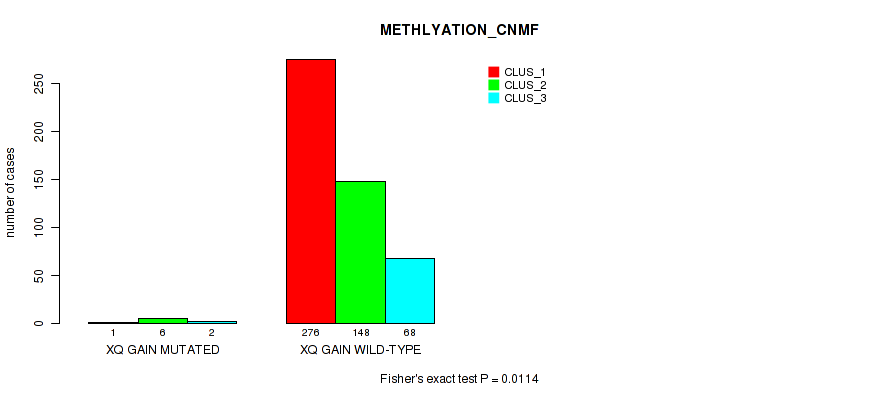
P value = 1e-04 (Fisher's exact test), Q value = 0.00063
Table S152. Gene #33: '1p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 1P LOSS MUTATED | 4 | 0 | 0 |
| 1P LOSS WILD-TYPE | 52 | 356 | 89 |
Figure S152. Get High-res Image Gene #33: '1p loss' versus Molecular Subtype #1: 'CN_CNMF'
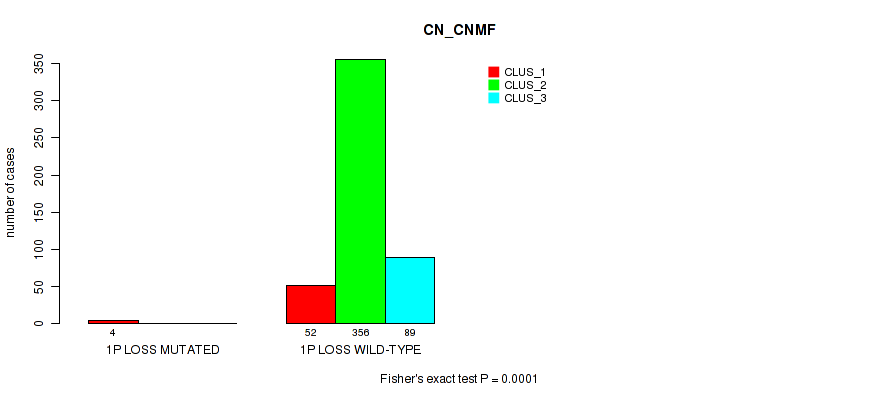
P value = 0.00132 (Fisher's exact test), Q value = 0.0051
Table S153. Gene #34: '1q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 1Q LOSS MUTATED | 3 | 0 | 0 |
| 1Q LOSS WILD-TYPE | 53 | 356 | 89 |
Figure S153. Get High-res Image Gene #34: '1q loss' versus Molecular Subtype #1: 'CN_CNMF'
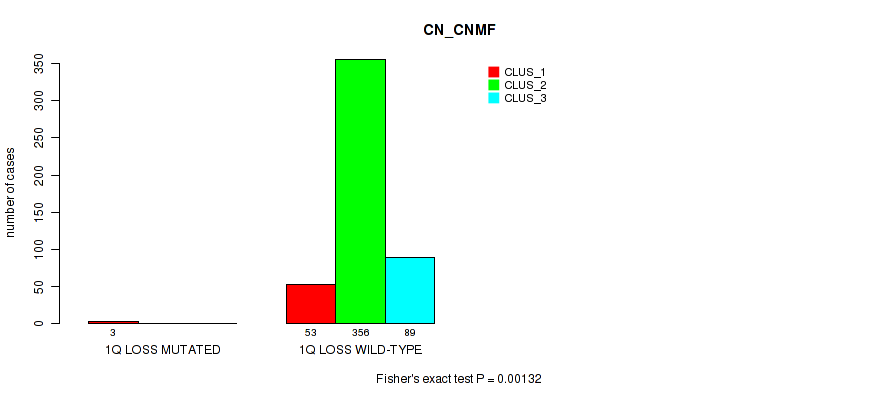
P value = 0.0478 (Fisher's exact test), Q value = 0.1
Table S154. Gene #34: '1q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 1Q LOSS MUTATED | 0 | 3 | 0 |
| 1Q LOSS WILD-TYPE | 277 | 151 | 70 |
Figure S154. Get High-res Image Gene #34: '1q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
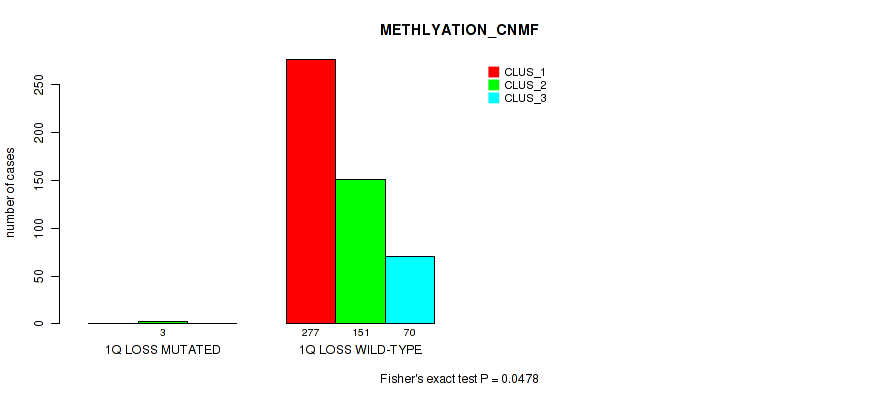
P value = 0.0266 (Fisher's exact test), Q value = 0.062
Table S155. Gene #34: '1q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 1Q LOSS MUTATED | 0 | 3 | 0 |
| 1Q LOSS WILD-TYPE | 182 | 148 | 167 |
Figure S155. Get High-res Image Gene #34: '1q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
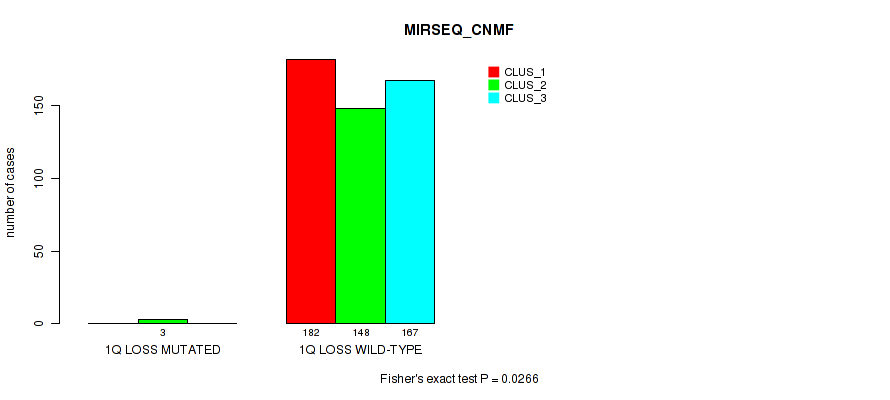
P value = 0.0167 (Fisher's exact test), Q value = 0.042
Table S156. Gene #34: '1q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 1Q LOSS MUTATED | 0 | 3 | 0 |
| 1Q LOSS WILD-TYPE | 179 | 125 | 193 |
Figure S156. Get High-res Image Gene #34: '1q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
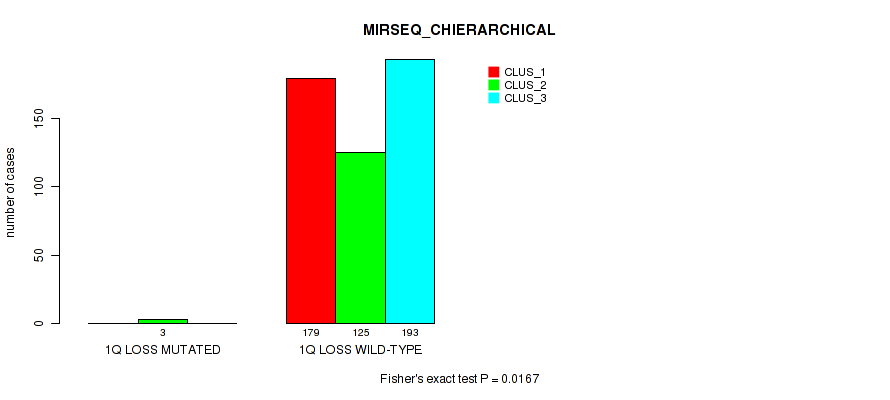
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S157. Gene #35: '2p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 2P LOSS MUTATED | 9 | 0 | 1 |
| 2P LOSS WILD-TYPE | 47 | 356 | 88 |
Figure S157. Get High-res Image Gene #35: '2p loss' versus Molecular Subtype #1: 'CN_CNMF'
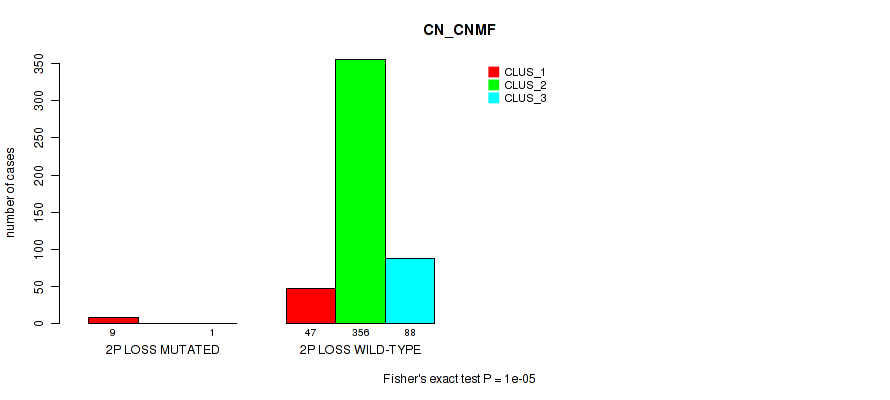
P value = 4e-05 (Fisher's exact test), Q value = 0.00029
Table S158. Gene #35: '2p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 2P LOSS MUTATED | 0 | 10 | 0 |
| 2P LOSS WILD-TYPE | 277 | 144 | 70 |
Figure S158. Get High-res Image Gene #35: '2p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
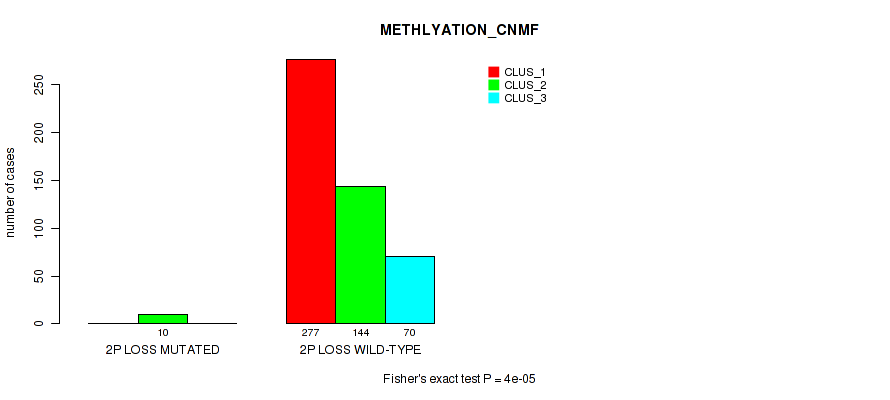
P value = 7e-05 (Fisher's exact test), Q value = 0.00046
Table S159. Gene #35: '2p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 2P LOSS MUTATED | 0 | 10 | 0 | 0 |
| 2P LOSS WILD-TYPE | 168 | 145 | 59 | 117 |
Figure S159. Get High-res Image Gene #35: '2p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
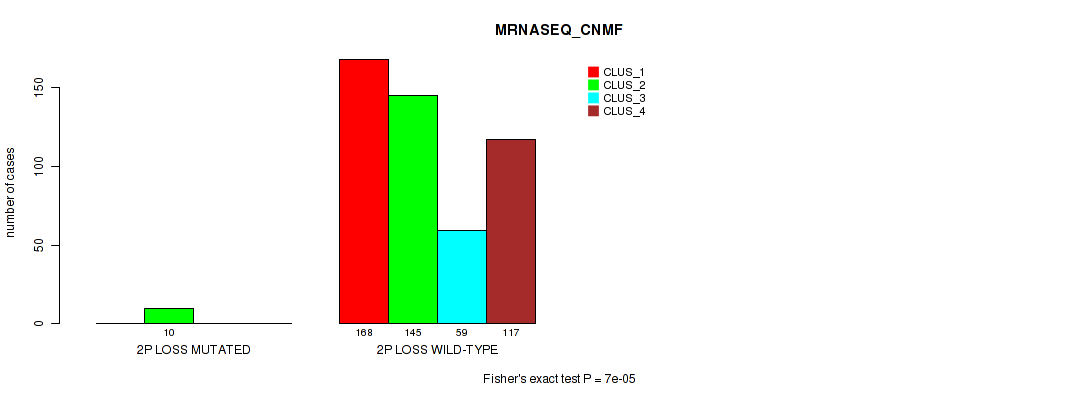
P value = 3e-05 (Fisher's exact test), Q value = 0.00024
Table S160. Gene #35: '2p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 2P LOSS MUTATED | 0 | 10 | 0 | 0 | 0 |
| 2P LOSS WILD-TYPE | 105 | 124 | 86 | 96 | 78 |
Figure S160. Get High-res Image Gene #35: '2p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
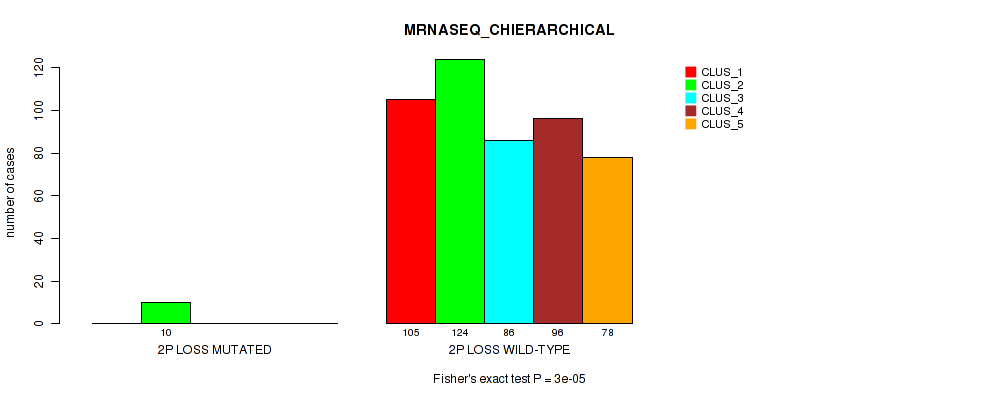
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S161. Gene #35: '2p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 2P LOSS MUTATED | 0 | 10 | 0 |
| 2P LOSS WILD-TYPE | 182 | 141 | 167 |
Figure S161. Get High-res Image Gene #35: '2p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'

P value = 2e-05 (Fisher's exact test), Q value = 0.00018
Table S162. Gene #35: '2p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 2P LOSS MUTATED | 0 | 10 | 0 |
| 2P LOSS WILD-TYPE | 179 | 118 | 193 |
Figure S162. Get High-res Image Gene #35: '2p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
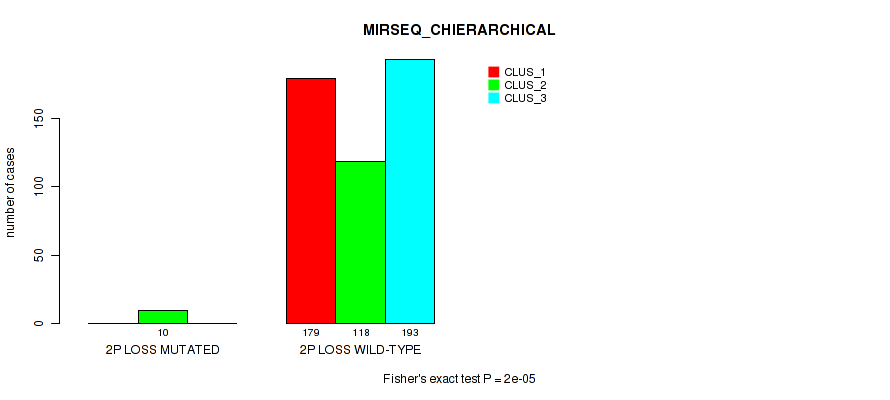
P value = 0.00044 (Fisher's exact test), Q value = 0.002
Table S163. Gene #35: '2p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 2P LOSS MUTATED | 1 | 7 | 0 |
| 2P LOSS WILD-TYPE | 164 | 119 | 179 |
Figure S163. Get High-res Image Gene #35: '2p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
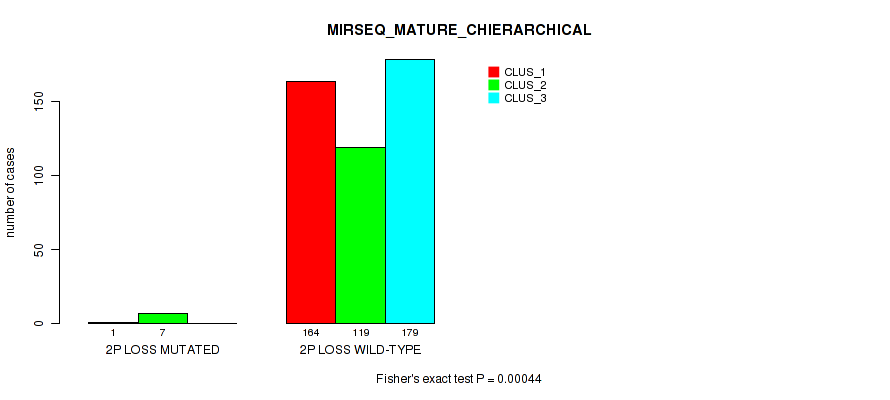
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S164. Gene #36: '2q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 2Q LOSS MUTATED | 10 | 0 | 0 |
| 2Q LOSS WILD-TYPE | 46 | 356 | 89 |
Figure S164. Get High-res Image Gene #36: '2q loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 3e-05 (Fisher's exact test), Q value = 0.00024
Table S165. Gene #36: '2q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 2Q LOSS MUTATED | 0 | 10 | 0 |
| 2Q LOSS WILD-TYPE | 277 | 144 | 70 |
Figure S165. Get High-res Image Gene #36: '2q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
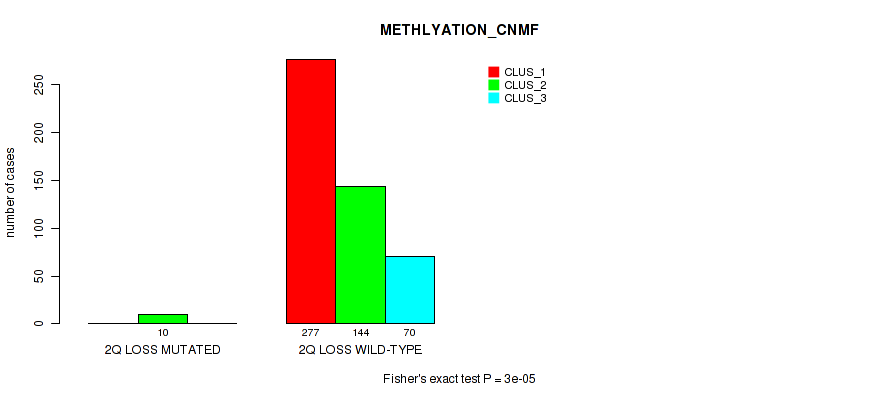
P value = 6e-05 (Fisher's exact test), Q value = 0.00041
Table S166. Gene #36: '2q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 2Q LOSS MUTATED | 0 | 10 | 0 | 0 |
| 2Q LOSS WILD-TYPE | 168 | 145 | 59 | 117 |
Figure S166. Get High-res Image Gene #36: '2q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
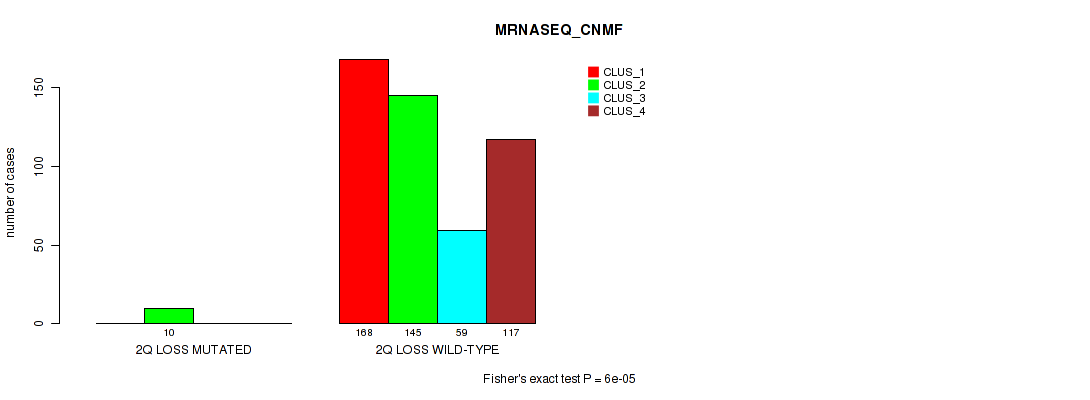
P value = 6e-05 (Fisher's exact test), Q value = 0.00041
Table S167. Gene #36: '2q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 2Q LOSS MUTATED | 0 | 10 | 0 | 0 | 0 |
| 2Q LOSS WILD-TYPE | 105 | 124 | 86 | 96 | 78 |
Figure S167. Get High-res Image Gene #36: '2q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
