This pipeline computes the correlation between significant copy number variation (cnv focal) genes and molecular subtypes.
Testing the association between copy number variation 20 focal events and 6 molecular subtypes across 191 patients, 69 significant findings detected with P value < 0.05 and Q value < 0.25.
-
amp_1p33 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
amp_1q43 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
amp_11q23.3 cnv correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
amp_13q31.3 cnv correlated to 'METHLYATION_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
amp_19p13.2 cnv correlated to 'CN_CNMF'.
-
amp_20q11.21 cnv correlated to 'CN_CNMF'.
-
amp_21q22.2 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
del_3p13 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
del_5q31.2 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
del_7p12.1 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
del_7q34 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
del_7q34 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
del_12p13.2 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
del_12q21.33 cnv correlated to 'CN_CNMF', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
del_16q23.1 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MIRSEQ_CNMF'.
-
del_17p13.2 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
del_17q11.2 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
del_18p11.21 cnv correlated to 'CN_CNMF', 'METHLYATION_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
del_20q13.13 cnv correlated to 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 20 focal events and 6 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 69 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| del 5q31 2 | 18 (9%) | 173 |
1e-05 (0.000109) |
1e-05 (0.000109) |
0.00535 (0.014) |
0.00083 (0.00383) |
0.00069 (0.00345) |
0.00119 (0.00468) |
| del 7p12 1 | 16 (8%) | 175 |
0.00021 (0.00157) |
0.00037 (0.00234) |
0.00183 (0.00627) |
1e-05 (0.000109) |
0.00568 (0.0145) |
0.00684 (0.0161) |
| del 7q34 | 24 (13%) | 167 |
1e-05 (0.000109) |
0.00011 (0.000943) |
0.023 (0.0468) |
1e-05 (0.000109) |
0.00087 (0.00386) |
0.00068 (0.00345) |
| del 7q34 | 24 (13%) | 167 |
1e-05 (0.000109) |
0.00012 (0.00096) |
0.0217 (0.0457) |
1e-05 (0.000109) |
0.0009 (0.00386) |
0.00072 (0.00346) |
| amp 21q22 2 | 14 (7%) | 177 |
1e-05 (0.000109) |
0.00054 (0.00309) |
0.00335 (0.00966) |
0.0365 (0.0682) |
0.00164 (0.00579) |
0.0667 (0.108) |
| del 17p13 2 | 15 (8%) | 176 |
1e-05 (0.000109) |
0.00025 (0.00167) |
0.0111 (0.0242) |
0.00023 (0.00162) |
0.086 (0.131) |
0.0012 (0.00468) |