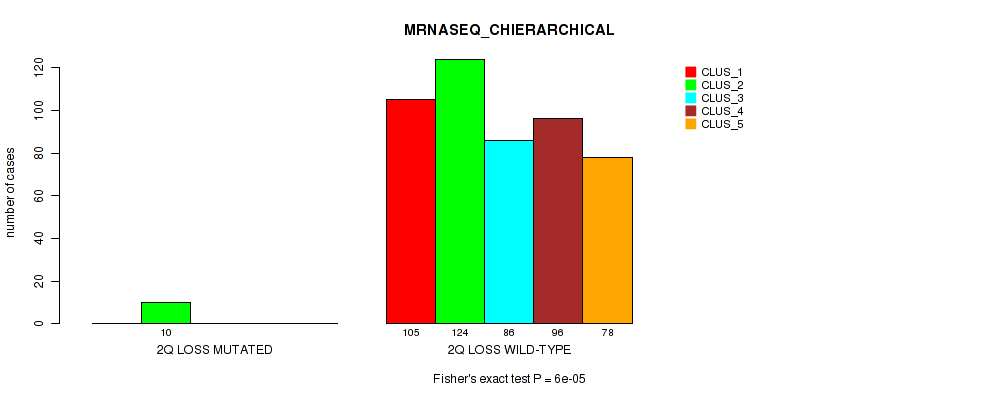
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S168. Gene #36: '2q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 2Q LOSS MUTATED | 0 | 10 | 0 |
| 2Q LOSS WILD-TYPE | 182 | 141 | 167 |
Figure S168. Get High-res Image Gene #36: '2q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S169. Gene #36: '2q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 2Q LOSS MUTATED | 0 | 10 | 0 |
| 2Q LOSS WILD-TYPE | 179 | 118 | 193 |
Figure S169. Get High-res Image Gene #36: '2q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
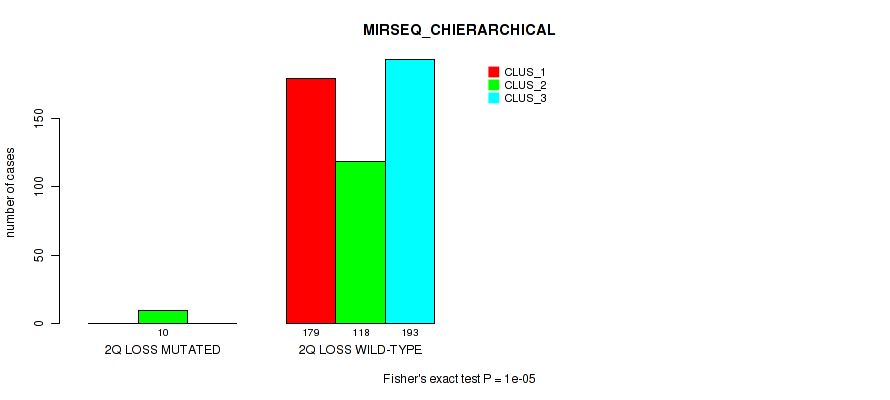
P value = 0.00056 (Fisher's exact test), Q value = 0.0026
Table S170. Gene #36: '2q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 2Q LOSS MUTATED | 1 | 7 | 0 |
| 2Q LOSS WILD-TYPE | 164 | 119 | 179 |
Figure S170. Get High-res Image Gene #36: '2q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 0.00123 (Fisher's exact test), Q value = 0.0048
Table S171. Gene #37: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 3P LOSS MUTATED | 3 | 0 | 0 |
| 3P LOSS WILD-TYPE | 53 | 356 | 89 |
Figure S171. Get High-res Image Gene #37: '3p loss' versus Molecular Subtype #1: 'CN_CNMF'
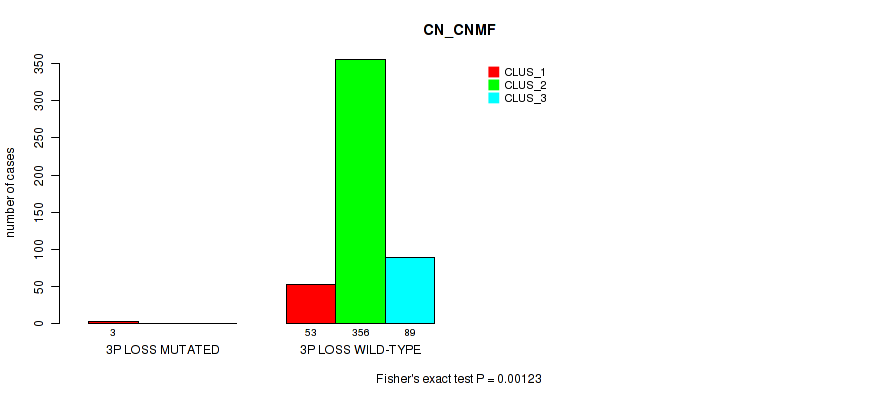
P value = 0.0494 (Fisher's exact test), Q value = 0.1
Table S172. Gene #37: '3p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 3P LOSS MUTATED | 0 | 3 | 0 |
| 3P LOSS WILD-TYPE | 277 | 151 | 70 |
Figure S172. Get High-res Image Gene #37: '3p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
P value = 0.0267 (Fisher's exact test), Q value = 0.062
Table S173. Gene #37: '3p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 3P LOSS MUTATED | 0 | 3 | 0 |
| 3P LOSS WILD-TYPE | 182 | 148 | 167 |
Figure S173. Get High-res Image Gene #37: '3p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
P value = 0.0168 (Fisher's exact test), Q value = 0.042
Table S174. Gene #37: '3p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 3P LOSS MUTATED | 0 | 3 | 0 |
| 3P LOSS WILD-TYPE | 179 | 125 | 193 |
Figure S174. Get High-res Image Gene #37: '3p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
P value = 0.00022 (Fisher's exact test), Q value = 0.0011
Table S175. Gene #38: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 3Q LOSS MUTATED | 4 | 0 | 0 |
| 3Q LOSS WILD-TYPE | 52 | 356 | 89 |
Figure S175. Get High-res Image Gene #38: '3q loss' versus Molecular Subtype #1: 'CN_CNMF'
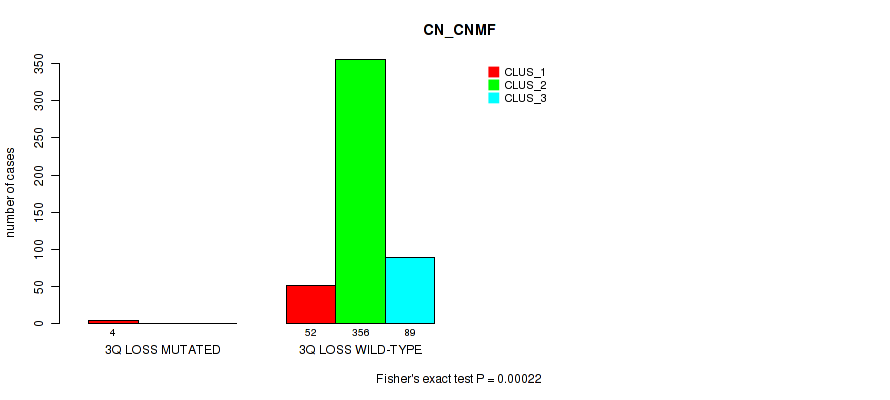
P value = 0.0174 (Fisher's exact test), Q value = 0.043
Table S176. Gene #38: '3q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 3Q LOSS MUTATED | 0 | 4 | 0 |
| 3Q LOSS WILD-TYPE | 277 | 150 | 70 |
Figure S176. Get High-res Image Gene #38: '3q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
P value = 0.0358 (Fisher's exact test), Q value = 0.081
Table S177. Gene #38: '3q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 3Q LOSS MUTATED | 0 | 4 | 0 | 0 |
| 3Q LOSS WILD-TYPE | 168 | 151 | 59 | 117 |
Figure S177. Get High-res Image Gene #38: '3q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
P value = 0.0459 (Fisher's exact test), Q value = 0.099
Table S178. Gene #38: '3q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 3Q LOSS MUTATED | 0 | 4 | 0 | 0 | 0 |
| 3Q LOSS WILD-TYPE | 105 | 130 | 86 | 96 | 78 |
Figure S178. Get High-res Image Gene #38: '3q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
P value = 0.00794 (Fisher's exact test), Q value = 0.023
Table S179. Gene #38: '3q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 3Q LOSS MUTATED | 0 | 4 | 0 |
| 3Q LOSS WILD-TYPE | 182 | 147 | 167 |
Figure S179. Get High-res Image Gene #38: '3q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
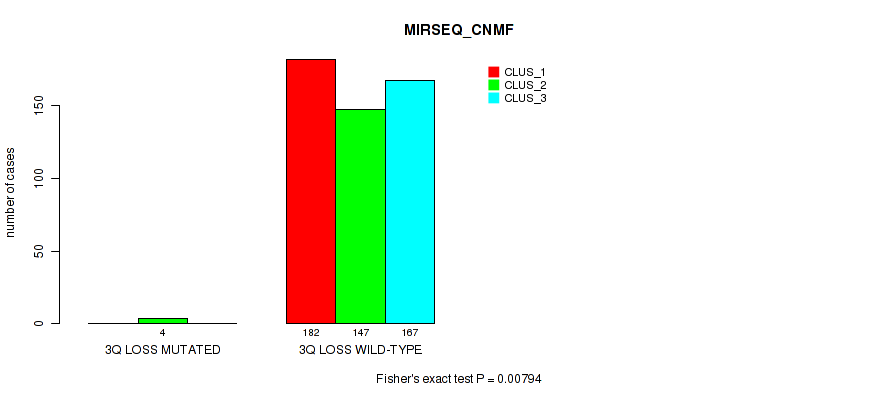
P value = 0.00394 (Fisher's exact test), Q value = 0.013
Table S180. Gene #38: '3q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 3Q LOSS MUTATED | 0 | 4 | 0 |
| 3Q LOSS WILD-TYPE | 179 | 124 | 193 |
Figure S180. Get High-res Image Gene #38: '3q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'

P value = 0.00018 (Fisher's exact test), Q value = 0.00096
Table S181. Gene #39: '6p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 6P LOSS MUTATED | 4 | 0 | 0 |
| 6P LOSS WILD-TYPE | 52 | 356 | 89 |
Figure S181. Get High-res Image Gene #39: '6p loss' versus Molecular Subtype #1: 'CN_CNMF'
P value = 0.0463 (Fisher's exact test), Q value = 0.1
Table S182. Gene #39: '6p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 6P LOSS MUTATED | 0 | 3 | 1 |
| 6P LOSS WILD-TYPE | 277 | 151 | 69 |
Figure S182. Get High-res Image Gene #39: '6p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
P value = 4e-05 (Fisher's exact test), Q value = 0.00029
Table S183. Gene #40: '6q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 6Q LOSS MUTATED | 6 | 1 | 0 |
| 6Q LOSS WILD-TYPE | 50 | 355 | 89 |
Figure S183. Get High-res Image Gene #40: '6q loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00234 (Fisher's exact test), Q value = 0.0083
Table S184. Gene #40: '6q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 6Q LOSS MUTATED | 0 | 6 | 1 |
| 6Q LOSS WILD-TYPE | 277 | 148 | 69 |
Figure S184. Get High-res Image Gene #40: '6q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
P value = 0.0124 (Fisher's exact test), Q value = 0.034
Table S185. Gene #40: '6q loss' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 61 | 79 | 81 |