| amp 1p33 | 7 (4%) | 184 |
0.00049 (0.00294) |
0.00246 (0.00757) |
0.125 (0.184) |
0.132 (0.19) |
0.00946 (0.0214) |
0.0475 (0.0838) |
| amp 1q43 | 7 (4%) | 184 |
0.00338 (0.00966) |
0.00237 (0.00757) |
0.373 (0.444) |
0.036 (0.0682) |
0.139 (0.196) |
0.0151 (0.0323) |
| del 3p13 | 8 (4%) | 183 |
0.00062 (0.00338) |
0.0457 (0.0819) |
0.066 (0.108) |
0.235 (0.294) |
0.0368 (0.0682) |
0.0484 (0.0841) |
| del 17q11 2 | 13 (7%) | 178 |
1e-05 (0.000109) |
0.00512 (0.0137) |
0.156 (0.217) |
0.00658 (0.0158) |
0.135 (0.193) |
0.00383 (0.0106) |
| del 12p13 2 | 10 (5%) | 181 |
3e-05 (0.000277) |
0.00158 (0.00575) |
0.196 (0.259) |
0.0732 (0.114) |
0.0695 (0.11) |
0.0101 (0.0224) |
| del 12q21 33 | 3 (2%) | 188 |
0.00121 (0.00468) |
0.701 (0.765) |
0.188 (0.25) |
0.586 (0.659) |
0.0086 (0.0198) |
0.00611 (0.0153) |
| del 16q23 1 | 9 (5%) | 182 |
0.00152 (0.0057) |
0.0226 (0.0467) |
0.331 (0.403) |
0.493 (0.563) |
0.0451 (0.0819) |
0.065 (0.108) |
| del 18p11 21 | 9 (5%) | 182 |
2e-05 (2e-04) |
0.00387 (0.0106) |
0.381 (0.448) |
0.333 (0.403) |
0.214 (0.273) |
0.0318 (0.0625) |
| amp 11q23 3 | 17 (9%) | 174 |
1e-05 (0.000109) |
0.00217 (0.00723) |
0.371 (0.444) |
0.201 (0.262) |
0.747 (0.8) |
0.184 (0.249) |
| amp 13q31 3 | 7 (4%) | 184 |
0.0813 (0.125) |
0.00245 (0.00757) |
0.0674 (0.108) |
0.037 (0.0682) |
0.736 (0.795) |
0.0571 (0.0964) |
| amp 19p13 2 | 6 (3%) | 185 |
0.0032 (0.0096) |
0.254 (0.314) |
0.856 (0.901) |
0.633 (0.696) |
0.893 (0.932) |
1 (1.00) |
| amp 20q11 21 | 3 (2%) | 188 |
0.00656 (0.0158) |
0.211 (0.273) |
||||
| del 20q13 13 | 4 (2%) | 187 |
0.029 (0.058) |
0.056 (0.096) |
0.785 (0.834) |
0.588 (0.659) |
0.125 (0.184) |
0.41 (0.478) |
| del 9q21 32 | 5 (3%) | 186 |
0.231 (0.292) |
0.465 (0.536) |
0.175 (0.242) |
0.602 (0.669) |
0.183 (0.249) |
0.106 (0.159) |
P value = 0.00049 (Fisher's exact test), Q value = 0.0029
Table S1. Gene #1: 'amp_1p33' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| AMP PEAK 1(1P33) MUTATED | 1 | 3 | 3 | 0 |
| AMP PEAK 1(1P33) WILD-TYPE | 147 | 11 | 24 | 2 |
Figure S1. Get High-res Image Gene #1: 'amp_1p33' versus Molecular Subtype #1: 'CN_CNMF'
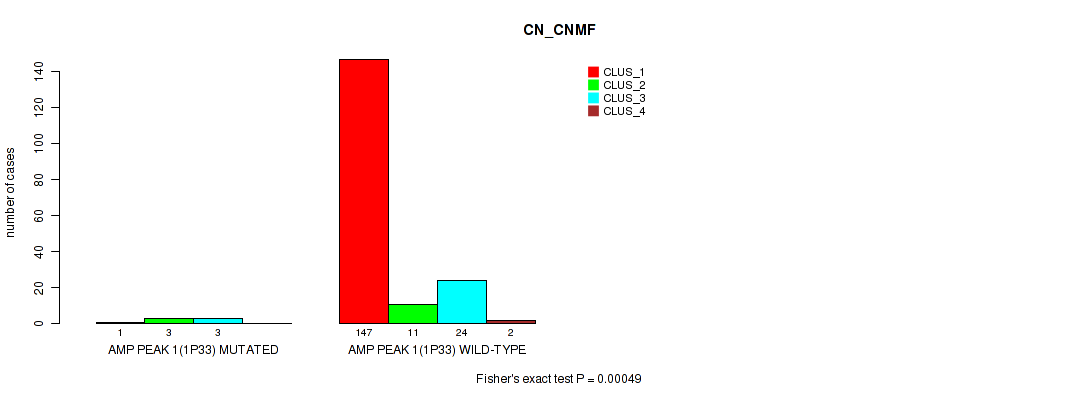
P value = 0.00246 (Fisher's exact test), Q value = 0.0076
Table S2. Gene #1: 'amp_1p33' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| AMP PEAK 1(1P33) MUTATED | 0 | 7 | 0 | 0 |
| AMP PEAK 1(1P33) WILD-TYPE | 33 | 54 | 47 | 44 |
Figure S2. Get High-res Image Gene #1: 'amp_1p33' versus Molecular Subtype #2: 'METHLYATION_CNMF'
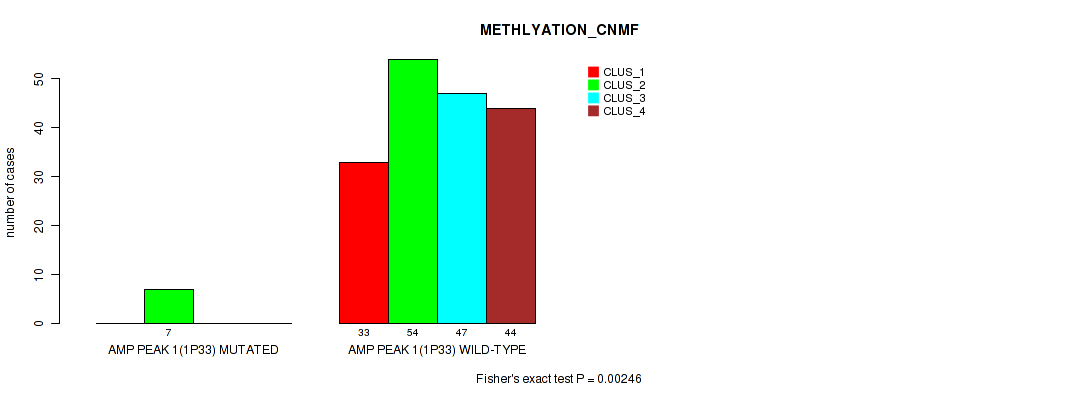
P value = 0.00946 (Fisher's exact test), Q value = 0.021