| 6Q LOSS MUTATED | 0 | 0 | 5 |
| 6Q LOSS WILD-TYPE | 61 | 79 | 76 |
Figure S185. Get High-res Image Gene #40: '6q loss' versus Molecular Subtype #3: 'RPPA_CNMF'
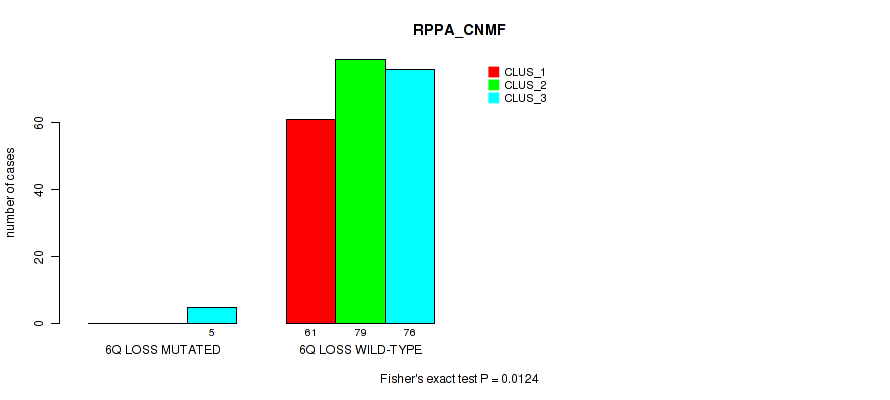
P value = 0.028 (Fisher's exact test), Q value = 0.064
Table S186. Gene #40: '6q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 6Q LOSS MUTATED | 1 | 6 | 0 | 0 |
| 6Q LOSS WILD-TYPE | 167 | 149 | 59 | 117 |
Figure S186. Get High-res Image Gene #40: '6q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
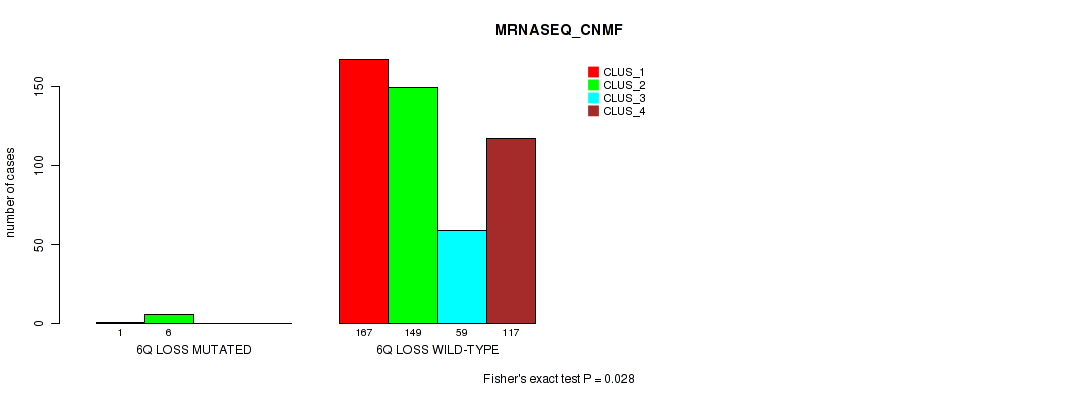
P value = 0.018 (Fisher's exact test), Q value = 0.044
Table S187. Gene #40: '6q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 6Q LOSS MUTATED | 1 | 6 | 0 | 0 | 0 |
| 6Q LOSS WILD-TYPE | 104 | 128 | 86 | 96 | 78 |
Figure S187. Get High-res Image Gene #40: '6q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
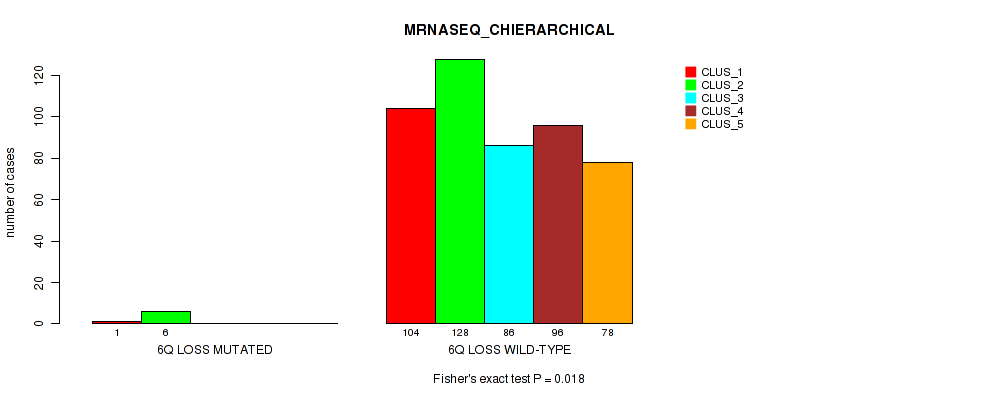
P value = 0.00489 (Fisher's exact test), Q value = 0.016
Table S188. Gene #40: '6q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 6Q LOSS MUTATED | 1 | 6 | 0 |
| 6Q LOSS WILD-TYPE | 181 | 145 | 167 |
Figure S188. Get High-res Image Gene #40: '6q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
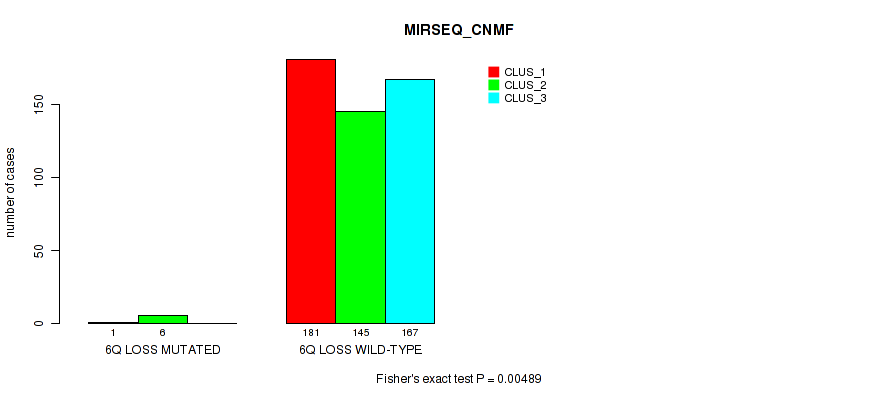
P value = 0.00064 (Fisher's exact test), Q value = 0.0028
Table S189. Gene #40: '6q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 6Q LOSS MUTATED | 1 | 6 | 0 |
| 6Q LOSS WILD-TYPE | 178 | 122 | 193 |
Figure S189. Get High-res Image Gene #40: '6q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
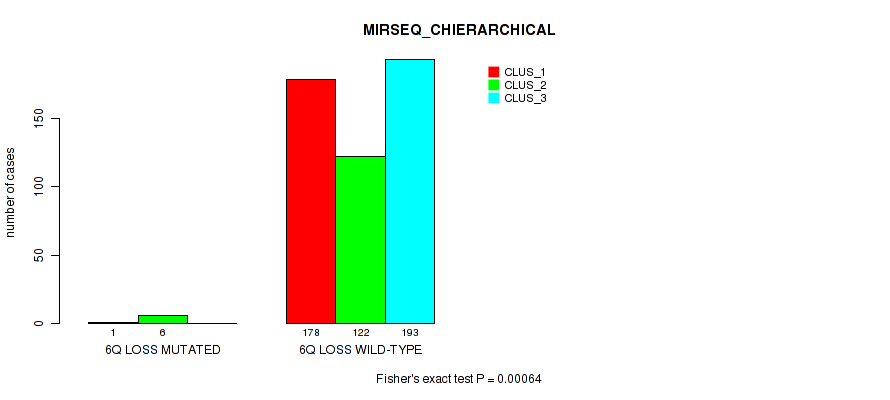
P value = 0.00979 (Fisher's exact test), Q value = 0.028
Table S190. Gene #40: '6q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 6Q LOSS MUTATED | 2 | 5 | 0 |
| 6Q LOSS WILD-TYPE | 163 | 121 | 179 |
Figure S190. Get High-res Image Gene #40: '6q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
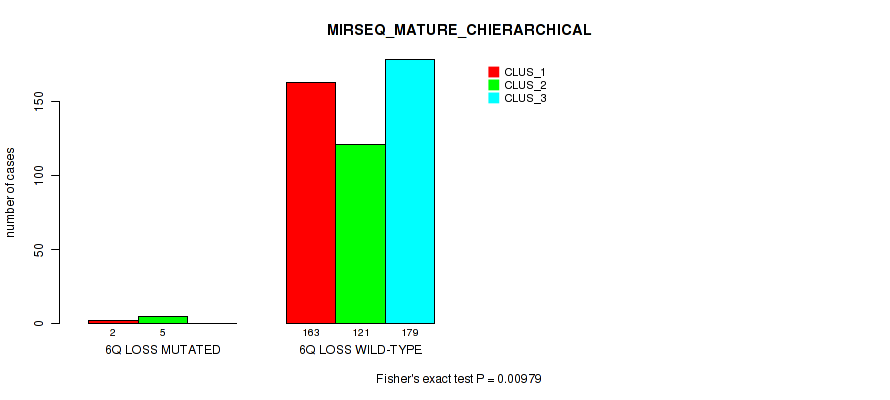
P value = 1e-04 (Fisher's exact test), Q value = 0.00063
Table S191. Gene #41: '8p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 8P LOSS MUTATED | 4 | 0 | 0 |
| 8P LOSS WILD-TYPE | 52 | 356 | 89 |
Figure S191. Get High-res Image Gene #41: '8p loss' versus Molecular Subtype #1: 'CN_CNMF'
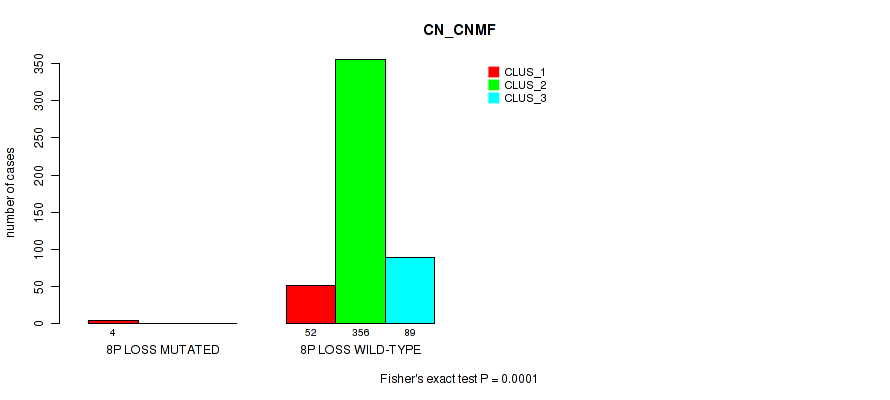
P value = 0.0173 (Fisher's exact test), Q value = 0.043
Table S192. Gene #41: '8p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 8P LOSS MUTATED | 0 | 4 | 0 |
| 8P LOSS WILD-TYPE | 277 | 150 | 70 |
Figure S192. Get High-res Image Gene #41: '8p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
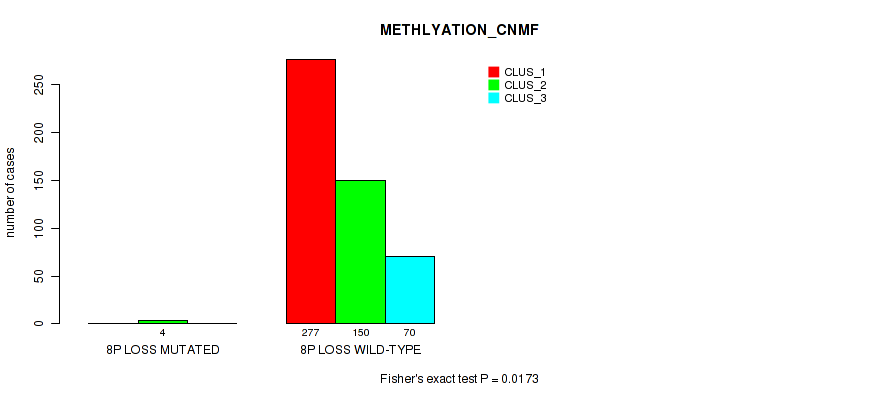
P value = 0.0364 (Fisher's exact test), Q value = 0.081
Table S193. Gene #41: '8p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 8P LOSS MUTATED | 0 | 4 | 0 | 0 |
| 8P LOSS WILD-TYPE | 168 | 151 | 59 | 117 |
Figure S193. Get High-res Image Gene #41: '8p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
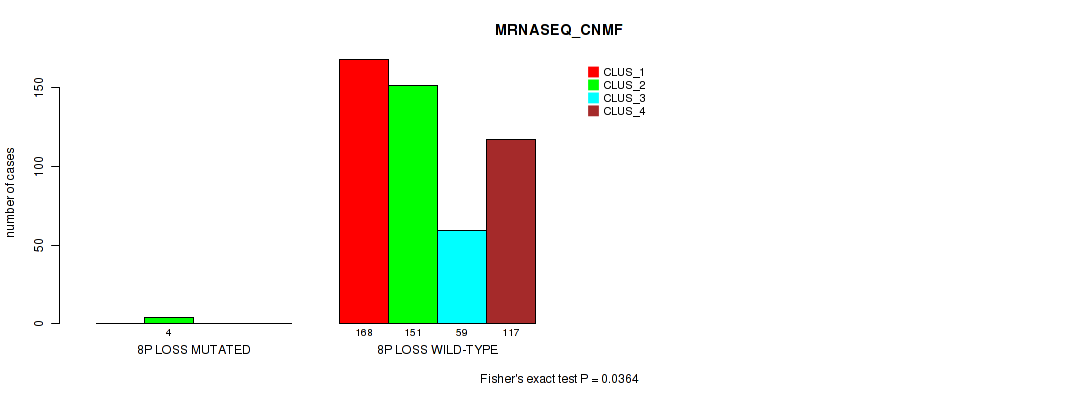
P value = 0.0494 (Fisher's exact test), Q value = 0.1
Table S194. Gene #41: '8p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 8P LOSS MUTATED | 0 | 4 | 0 | 0 | 0 |
| 8P LOSS WILD-TYPE | 105 | 130 | 86 | 96 | 78 |
Figure S194. Get High-res Image Gene #41: '8p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
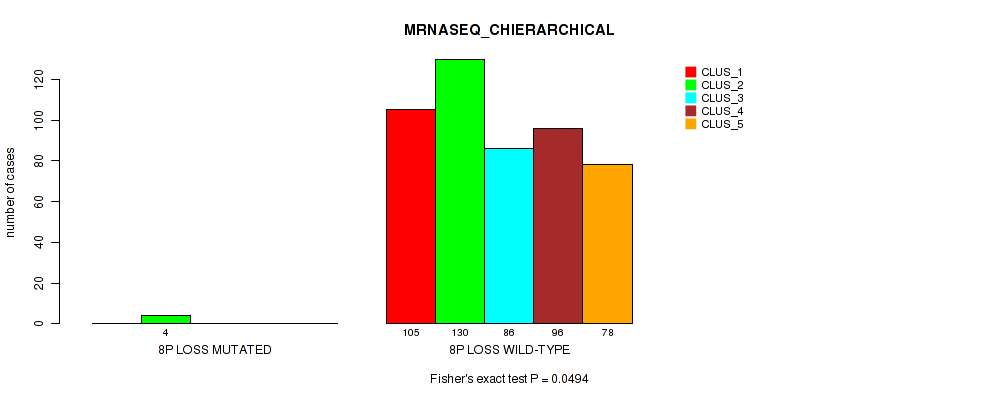
P value = 0.00777 (Fisher's exact test), Q value = 0.023
Table S195. Gene #41: '8p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 8P LOSS MUTATED | 0 | 4 | 0 |
| 8P LOSS WILD-TYPE | 182 | 147 | 167 |
Figure S195. Get High-res Image Gene #41: '8p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
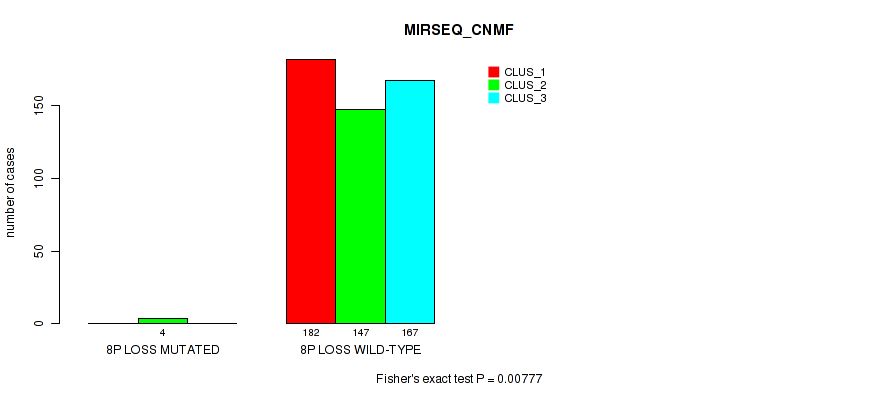
P value = 0.00422 (Fisher's exact test), Q value = 0.014
Table S196. Gene #41: '8p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 8P LOSS MUTATED | 0 | 4 | 0 |
| 8P LOSS WILD-TYPE | 179 | 124 | 193 |
Figure S196. Get High-res Image Gene #41: '8p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
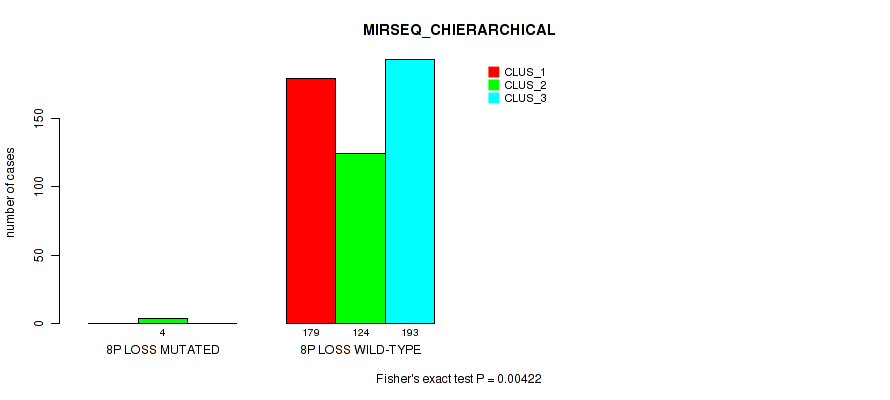
P value = 0.0189 (Fisher's exact test), Q value = 0.045
Table S197. Gene #41: '8p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 8P LOSS MUTATED | 0 | 3 | 0 |
| 8P LOSS WILD-TYPE | 165 | 123 | 179 |
Figure S197. Get High-res Image Gene #41: '8p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
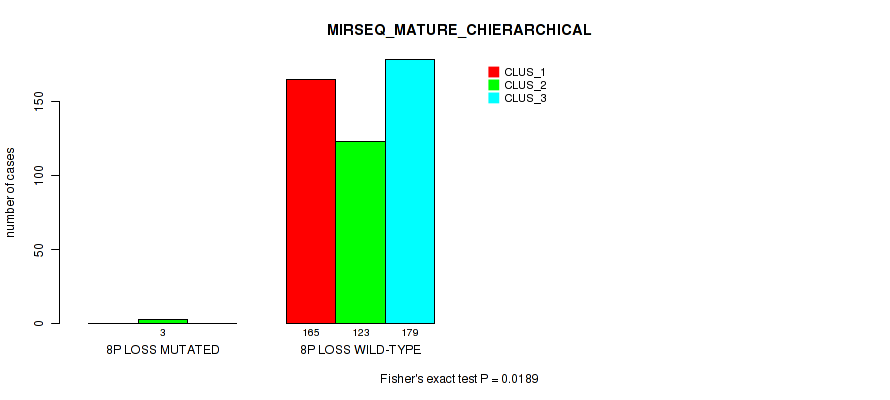
P value = 0.00013 (Fisher's exact test), Q value = 0.00074
Table S198. Gene #42: '8q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 8Q LOSS MUTATED | 4 | 0 | 0 |
| 8Q LOSS WILD-TYPE | 52 | 356 | 89 |
Figure S198. Get High-res Image Gene #42: '8q loss' versus Molecular Subtype #1: 'CN_CNMF'
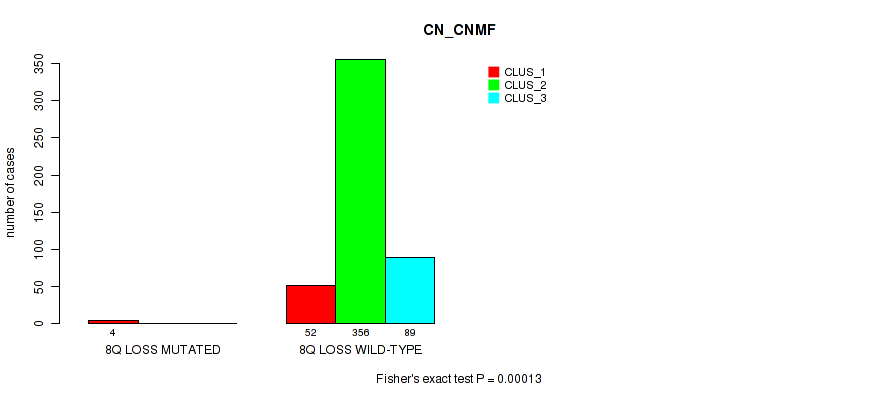
P value = 0.0174 (Fisher's exact test), Q value = 0.043
Table S199. Gene #42: '8q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 8Q LOSS MUTATED | 0 | 4 | 0 |
| 8Q LOSS WILD-TYPE | 277 | 150 | 70 |
Figure S199. Get High-res Image Gene #42: '8q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
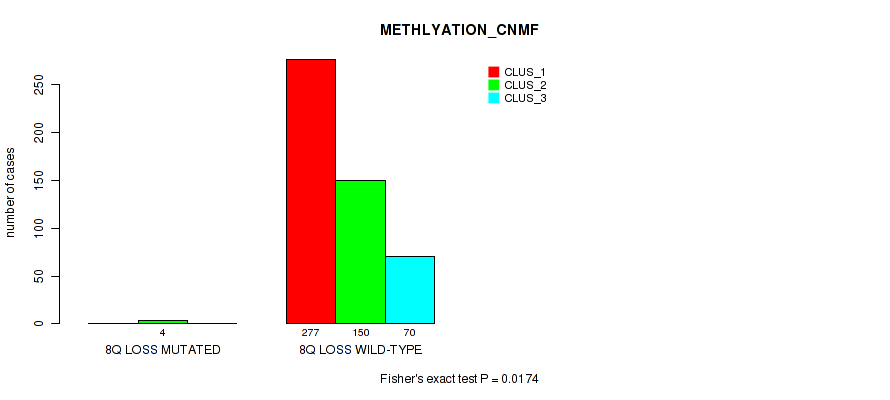
P value = 0.0362 (Fisher's exact test), Q value = 0.081
Table S200. Gene #42: '8q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 8Q LOSS MUTATED | 0 | 4 | 0 | 0 |
| 8Q LOSS WILD-TYPE | 168 | 151 | 59 | 117 |
Figure S200. Get High-res Image Gene #42: '8q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
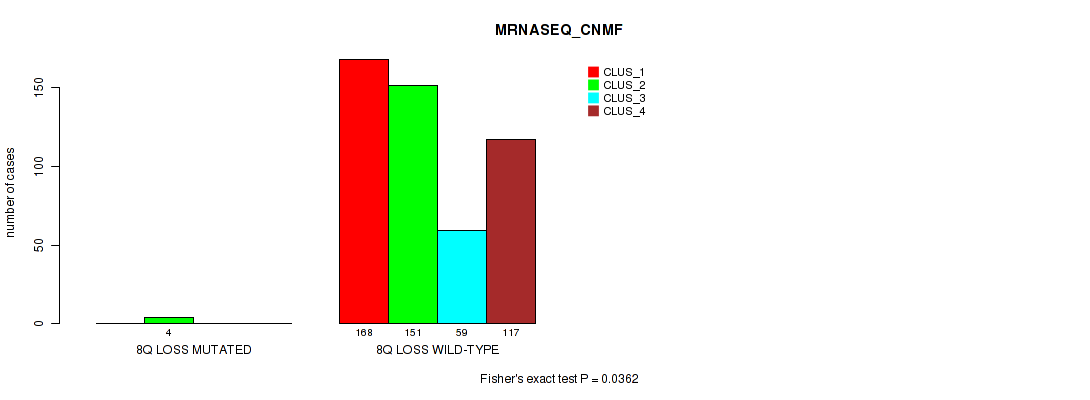
P value = 0.0458 (Fisher's exact test), Q value = 0.099