Table S3. Gene #1: 'amp_1p33' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| AMP PEAK 1(1P33) MUTATED | 0 | 3 | 0 | 4 |
| AMP PEAK 1(1P33) WILD-TYPE | 58 | 34 | 41 | 39 |
Figure S3. Get High-res Image Gene #1: 'amp_1p33' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
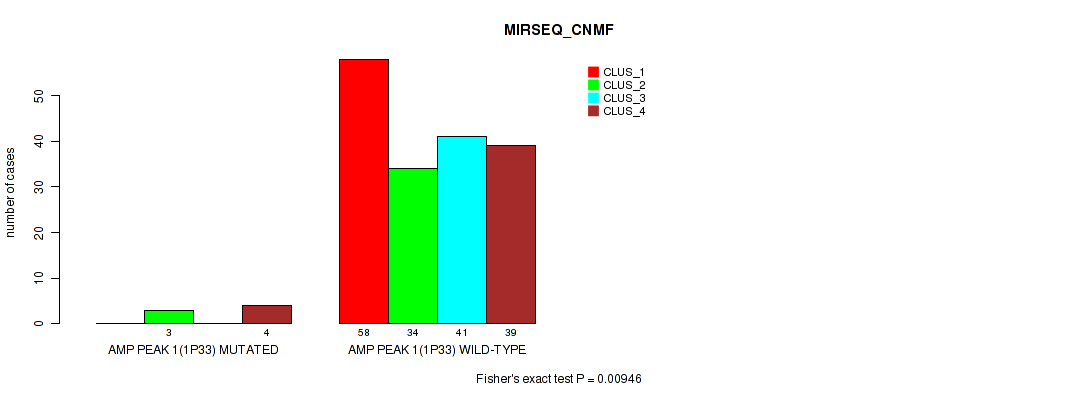
P value = 0.0475 (Fisher's exact test), Q value = 0.084
Table S4. Gene #1: 'amp_1p33' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| AMP PEAK 1(1P33) MUTATED | 0 | 3 | 4 |
| AMP PEAK 1(1P33) WILD-TYPE | 67 | 31 | 74 |
Figure S4. Get High-res Image Gene #1: 'amp_1p33' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
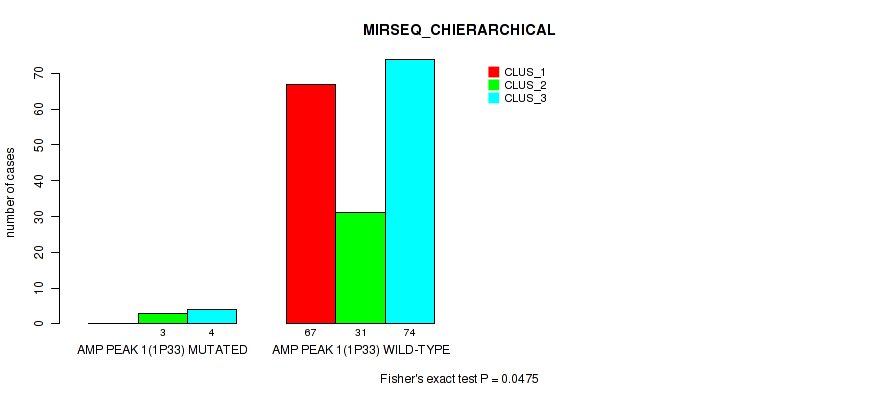
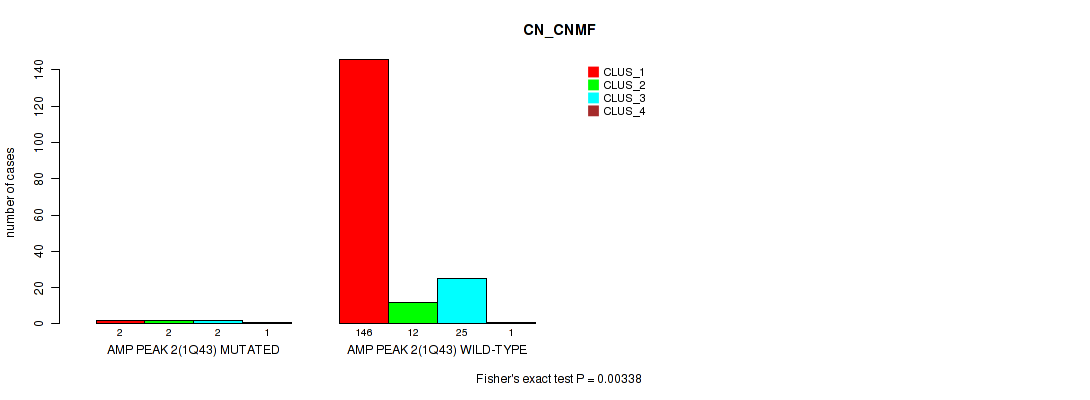
P value = 0.00338 (Fisher's exact test), Q value = 0.0097
Table S5. Gene #2: 'amp_1q43' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| AMP PEAK 2(1Q43) MUTATED | 2 | 2 | 2 | 1 |
| AMP PEAK 2(1Q43) WILD-TYPE | 146 | 12 | 25 | 1 |
Figure S5. Get High-res Image Gene #2: 'amp_1q43' versus Molecular Subtype #1: 'CN_CNMF'
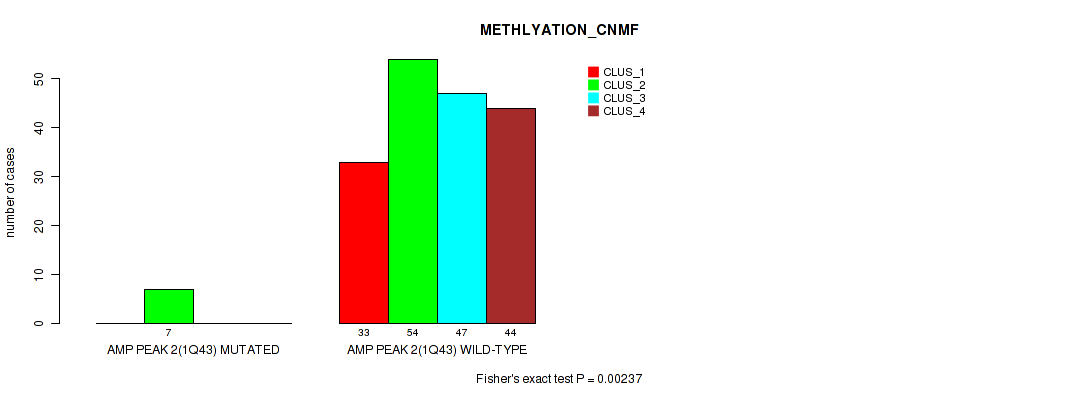
P value = 0.00237 (Fisher's exact test), Q value = 0.0076
Table S6. Gene #2: 'amp_1q43' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| AMP PEAK 2(1Q43) MUTATED | 0 | 7 | 0 | 0 |
| AMP PEAK 2(1Q43) WILD-TYPE | 33 | 54 | 47 | 44 |