Table S201. Gene #42: '8q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 8Q LOSS MUTATED | 0 | 4 | 0 | 0 | 0 |
| 8Q LOSS WILD-TYPE | 105 | 130 | 86 | 96 | 78 |
Figure S201. Get High-res Image Gene #42: '8q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
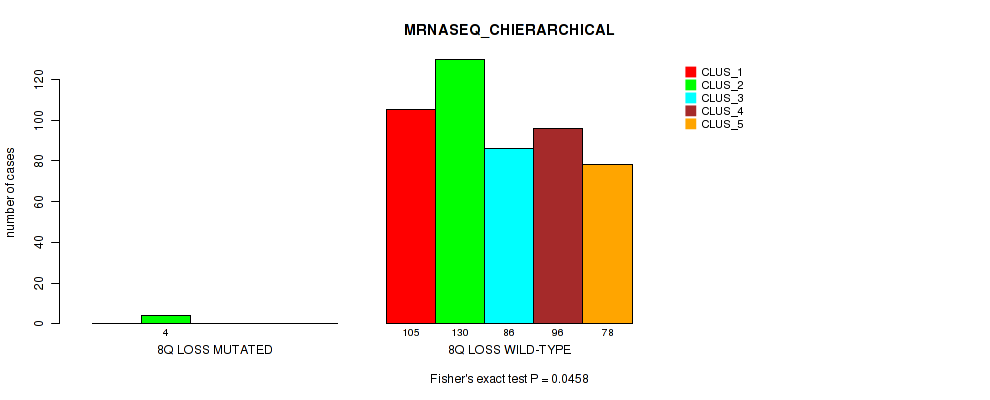
P value = 0.00774 (Fisher's exact test), Q value = 0.023
Table S202. Gene #42: '8q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 8Q LOSS MUTATED | 0 | 4 | 0 |
| 8Q LOSS WILD-TYPE | 182 | 147 | 167 |
Figure S202. Get High-res Image Gene #42: '8q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
P value = 0.00421 (Fisher's exact test), Q value = 0.014
Table S203. Gene #42: '8q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 8Q LOSS MUTATED | 0 | 4 | 0 |
| 8Q LOSS WILD-TYPE | 179 | 124 | 193 |
Figure S203. Get High-res Image Gene #42: '8q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
P value = 0.0187 (Fisher's exact test), Q value = 0.045
Table S204. Gene #42: '8q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 8Q LOSS MUTATED | 0 | 3 | 0 |
| 8Q LOSS WILD-TYPE | 165 | 123 | 179 |
Figure S204. Get High-res Image Gene #42: '8q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S205. Gene #43: '9p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 9P LOSS MUTATED | 15 | 2 | 2 |
| 9P LOSS WILD-TYPE | 41 | 354 | 87 |
Figure S205. Get High-res Image Gene #43: '9p loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.0379 (Fisher's exact test), Q value = 0.084
Table S206. Gene #43: '9p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 9P LOSS MUTATED | 6 | 11 | 2 |
| 9P LOSS WILD-TYPE | 271 | 143 | 68 |
Figure S206. Get High-res Image Gene #43: '9p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
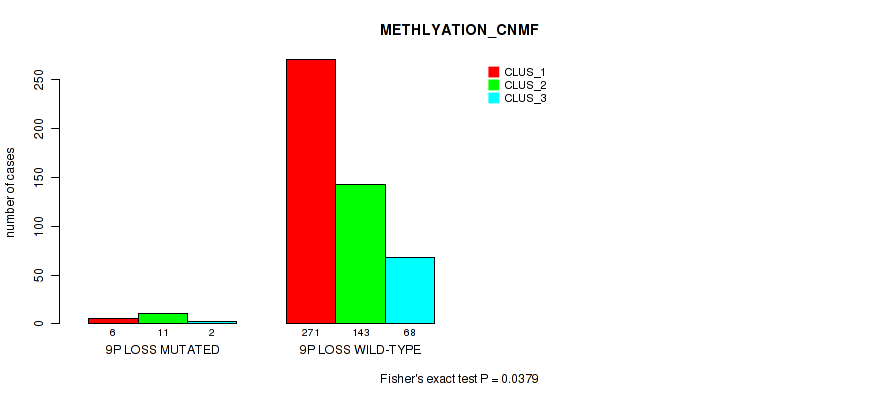
P value = 0.031 (Fisher's exact test), Q value = 0.07
Table S207. Gene #43: '9p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 9P LOSS MUTATED | 2 | 11 | 3 | 3 |
| 9P LOSS WILD-TYPE | 166 | 144 | 56 | 114 |
Figure S207. Get High-res Image Gene #43: '9p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
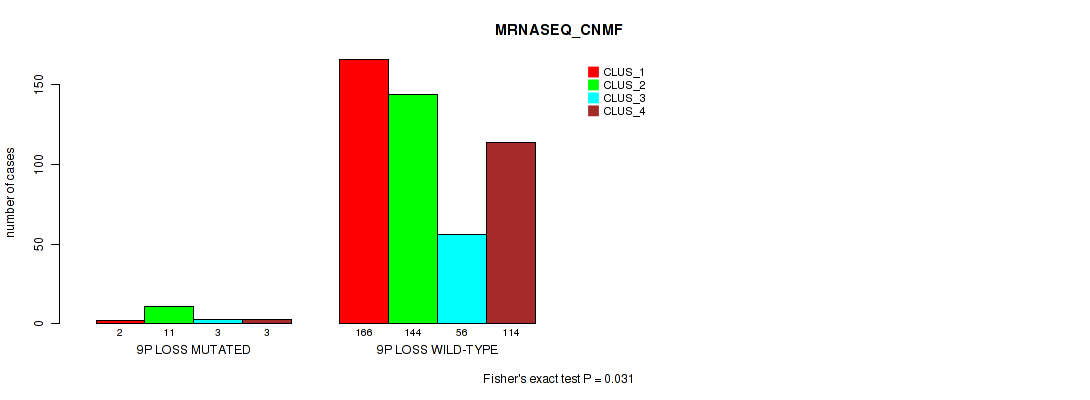
P value = 0.0087 (Fisher's exact test), Q value = 0.025
Table S208. Gene #43: '9p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 9P LOSS MUTATED | 8 | 10 | 1 |
| 9P LOSS WILD-TYPE | 174 | 141 | 166 |
Figure S208. Get High-res Image Gene #43: '9p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'

P value = 0.00712 (Fisher's exact test), Q value = 0.021
Table S209. Gene #43: '9p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 9P LOSS MUTATED | 7 | 10 | 2 |
| 9P LOSS WILD-TYPE | 172 | 118 | 191 |
Figure S209. Get High-res Image Gene #43: '9p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
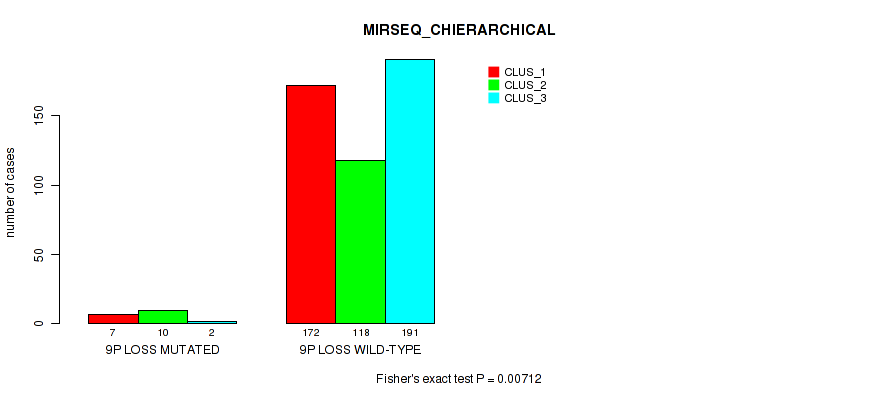
P value = 0.00422 (Fisher's exact test), Q value = 0.014
Table S210. Gene #43: '9p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 9P LOSS MUTATED | 7 | 9 | 1 |
| 9P LOSS WILD-TYPE | 158 | 117 | 178 |
Figure S210. Get High-res Image Gene #43: '9p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
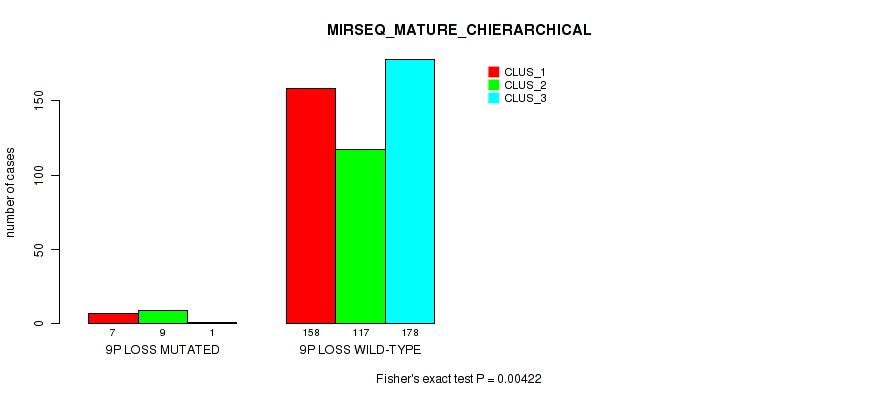
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S211. Gene #44: '9q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 9Q LOSS MUTATED | 20 | 2 | 3 |
| 9Q LOSS WILD-TYPE | 36 | 354 | 86 |
Figure S211. Get High-res Image Gene #44: '9q loss' versus Molecular Subtype #1: 'CN_CNMF'
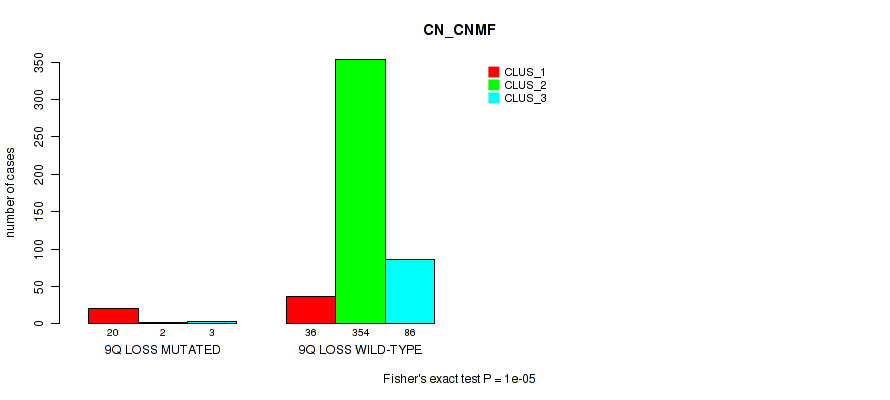
P value = 0.00933 (Fisher's exact test), Q value = 0.027
Table S212. Gene #44: '9q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 9Q LOSS MUTATED | 12 | 11 | 2 |
| 9Q LOSS WILD-TYPE | 170 | 140 | 165 |
Figure S212. Get High-res Image Gene #44: '9q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
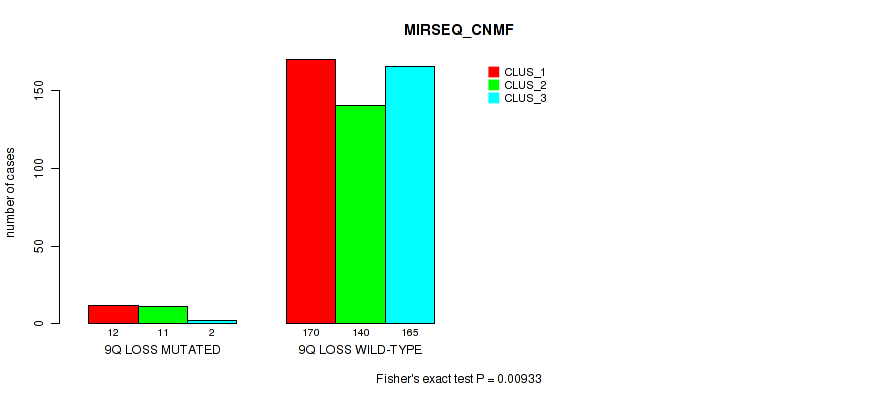
P value = 0.00704 (Fisher's exact test), Q value = 0.021
Table S213. Gene #44: '9q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 9Q LOSS MUTATED | 11 | 11 | 3 |
| 9Q LOSS WILD-TYPE | 168 | 117 | 190 |
Figure S213. Get High-res Image Gene #44: '9q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
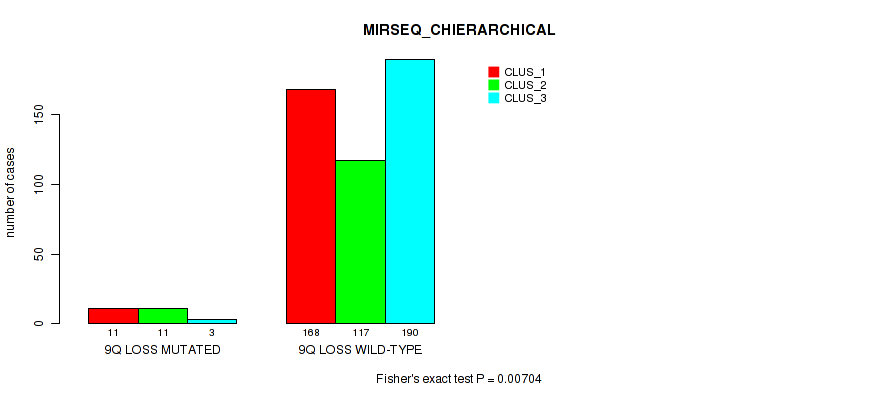
P value = 0.00488 (Fisher's exact test), Q value = 0.016
Table S214. Gene #44: '9q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 9Q LOSS MUTATED | 11 | 10 | 2 |
| 9Q LOSS WILD-TYPE | 154 | 116 | 177 |
Figure S214. Get High-res Image Gene #44: '9q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
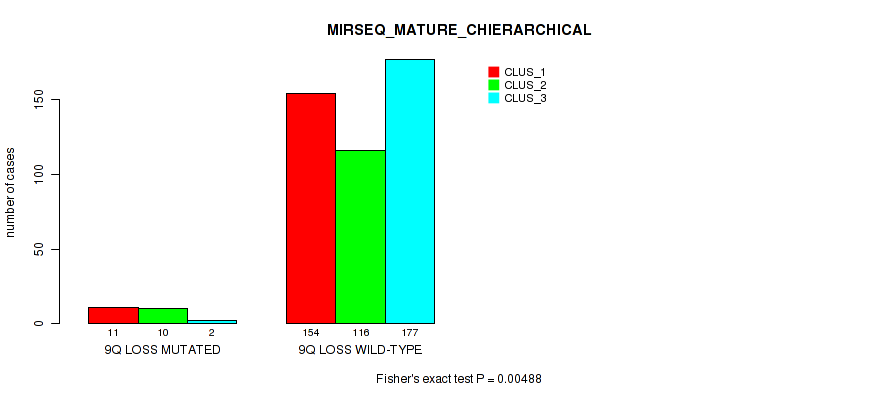
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S215. Gene #45: '10p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 10P LOSS MUTATED | 7 | 0 | 0 |
| 10P LOSS WILD-TYPE | 49 | 356 | 89 |
Figure S215. Get High-res Image Gene #45: '10p loss' versus Molecular Subtype #1: 'CN_CNMF'
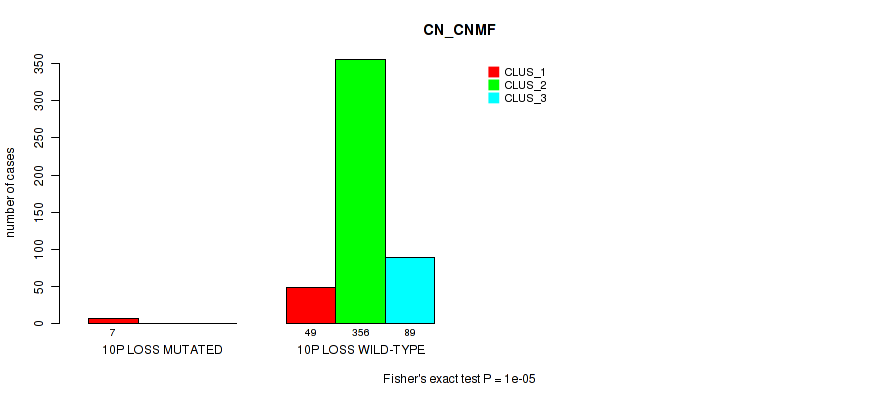
P value = 0.0116 (Fisher's exact test), Q value = 0.032
Table S216. Gene #45: '10p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 10P LOSS MUTATED | 1 | 6 | 0 |
| 10P LOSS WILD-TYPE | 276 | 148 | 70 |
Figure S216. Get High-res Image Gene #45: '10p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
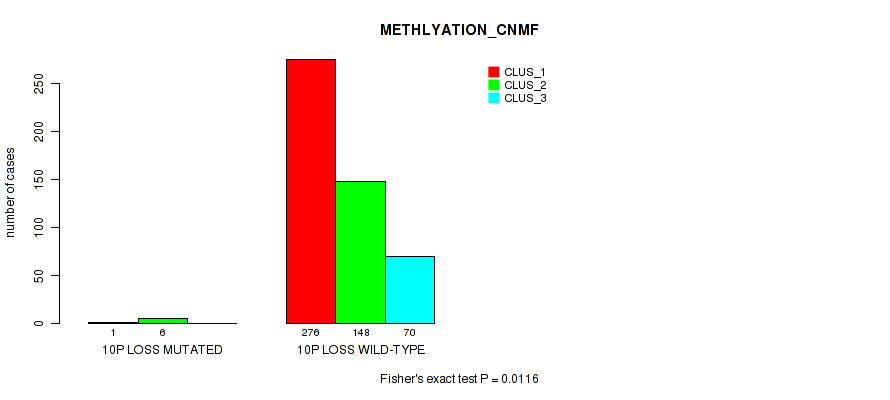
P value = 0.0206 (Fisher's exact test), Q value = 0.049
Table S217. Gene #45: '10p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 10P LOSS MUTATED | 0 | 6 | 0 | 1 |
| 10P LOSS WILD-TYPE | 168 | 149 | 59 | 116 |
Figure S217. Get High-res Image Gene #45: '10p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
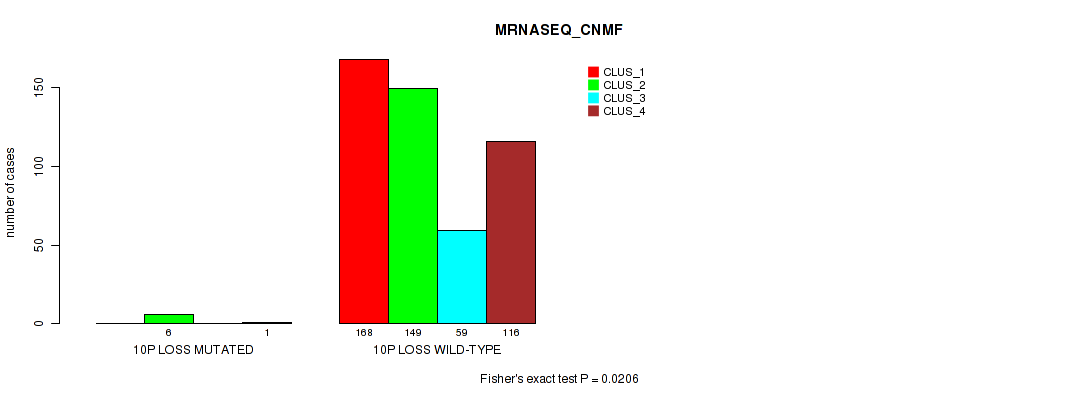
P value = 0.0154 (Fisher's exact test), Q value = 0.04
Table S218. Gene #45: '10p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 10P LOSS MUTATED | 0 | 6 | 0 | 1 | 0 |
| 10P LOSS WILD-TYPE | 105 | 128 | 86 | 95 | 78 |
Figure S218. Get High-res Image Gene #45: '10p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
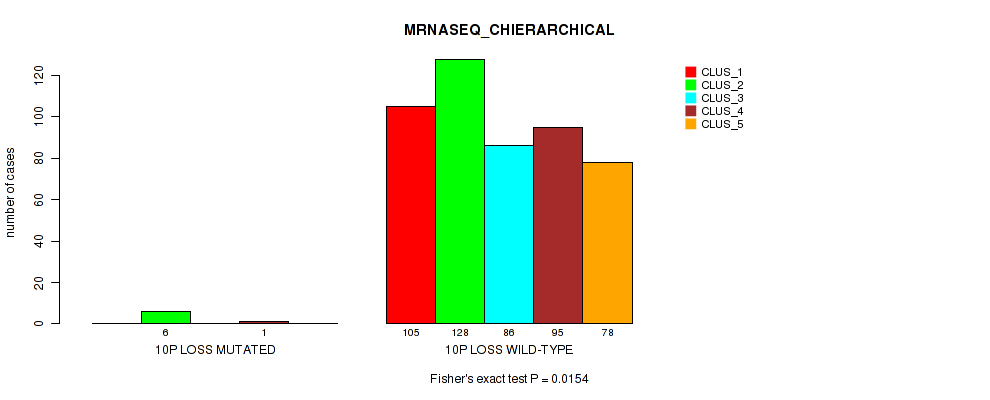
P value = 0.00474 (Fisher's exact test), Q value = 0.015
Table S219. Gene #45: '10p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 10P LOSS MUTATED | 1 | 6 | 0 |
| 10P LOSS WILD-TYPE | 181 | 145 | 167 |
Figure S219. Get High-res Image Gene #45: '10p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
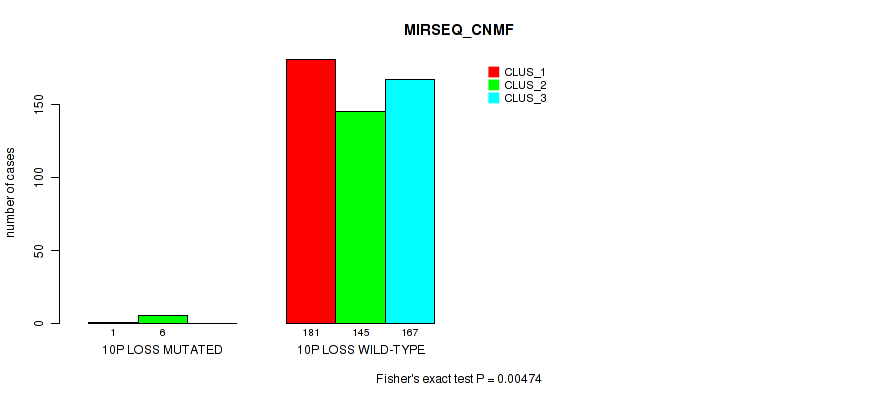
P value = 0.00063 (Fisher's exact test), Q value = 0.0028
Table S220. Gene #45: '10p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 10P LOSS MUTATED | 1 | 6 | 0 |
| 10P LOSS WILD-TYPE | 178 | 122 | 193 |
Figure S220. Get High-res Image Gene #45: '10p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
P value = 0.0112 (Fisher's exact test), Q value = 0.031
Table S221. Gene #45: '10p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 10P LOSS MUTATED | 2 | 5 | 0 |
| 10P LOSS WILD-TYPE | 163 | 121 | 179 |
Figure S221. Get High-res Image Gene #45: '10p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S222. Gene #46: '10q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 10Q LOSS MUTATED | 6 | 0 | 0 |
| 10Q LOSS WILD-TYPE | 50 | 356 | 89 |
Figure S222. Get High-res Image Gene #46: '10q loss' versus Molecular Subtype #1: 'CN_CNMF'
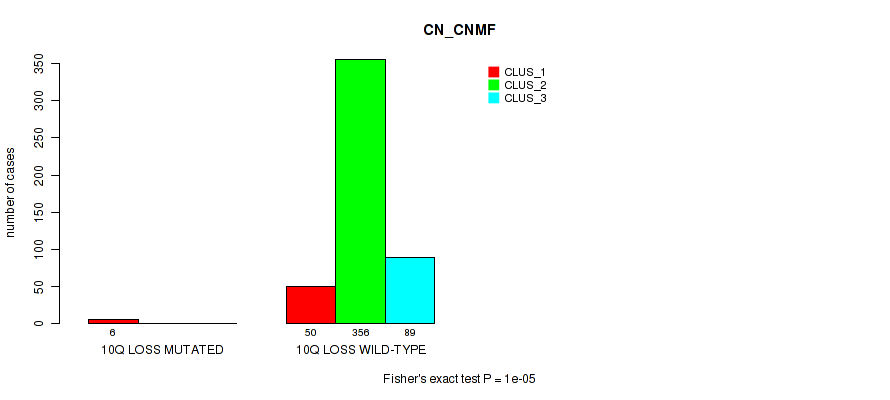
P value = 0.0292 (Fisher's exact test), Q value = 0.066
Table S223. Gene #46: '10q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 10Q LOSS MUTATED | 1 | 5 | 0 |
| 10Q LOSS WILD-TYPE | 276 | 149 | 70 |
Figure S223. Get High-res Image Gene #46: '10q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
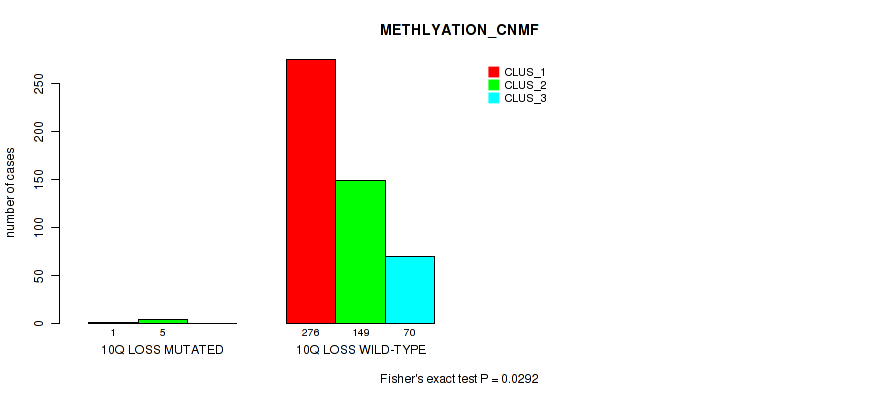
P value = 0.0456 (Fisher's exact test), Q value = 0.099
Table S224. Gene #46: '10q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 10Q LOSS MUTATED | 0 | 5 | 0 | 1 | 0 |
| 10Q LOSS WILD-TYPE | 105 | 129 | 86 | 95 | 78 |
Figure S224. Get High-res Image Gene #46: '10q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
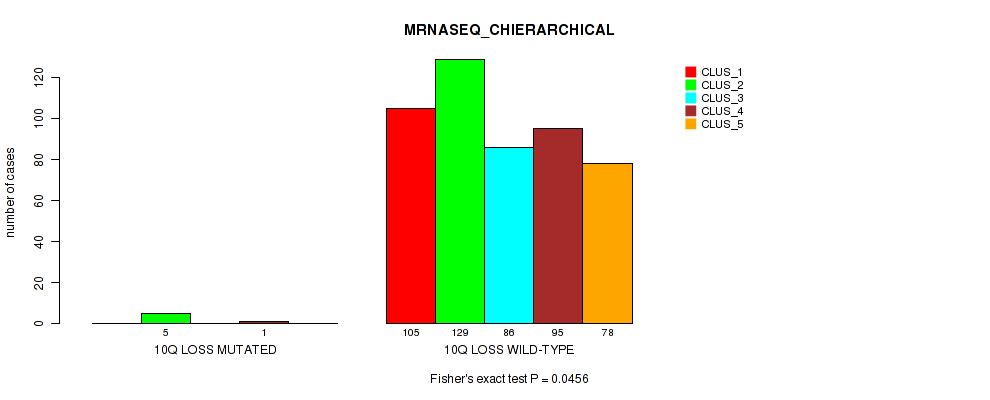
P value = 0.0144 (Fisher's exact test), Q value = 0.038
Table S225. Gene #46: '10q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 10Q LOSS MUTATED | 1 | 5 | 0 |
| 10Q LOSS WILD-TYPE | 181 | 146 | 167 |
Figure S225. Get High-res Image Gene #46: '10q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
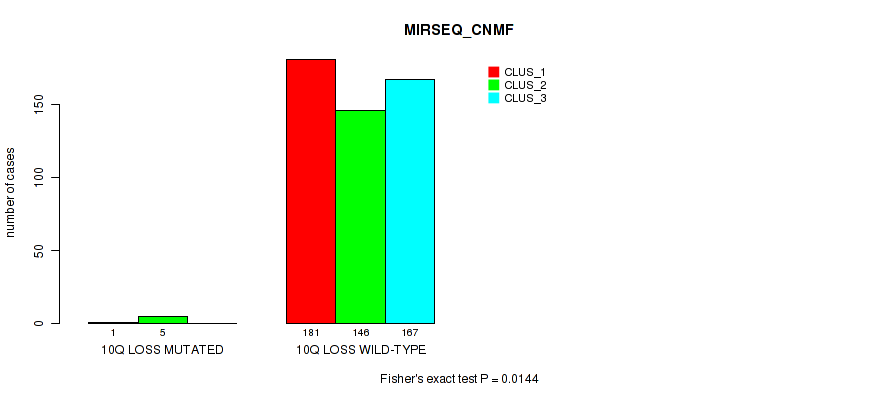
P value = 0.00445 (Fisher's exact test), Q value = 0.014
Table S226. Gene #46: '10q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 10Q LOSS MUTATED | 1 | 5 | 0 |
| 10Q LOSS WILD-TYPE | 178 | 123 | 193 |
Figure S226. Get High-res Image Gene #46: '10q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'

P value = 0.028 (Fisher's exact test), Q value = 0.064
Table S227. Gene #46: '10q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 10Q LOSS MUTATED | 2 | 4 | 0 |
| 10Q LOSS WILD-TYPE | 163 | 122 | 179 |
Figure S227. Get High-res Image Gene #46: '10q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S228. Gene #47: '11p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 11P LOSS MUTATED | 6 | 0 | 1 |
| 11P LOSS WILD-TYPE | 50 | 356 | 88 |
Figure S228. Get High-res Image Gene #47: '11p loss' versus Molecular Subtype #1: 'CN_CNMF'
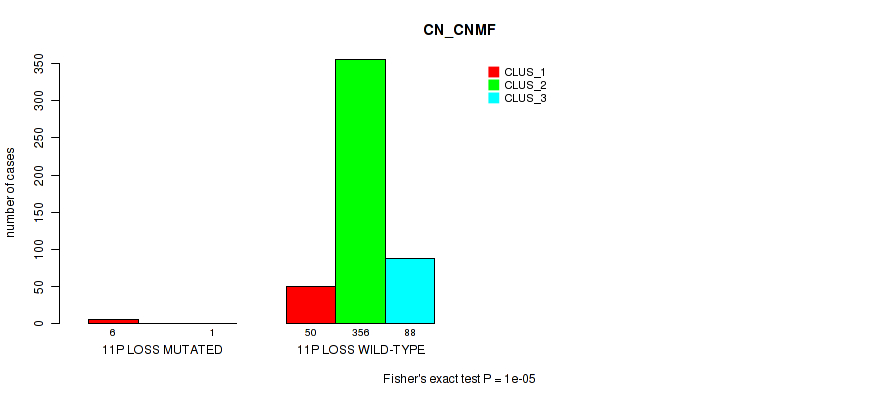
P value = 0.011 (Fisher's exact test), Q value = 0.031
Table S229. Gene #47: '11p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 11P LOSS MUTATED | 1 | 6 | 0 |
| 11P LOSS WILD-TYPE | 276 | 148 | 70 |
Figure S229. Get High-res Image Gene #47: '11p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
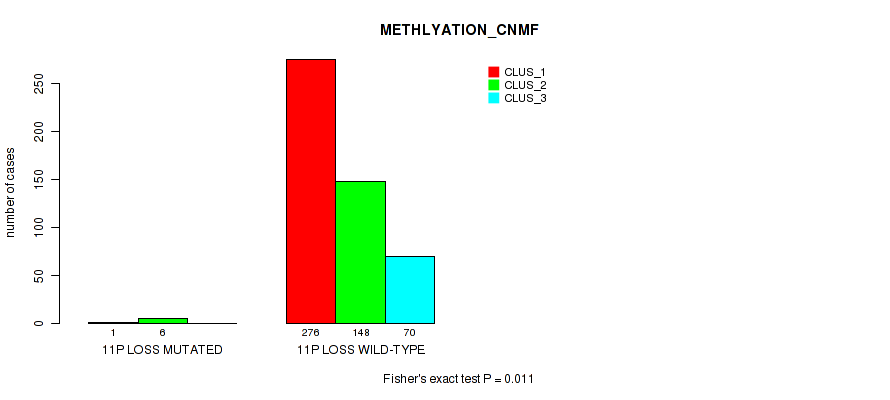
P value = 0.00648 (Fisher's exact test), Q value = 0.02
Table S230. Gene #47: '11p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 11P LOSS MUTATED | 0 | 6 | 1 | 0 |
| 11P LOSS WILD-TYPE | 168 | 149 | 58 | 117 |
Figure S230. Get High-res Image Gene #47: '11p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
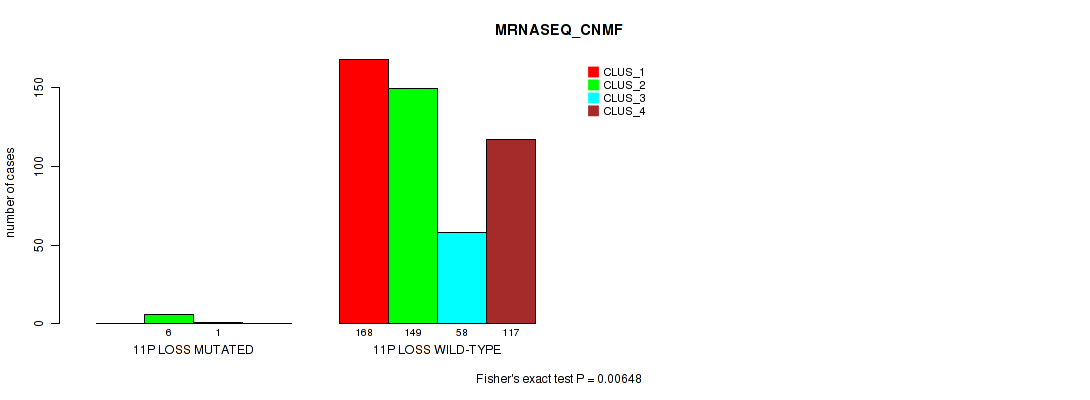
P value = 0.0175 (Fisher's exact test), Q value = 0.043
Table S231. Gene #47: '11p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 11P LOSS MUTATED | 1 | 6 | 0 | 0 | 0 |
| 11P LOSS WILD-TYPE | 104 | 128 | 86 | 96 | 78 |
Figure S231. Get High-res Image Gene #47: '11p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
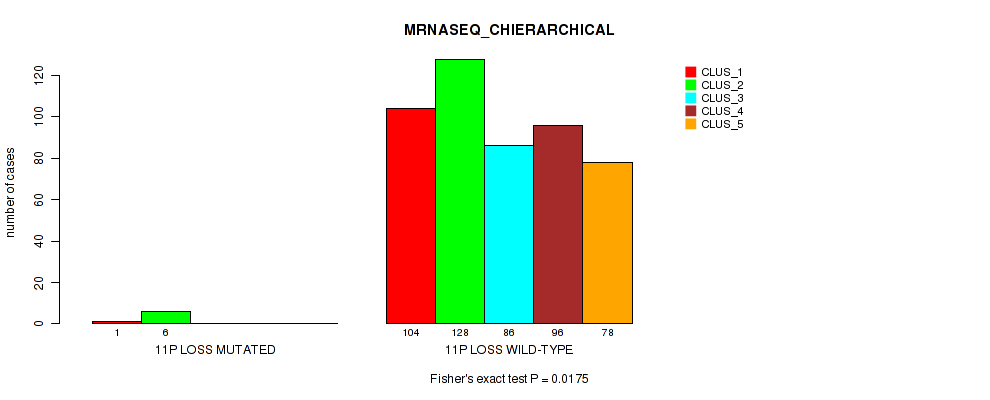
P value = 0.00494 (Fisher's exact test), Q value = 0.016
Table S232. Gene #47: '11p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 11P LOSS MUTATED | 1 | 6 | 0 |
| 11P LOSS WILD-TYPE | 181 | 145 | 167 |
Figure S232. Get High-res Image Gene #47: '11p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
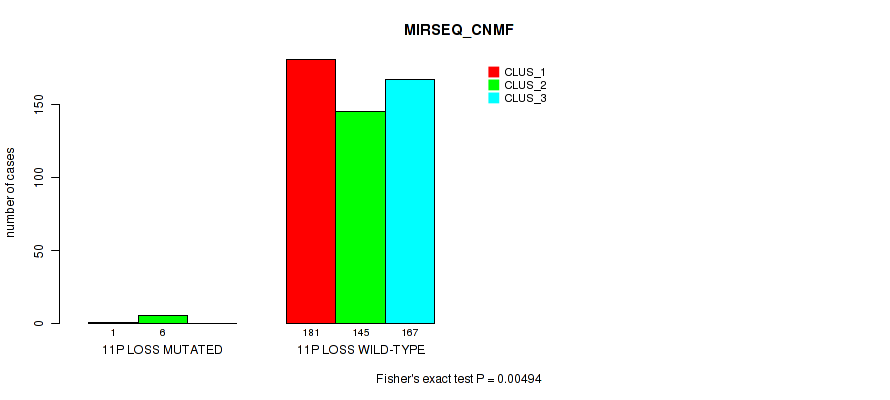
P value = 0.00059 (Fisher's exact test), Q value = 0.0026
Table S233. Gene #47: '11p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 11P LOSS MUTATED | 1 | 6 | 0 |
| 11P LOSS WILD-TYPE | 178 | 122 | 193 |
Figure S233. Get High-res Image Gene #47: '11p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'