Figure S6. Get High-res Image Gene #2: 'amp_1q43' versus Molecular Subtype #2: 'METHLYATION_CNMF'
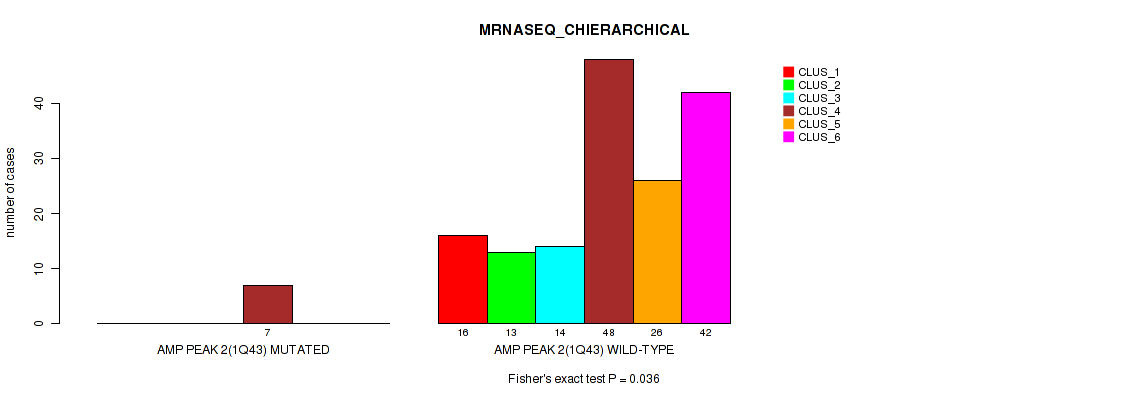
P value = 0.036 (Fisher's exact test), Q value = 0.068
Table S7. Gene #2: 'amp_1q43' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| AMP PEAK 2(1Q43) MUTATED | 0 | 0 | 0 | 7 | 0 | 0 |
| AMP PEAK 2(1Q43) WILD-TYPE | 16 | 13 | 14 | 48 | 26 | 42 |
Figure S7. Get High-res Image Gene #2: 'amp_1q43' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
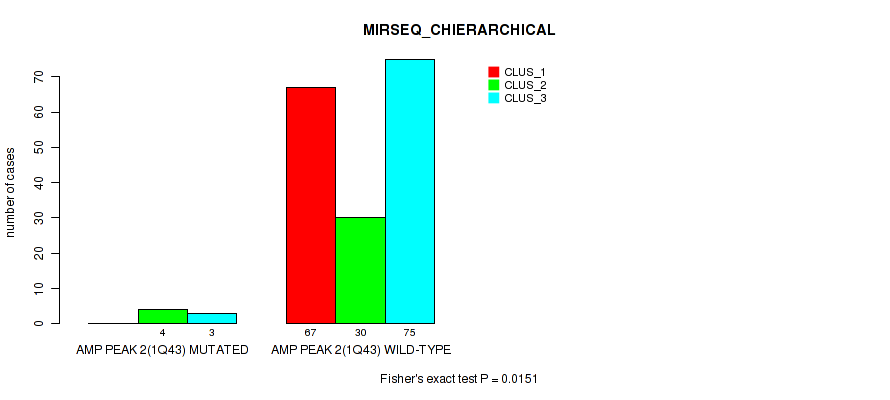
P value = 0.0151 (Fisher's exact test), Q value = 0.032
Table S8. Gene #2: 'amp_1q43' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| AMP PEAK 2(1Q43) MUTATED | 0 | 4 | 3 |
| AMP PEAK 2(1Q43) WILD-TYPE | 67 | 30 | 75 |
Figure S8. Get High-res Image Gene #2: 'amp_1q43' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S9. Gene #3: 'amp_11q23.3' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| AMP PEAK 3(11Q23.3) MUTATED | 0 | 14 | 3 | 0 |
| AMP PEAK 3(11Q23.3) WILD-TYPE | 148 | 0 | 24 | 2 |
Figure S9. Get High-res Image Gene #3: 'amp_11q23.3' versus Molecular Subtype #1: 'CN_CNMF'
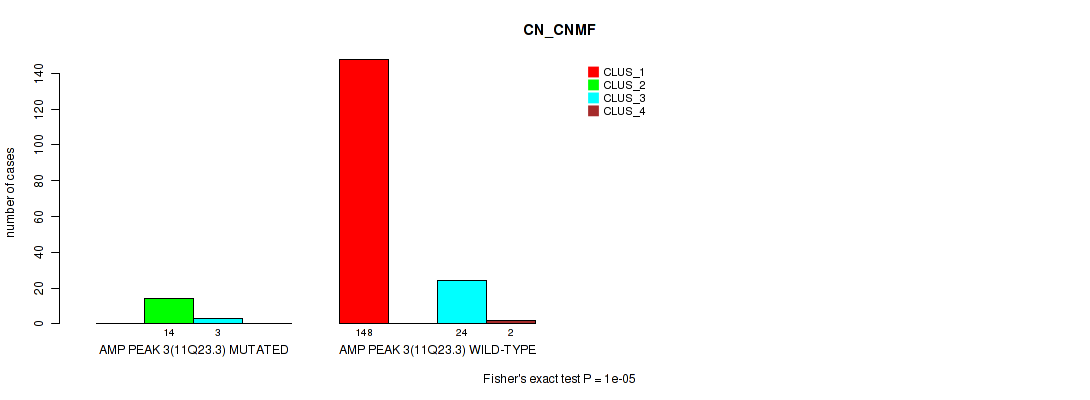
P value = 0.00217 (Fisher's exact test), Q value = 0.0072
Table S10. Gene #3: 'amp_11q23.3' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| AMP PEAK 3(11Q23.3) MUTATED | 0 | 12 | 1 | 4 |