P value = 0.0107 (Fisher's exact test), Q value = 0.03
Table S234. Gene #47: '11p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 11P LOSS MUTATED | 2 | 5 | 0 |
| 11P LOSS WILD-TYPE | 163 | 121 | 179 |
Figure S234. Get High-res Image Gene #47: '11p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
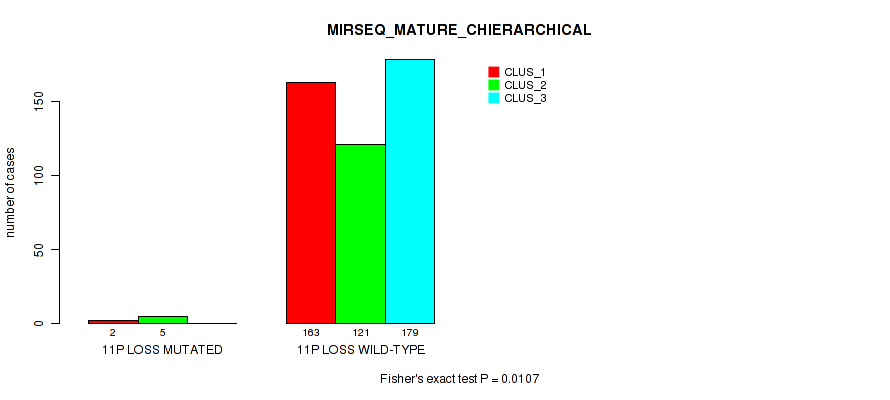
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S235. Gene #48: '11q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 11Q LOSS MUTATED | 7 | 1 | 1 |
| 11Q LOSS WILD-TYPE | 49 | 355 | 88 |
Figure S235. Get High-res Image Gene #48: '11q loss' versus Molecular Subtype #1: 'CN_CNMF'
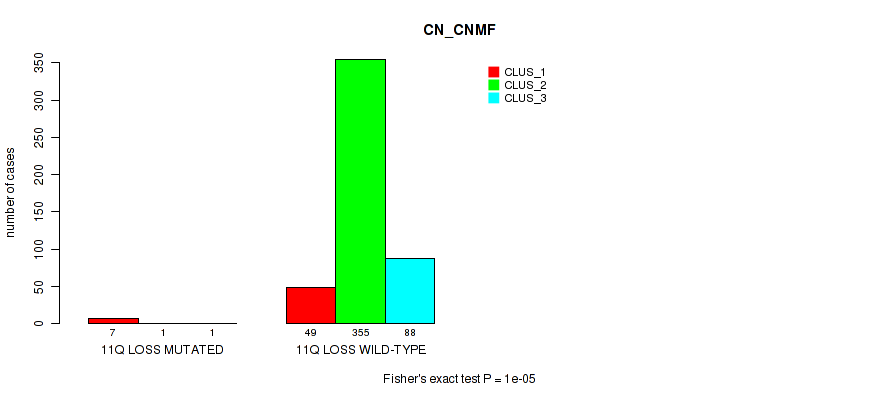
P value = 0.0173 (Fisher's exact test), Q value = 0.043
Table S236. Gene #48: '11q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 11Q LOSS MUTATED | 2 | 7 | 0 |
| 11Q LOSS WILD-TYPE | 275 | 147 | 70 |
Figure S236. Get High-res Image Gene #48: '11q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
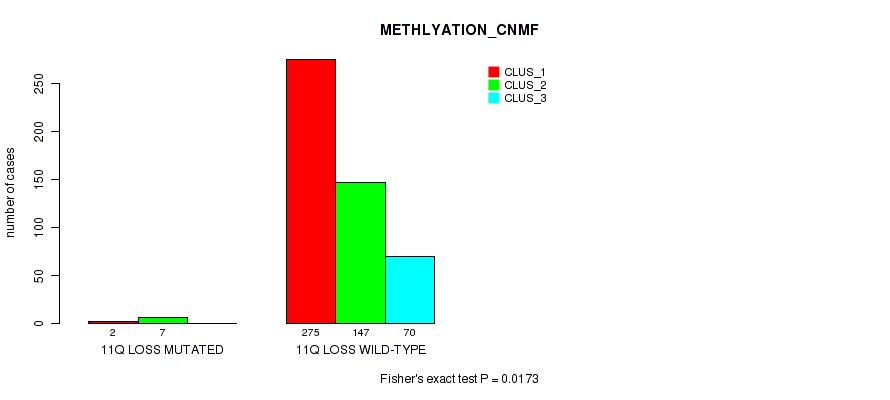
P value = 0.0158 (Fisher's exact test), Q value = 0.041
Table S237. Gene #48: '11q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 11Q LOSS MUTATED | 1 | 7 | 1 | 0 |
| 11Q LOSS WILD-TYPE | 167 | 148 | 58 | 117 |
Figure S237. Get High-res Image Gene #48: '11q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
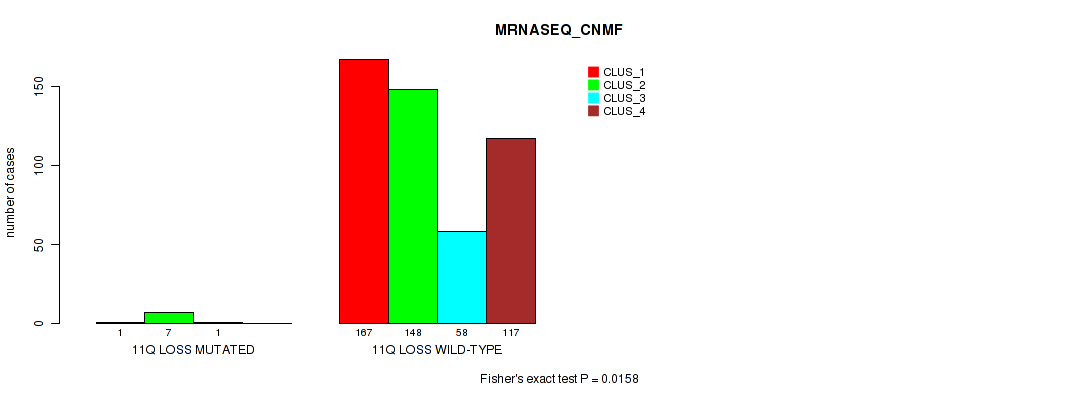
P value = 0.0184 (Fisher's exact test), Q value = 0.045
Table S238. Gene #48: '11q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 11Q LOSS MUTATED | 1 | 7 | 0 | 0 | 1 |
| 11Q LOSS WILD-TYPE | 104 | 127 | 86 | 96 | 77 |
Figure S238. Get High-res Image Gene #48: '11q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
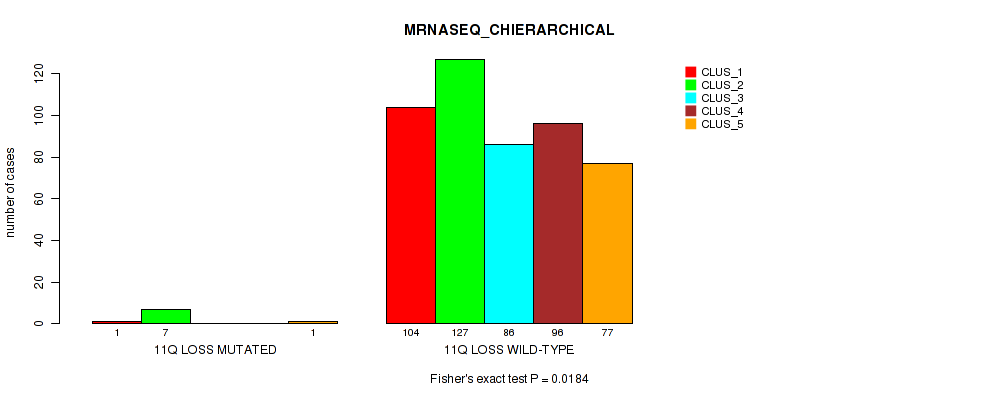
P value = 0.00057 (Fisher's exact test), Q value = 0.0026
Table S239. Gene #48: '11q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 11Q LOSS MUTATED | 1 | 8 | 0 |
| 11Q LOSS WILD-TYPE | 181 | 143 | 167 |
Figure S239. Get High-res Image Gene #48: '11q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
P value = 0.00247 (Fisher's exact test), Q value = 0.0086
Table S240. Gene #48: '11q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 11Q LOSS MUTATED | 1 | 7 | 1 |
| 11Q LOSS WILD-TYPE | 178 | 121 | 192 |
Figure S240. Get High-res Image Gene #48: '11q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
P value = 0.00255 (Fisher's exact test), Q value = 0.0088
Table S241. Gene #48: '11q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 11Q LOSS MUTATED | 2 | 6 | 0 |
| 11Q LOSS WILD-TYPE | 163 | 120 | 179 |
Figure S241. Get High-res Image Gene #48: '11q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
P value = 0.00161 (Fisher's exact test), Q value = 0.006
Table S242. Gene #49: '13q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 13Q LOSS MUTATED | 6 | 5 | 3 |
| 13Q LOSS WILD-TYPE | 50 | 351 | 86 |
Figure S242. Get High-res Image Gene #49: '13q loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.0148 (Fisher's exact test), Q value = 0.039
Table S243. Gene #49: '13q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 13Q LOSS MUTATED | 3 | 9 | 2 |
| 13Q LOSS WILD-TYPE | 274 | 145 | 68 |
Figure S243. Get High-res Image Gene #49: '13q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
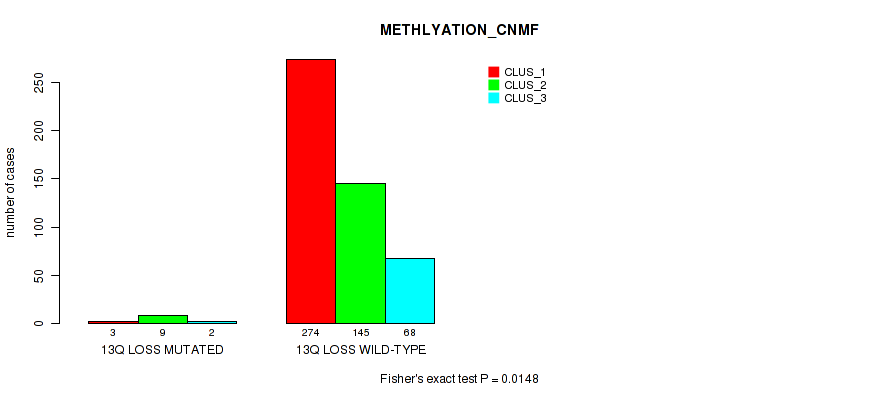
P value = 0.00534 (Fisher's exact test), Q value = 0.017
Table S244. Gene #49: '13q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 13Q LOSS MUTATED | 0 | 9 | 2 | 3 |
| 13Q LOSS WILD-TYPE | 168 | 146 | 57 | 114 |
Figure S244. Get High-res Image Gene #49: '13q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
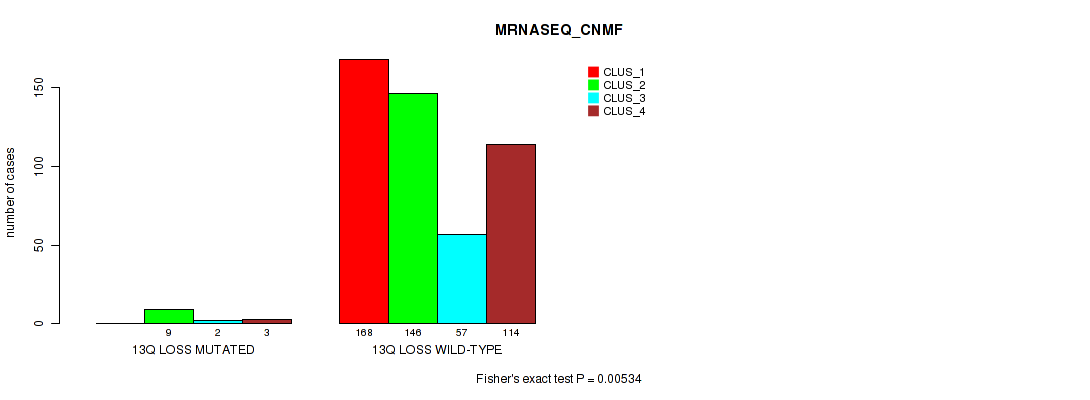
P value = 0.00799 (Fisher's exact test), Q value = 0.023
Table S245. Gene #49: '13q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 13Q LOSS MUTATED | 0 | 9 | 2 | 3 | 0 |
| 13Q LOSS WILD-TYPE | 105 | 125 | 84 | 93 | 78 |
Figure S245. Get High-res Image Gene #49: '13q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
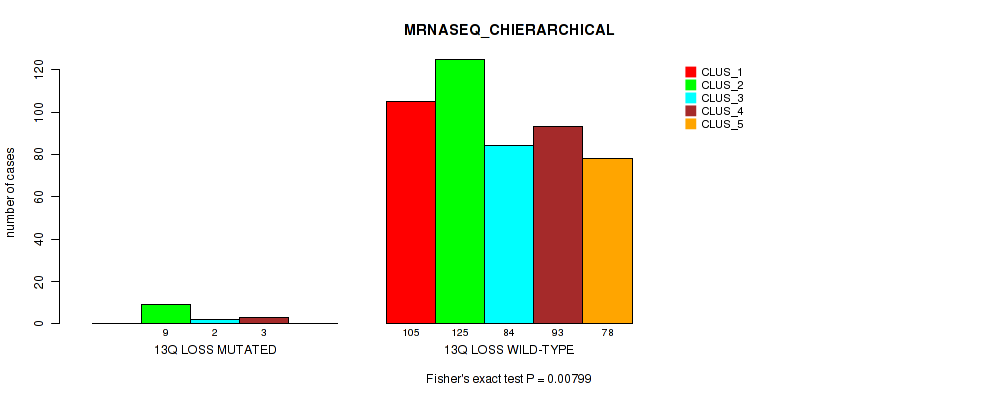
P value = 0.00105 (Fisher's exact test), Q value = 0.0042
Table S246. Gene #49: '13q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 13Q LOSS MUTATED | 0 | 9 | 5 |
| 13Q LOSS WILD-TYPE | 182 | 142 | 162 |
Figure S246. Get High-res Image Gene #49: '13q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
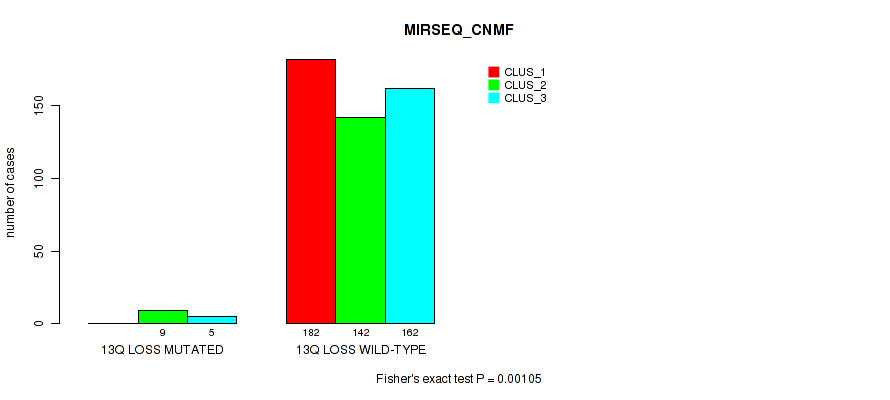
P value = 0.00041 (Fisher's exact test), Q value = 0.0019
Table S247. Gene #49: '13q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 13Q LOSS MUTATED | 0 | 9 | 5 |
| 13Q LOSS WILD-TYPE | 179 | 119 | 188 |
Figure S247. Get High-res Image Gene #49: '13q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
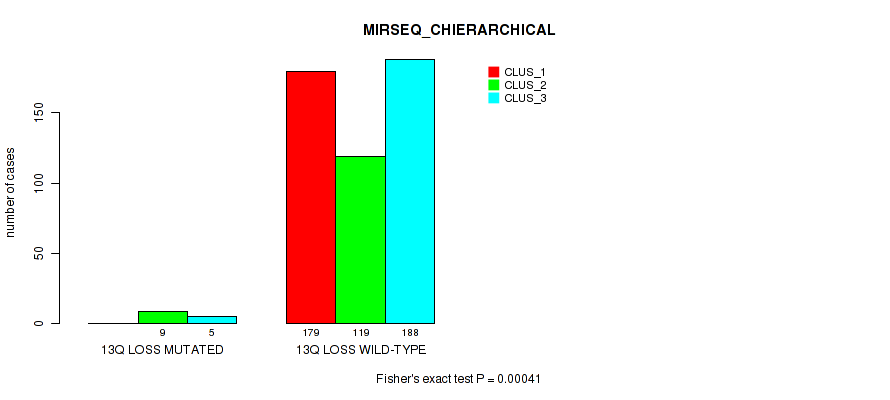
P value = 0.0127 (Fisher's exact test), Q value = 0.035
Table S248. Gene #49: '13q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 13Q LOSS MUTATED | 1 | 8 | 4 |
| 13Q LOSS WILD-TYPE | 164 | 118 | 175 |
Figure S248. Get High-res Image Gene #49: '13q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
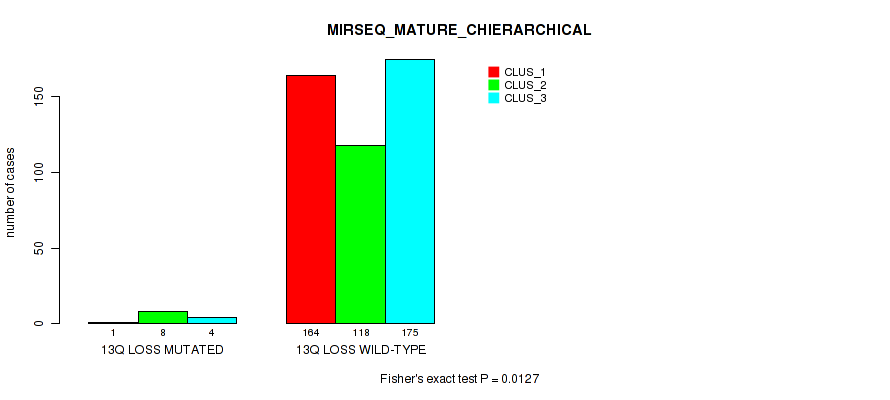
P value = 0.00014 (Fisher's exact test), Q value = 0.00079
Table S249. Gene #50: '15q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 15Q LOSS MUTATED | 5 | 1 | 2 |
| 15Q LOSS WILD-TYPE | 51 | 355 | 87 |
Figure S249. Get High-res Image Gene #50: '15q loss' versus Molecular Subtype #1: 'CN_CNMF'
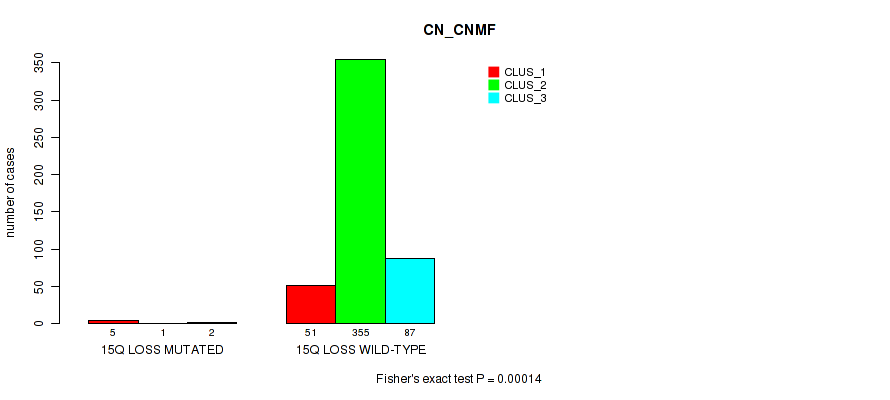
P value = 2e-04 (Fisher's exact test), Q value = 0.001
Table S250. Gene #50: '15q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 15Q LOSS MUTATED | 0 | 8 | 0 |
| 15Q LOSS WILD-TYPE | 277 | 146 | 70 |
Figure S250. Get High-res Image Gene #50: '15q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
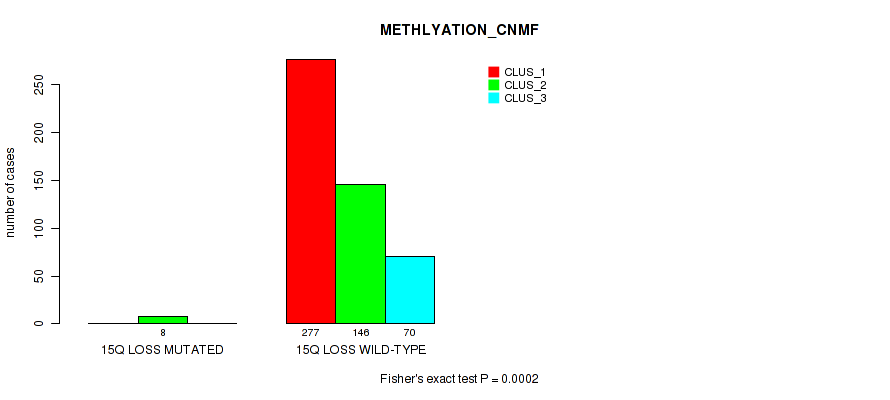
P value = 0.00059 (Fisher's exact test), Q value = 0.0026
Table S251. Gene #50: '15q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 15Q LOSS MUTATED | 0 | 8 | 0 | 0 |
| 15Q LOSS WILD-TYPE | 168 | 147 | 59 | 117 |
Figure S251. Get High-res Image Gene #50: '15q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
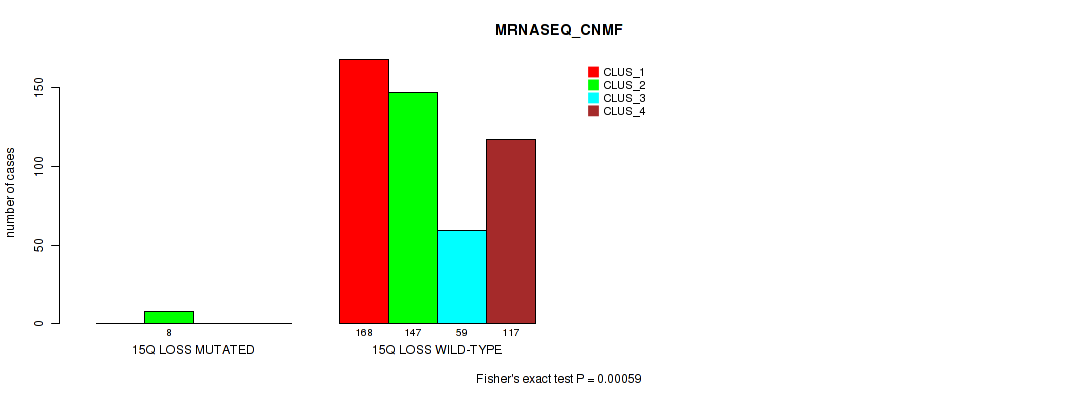
P value = 0.00026 (Fisher's exact test), Q value = 0.0013
Table S252. Gene #50: '15q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 15Q LOSS MUTATED | 0 | 8 | 0 | 0 | 0 |
| 15Q LOSS WILD-TYPE | 105 | 126 | 86 | 96 | 78 |
Figure S252. Get High-res Image Gene #50: '15q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
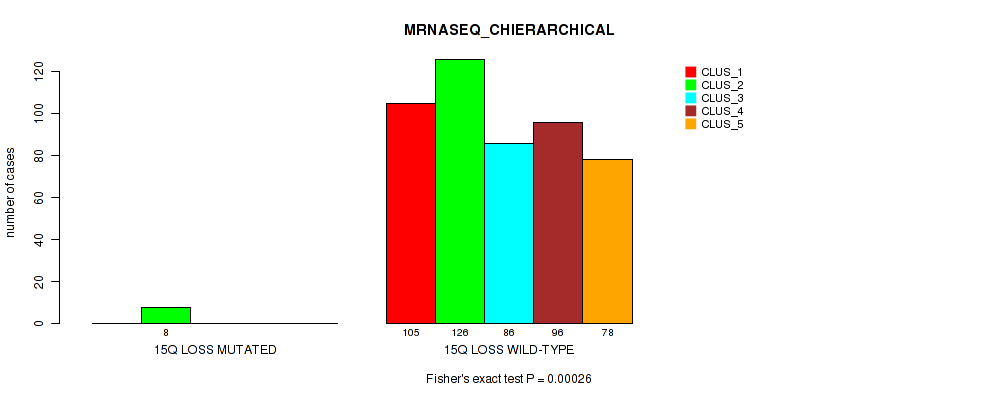
P value = 7e-05 (Fisher's exact test), Q value = 0.00046
Table S253. Gene #50: '15q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 15Q LOSS MUTATED | 0 | 8 | 0 |
| 15Q LOSS WILD-TYPE | 182 | 143 | 167 |
Figure S253. Get High-res Image Gene #50: '15q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
P value = 2e-05 (Fisher's exact test), Q value = 0.00018
Table S254. Gene #50: '15q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 15Q LOSS MUTATED | 0 | 8 | 0 |
| 15Q LOSS WILD-TYPE | 179 | 120 | 193 |
Figure S254. Get High-res Image Gene #50: '15q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
P value = 0.00195 (Fisher's exact test), Q value = 0.0071
Table S255. Gene #50: '15q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 15Q LOSS MUTATED | 1 | 6 | 0 |
| 15Q LOSS WILD-TYPE | 164 | 120 | 179 |
Figure S255. Get High-res Image Gene #50: '15q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
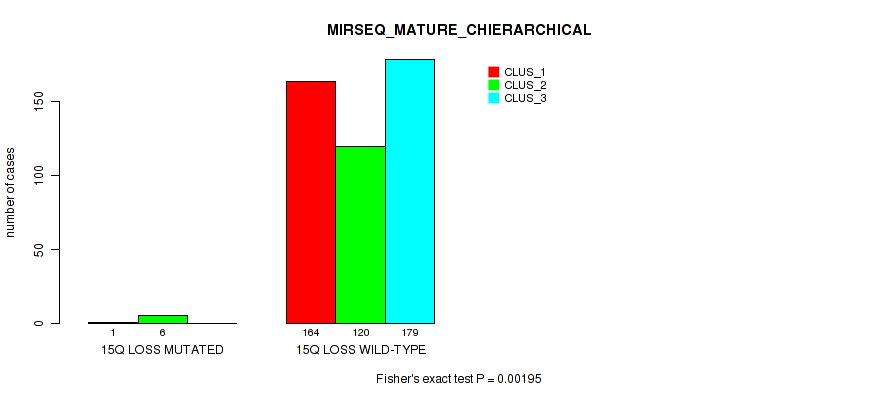
P value = 0.0144 (Fisher's exact test), Q value = 0.038
Table S256. Gene #51: '17p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 17P LOSS MUTATED | 4 | 3 | 1 |
| 17P LOSS WILD-TYPE | 52 | 353 | 88 |
Figure S256. Get High-res Image Gene #51: '17p loss' versus Molecular Subtype #1: 'CN_CNMF'
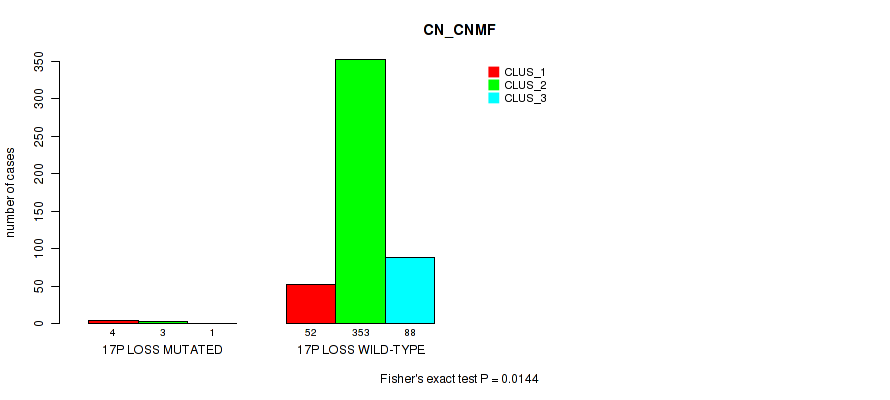
P value = 0.002 (Fisher's exact test), Q value = 0.0072
Table S257. Gene #53: '18p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 18P LOSS MUTATED | 3 | 0 | 1 |
| 18P LOSS WILD-TYPE | 53 | 356 | 88 |
Figure S257. Get High-res Image Gene #53: '18p loss' versus Molecular Subtype #1: 'CN_CNMF'
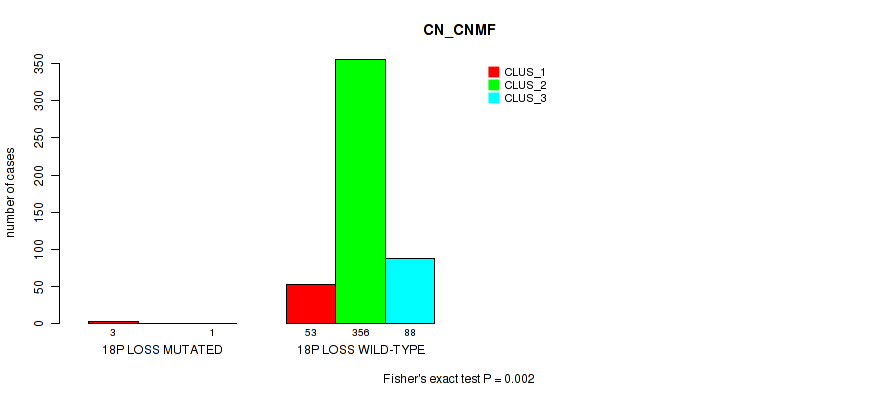
P value = 0.00214 (Fisher's exact test), Q value = 0.0077
Table S258. Gene #54: '18q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 18Q LOSS MUTATED | 3 | 0 | 1 |
| 18Q LOSS WILD-TYPE | 53 | 356 | 88 |
Figure S258. Get High-res Image Gene #54: '18q loss' versus Molecular Subtype #1: 'CN_CNMF'
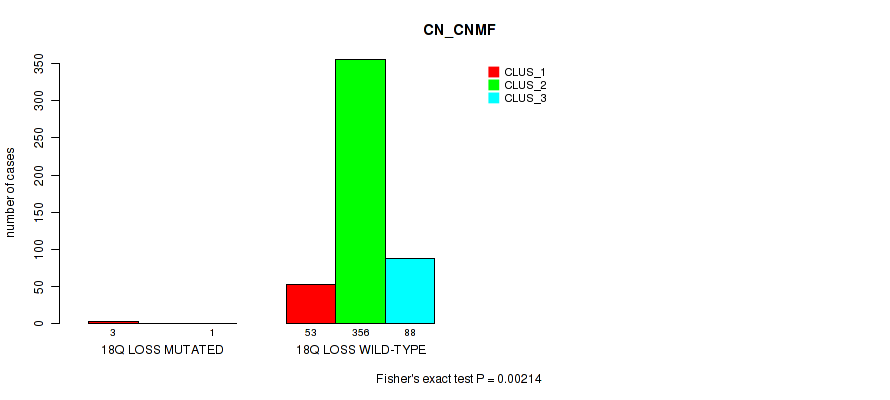
P value = 0.00932 (Fisher's exact test), Q value = 0.027
Table S259. Gene #55: '19p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 19P LOSS MUTATED | 3 | 1 | 1 |
| 19P LOSS WILD-TYPE | 53 | 355 | 88 |
Figure S259. Get High-res Image Gene #55: '19p loss' versus Molecular Subtype #1: 'CN_CNMF'
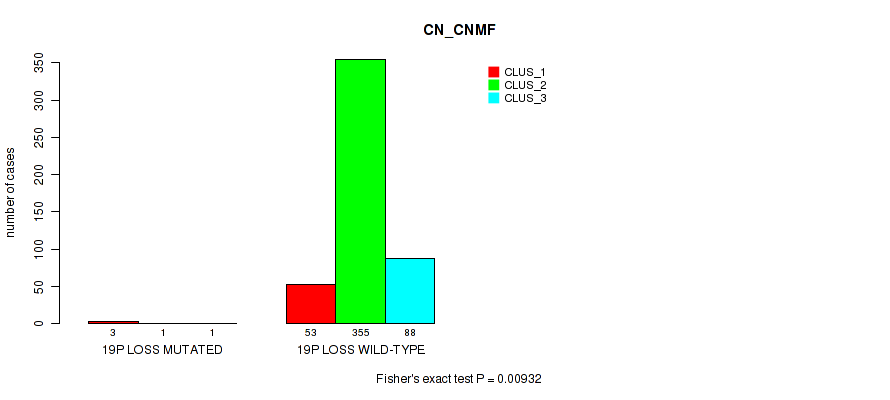
P value = 0.00657 (Fisher's exact test), Q value = 0.02
Table S260. Gene #55: '19p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 19P LOSS MUTATED | 0 | 5 | 0 |
| 19P LOSS WILD-TYPE | 277 | 149 | 70 |
Figure S260. Get High-res Image Gene #55: '19p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
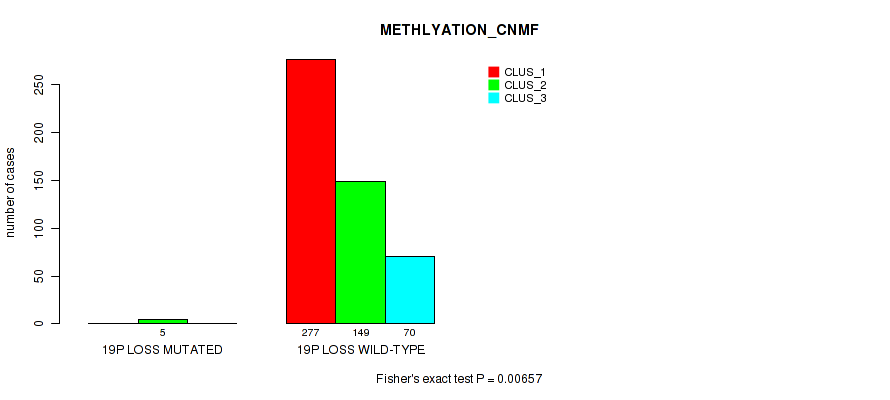
P value = 0.0167 (Fisher's exact test), Q value = 0.042
Table S261. Gene #55: '19p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 19P LOSS MUTATED | 0 | 5 | 0 | 0 |
| 19P LOSS WILD-TYPE | 168 | 150 | 59 | 117 |
Figure S261. Get High-res Image Gene #55: '19p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
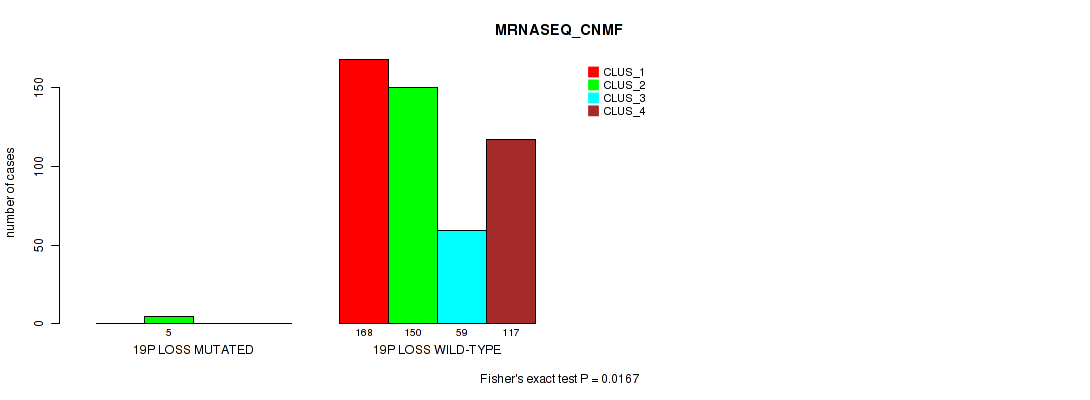
P value = 0.0129 (Fisher's exact test), Q value = 0.035
Table S262. Gene #55: '19p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 19P LOSS MUTATED | 0 | 5 | 0 | 0 | 0 |
| 19P LOSS WILD-TYPE | 105 | 129 | 86 | 96 | 78 |
Figure S262. Get High-res Image Gene #55: '19p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
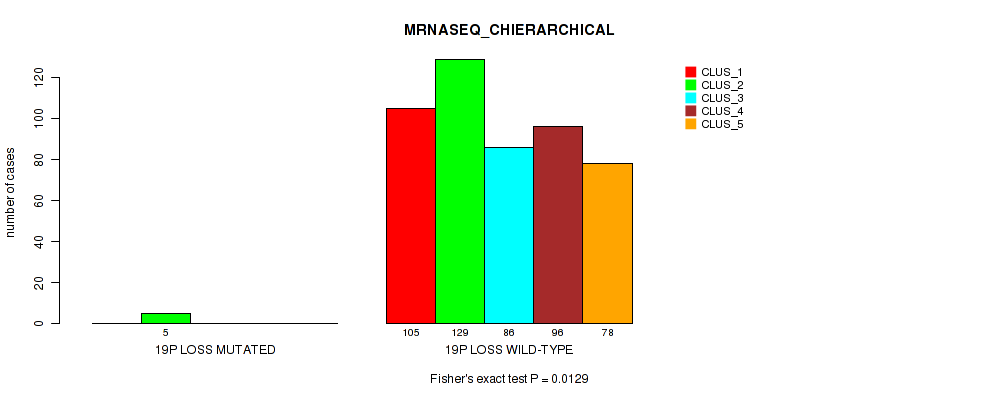
P value = 0.00239 (Fisher's exact test), Q value = 0.0084
Table S263. Gene #55: '19p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 19P LOSS MUTATED | 0 | 5 | 0 |
| 19P LOSS WILD-TYPE | 182 | 146 | 167 |
Figure S263. Get High-res Image Gene #55: '19p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'

P value = 0.00107 (Fisher's exact test), Q value = 0.0043
Table S264. Gene #55: '19p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 19P LOSS MUTATED | 0 | 5 | 0 |
| 19P LOSS WILD-TYPE | 179 | 123 | 193 |
Figure S264. Get High-res Image Gene #55: '19p loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
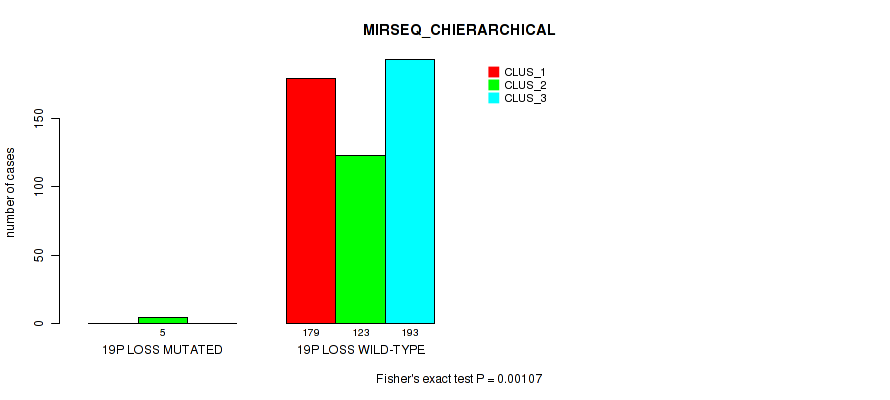
P value = 0.00145 (Fisher's exact test), Q value = 0.0055
Table S265. Gene #55: '19p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 19P LOSS MUTATED | 0 | 5 | 0 |
| 19P LOSS WILD-TYPE | 165 | 121 | 179 |
Figure S265. Get High-res Image Gene #55: '19p loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
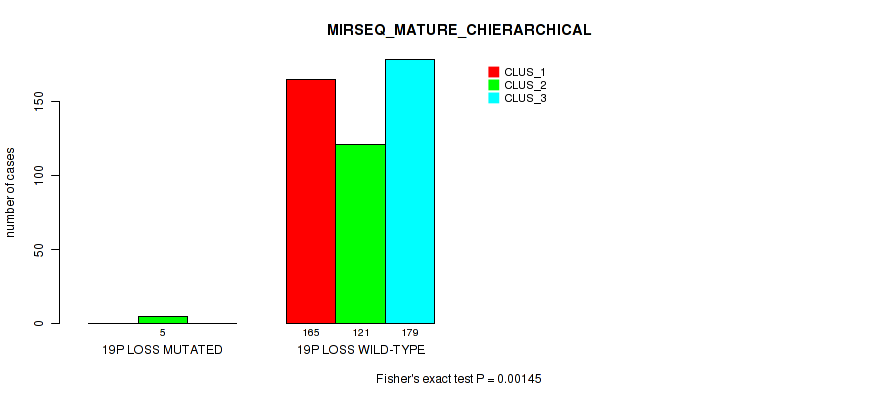
P value = 0.00065 (Fisher's exact test), Q value = 0.0028
Table S266. Gene #56: '21q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 21Q LOSS MUTATED | 5 | 2 | 1 |
| 21Q LOSS WILD-TYPE | 51 | 354 | 88 |
Figure S266. Get High-res Image Gene #56: '21q loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.0287 (Fisher's exact test), Q value = 0.065
Table S267. Gene #56: '21q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 21Q LOSS MUTATED | 4 | 4 | 0 |
| 21Q LOSS WILD-TYPE | 175 | 124 | 193 |
Figure S267. Get High-res Image Gene #56: '21q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
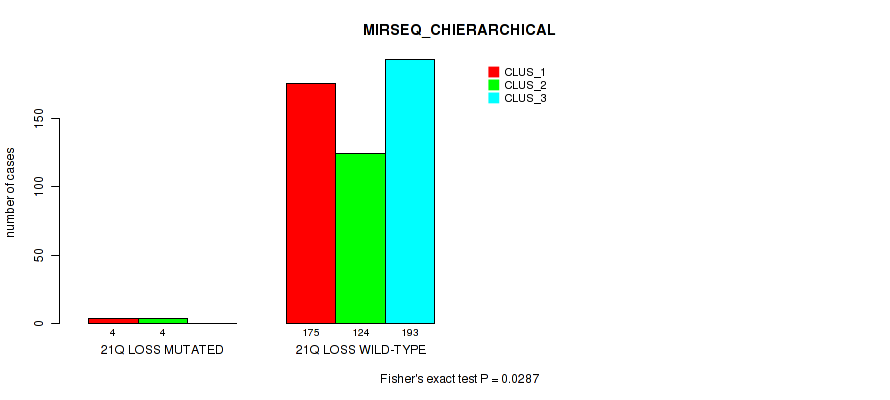
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S268. Gene #57: '22q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 56 | 356 | 89 |
| 22Q LOSS MUTATED | 13 | 0 | 76 |
| 22Q LOSS WILD-TYPE | 43 | 356 | 13 |
Figure S268. Get High-res Image Gene #57: '22q loss' versus Molecular Subtype #1: 'CN_CNMF'
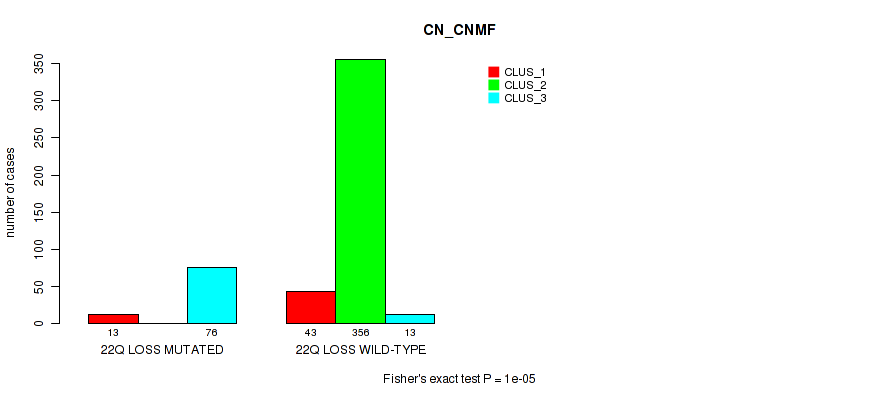
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S269. Gene #57: '22q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 277 | 154 | 70 |
| 22Q LOSS MUTATED | 36 | 47 | 6 |
| 22Q LOSS WILD-TYPE | 241 | 107 | 64 |
Figure S269. Get High-res Image Gene #57: '22q loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
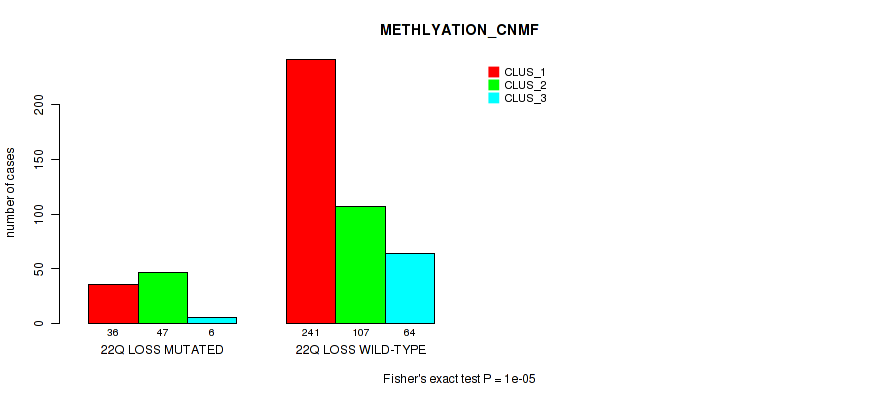
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S270. Gene #57: '22q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 168 | 155 | 59 | 117 |
| 22Q LOSS MUTATED | 8 | 47 | 8 | 26 |
| 22Q LOSS WILD-TYPE | 160 | 108 | 51 | 91 |
Figure S270. Get High-res Image Gene #57: '22q loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
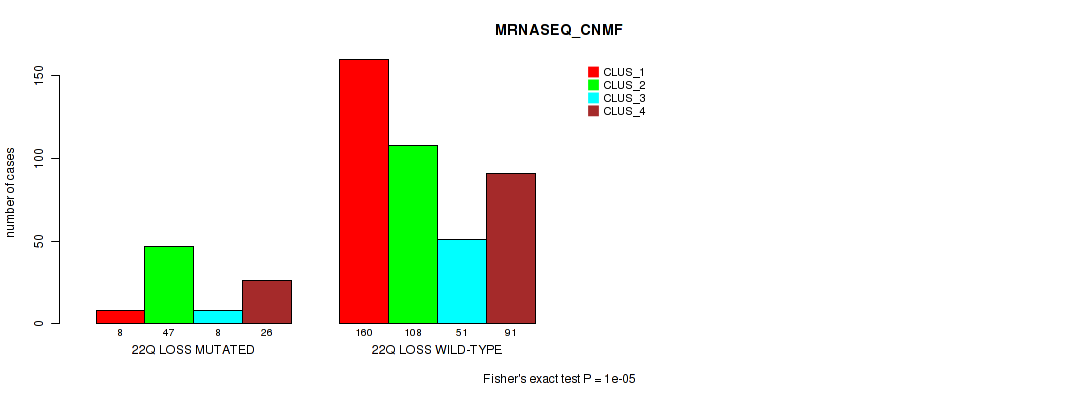
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S271. Gene #57: '22q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 105 | 134 | 86 | 96 | 78 |
| 22Q LOSS MUTATED | 9 | 42 | 13 | 23 | 2 |
| 22Q LOSS WILD-TYPE | 96 | 92 | 73 | 73 | 76 |
Figure S271. Get High-res Image Gene #57: '22q loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
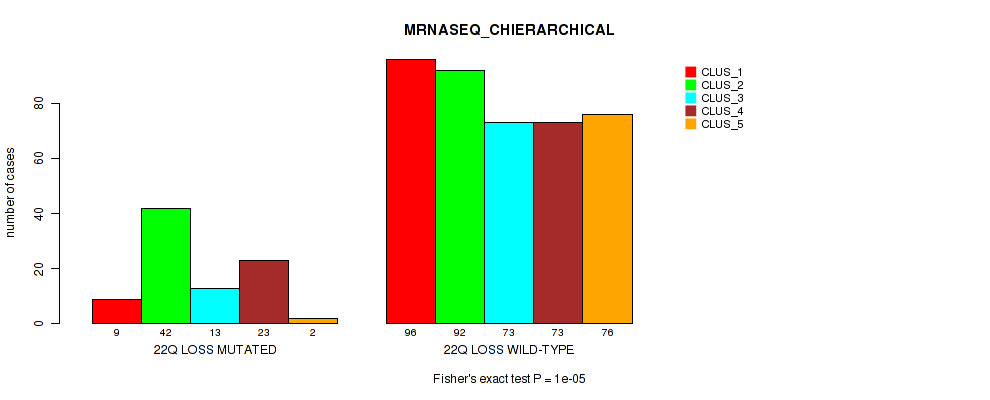
P value = 0.00042 (Fisher's exact test), Q value = 0.002
Table S272. Gene #57: '22q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| 22Q LOSS MUTATED | 23 | 43 | 23 |
| 22Q LOSS WILD-TYPE | 159 | 108 | 144 |
Figure S272. Get High-res Image Gene #57: '22q loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
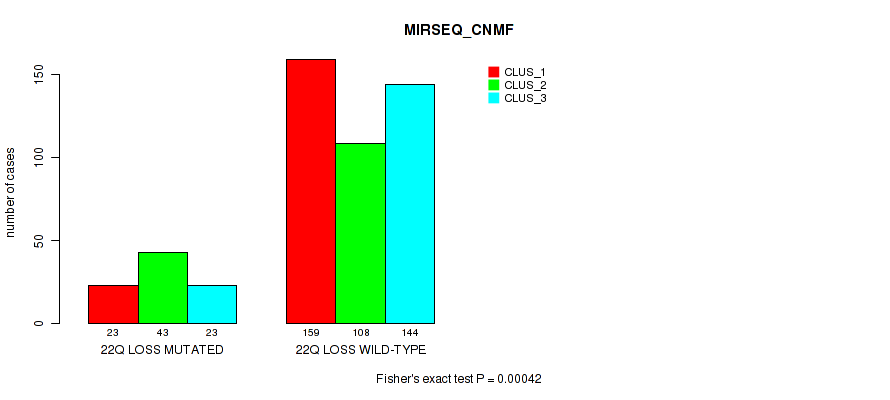
P value = 4e-05 (Fisher's exact test), Q value = 0.00029
Table S273. Gene #57: '22q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| 22Q LOSS MUTATED | 21 | 40 | 28 |
| 22Q LOSS WILD-TYPE | 158 | 88 | 165 |
Figure S273. Get High-res Image Gene #57: '22q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
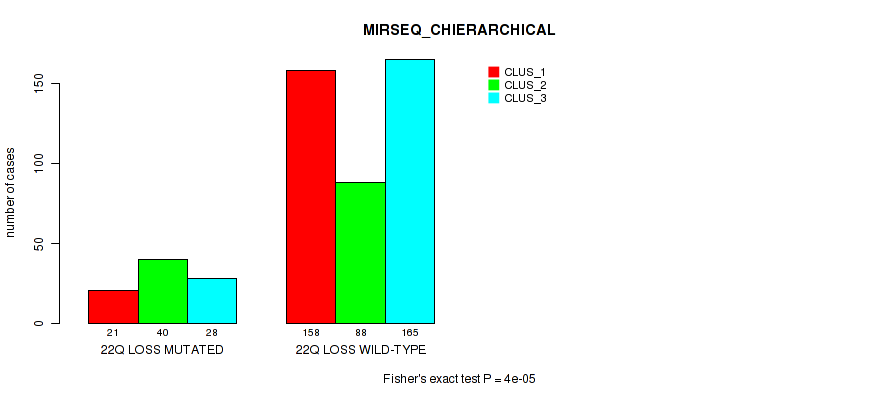
P value = 0.00018 (Fisher's exact test), Q value = 0.00096
Table S274. Gene #57: '22q loss' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 160 | 164 | 146 |
| 22Q LOSS MUTATED | 17 | 23 | 41 |
| 22Q LOSS WILD-TYPE | 143 | 141 | 105 |
Figure S274. Get High-res Image Gene #57: '22q loss' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
P value = 0.00026 (Fisher's exact test), Q value = 0.0013
Table S275. Gene #57: '22q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| 22Q LOSS MUTATED | 22 | 37 | 22 |
| 22Q LOSS WILD-TYPE | 143 | 89 | 157 |
Figure S275. Get High-res Image Gene #57: '22q loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
P value = 0.0375 (Fisher's exact test), Q value = 0.083
Table S276. Gene #58: 'xp loss' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 160 | 164 | 146 |
| XP LOSS MUTATED | 0 | 4 | 0 |
| XP LOSS WILD-TYPE | 160 | 160 | 146 |
Figure S276. Get High-res Image Gene #58: 'xp loss' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
P value = 0.0281 (Fisher's exact test), Q value = 0.064
Table S277. Gene #59: 'xq loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 182 | 151 | 167 |
| XQ LOSS MUTATED | 0 | 3 | 0 |
| XQ LOSS WILD-TYPE | 182 | 148 | 167 |
Figure S277. Get High-res Image Gene #59: 'xq loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
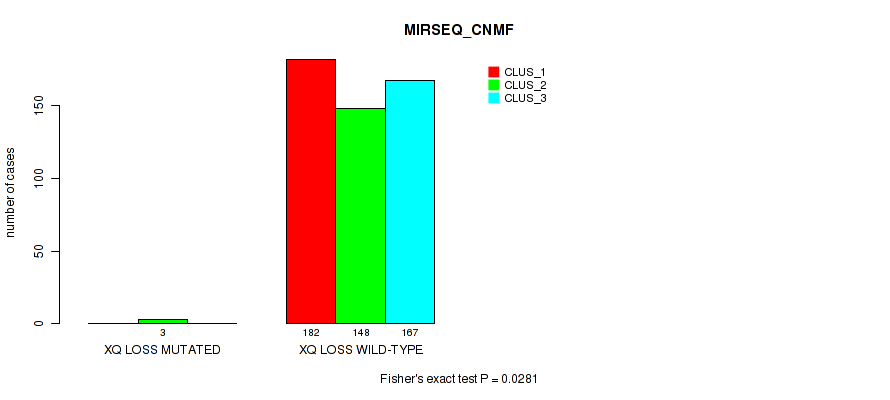
P value = 0.0166 (Fisher's exact test), Q value = 0.042
Table S278. Gene #59: 'xq loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 179 | 128 | 193 |
| XQ LOSS MUTATED | 0 | 3 | 0 |
| XQ LOSS WILD-TYPE | 179 | 125 | 193 |
Figure S278. Get High-res Image Gene #59: 'xq loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'

P value = 0.019 (Fisher's exact test), Q value = 0.045
Table S279. Gene #59: 'xq loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 165 | 126 | 179 |
| XQ LOSS MUTATED | 0 | 3 | 0 |
| XQ LOSS WILD-TYPE | 165 | 123 | 179 |
Figure S279. Get High-res Image Gene #59: 'xq loss' versus Molecular Subtype #10: 'MIRSEQ_MATURE_CHIERARCHICAL'

-
Copy number data file = broad_values_by_arm.txt from GISTIC pipeline
-
Processed Copy number data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/THCA-TP/15101916/transformed.cor.cli.txt
-
Molecular subtypes file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_mergedClustering/THCA-TP/15125146/THCA-TP.transferedmergedcluster.txt
-
Number of patients = 501
-
Number of significantly arm-level cnvs = 59
-
Number of molecular subtypes = 10
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.