| AMP PEAK 3(11Q23.3) WILD-TYPE | 33 | 49 | 46 | 40 |
Figure S10. Get High-res Image Gene #3: 'amp_11q23.3' versus Molecular Subtype #2: 'METHLYATION_CNMF'
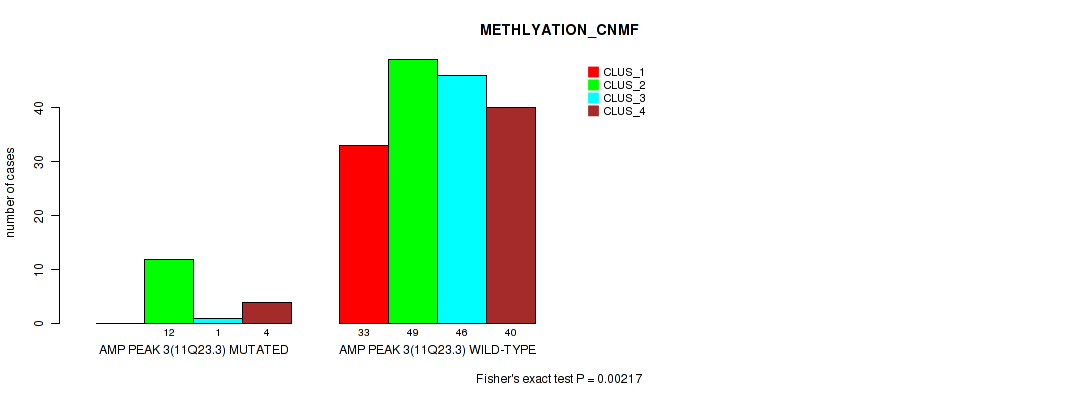
P value = 0.00245 (Fisher's exact test), Q value = 0.0076
Table S11. Gene #4: 'amp_13q31.3' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| AMP PEAK 4(13Q31.3) MUTATED | 0 | 7 | 0 | 0 |
| AMP PEAK 4(13Q31.3) WILD-TYPE | 33 | 54 | 47 | 44 |
Figure S11. Get High-res Image Gene #4: 'amp_13q31.3' versus Molecular Subtype #2: 'METHLYATION_CNMF'
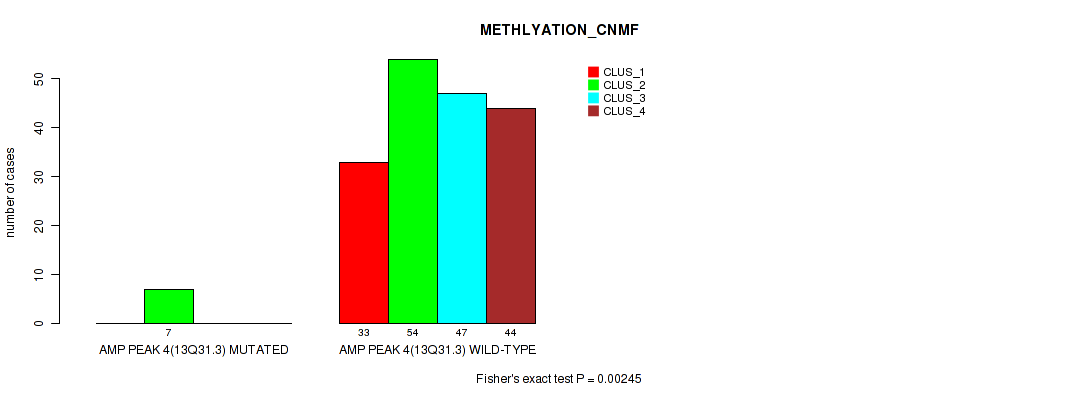
P value = 0.037 (Fisher's exact test), Q value = 0.068
Table S12. Gene #4: 'amp_13q31.3' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| AMP PEAK 4(13Q31.3) MUTATED | 0 | 0 | 0 | 7 | 0 | 0 |
| AMP PEAK 4(13Q31.3) WILD-TYPE | 16 | 13 | 14 | 48 | 26 | 42 |
Figure S12. Get High-res Image Gene #4: 'amp_13q31.3' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
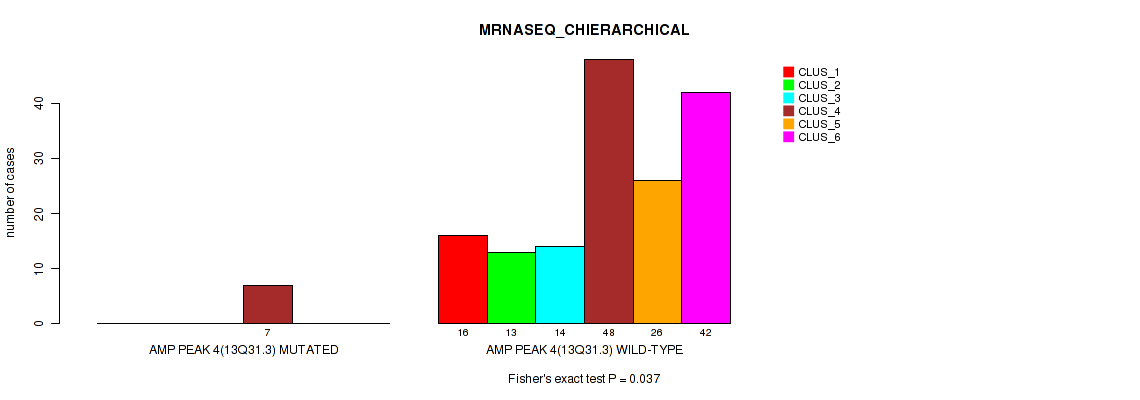
P value = 0.0032 (Fisher's exact test), Q value = 0.0096
Table S13. Gene #5: 'amp_19p13.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| AMP PEAK 5(19P13.2) MUTATED | 1 | 1 | 4 | 0 |
| AMP PEAK 5(19P13.2) WILD-TYPE | 147 | 13 | 23 | 2 |
Figure S13. Get High-res Image Gene #5: 'amp_19p13.2' versus Molecular Subtype #1: 'CN_CNMF'
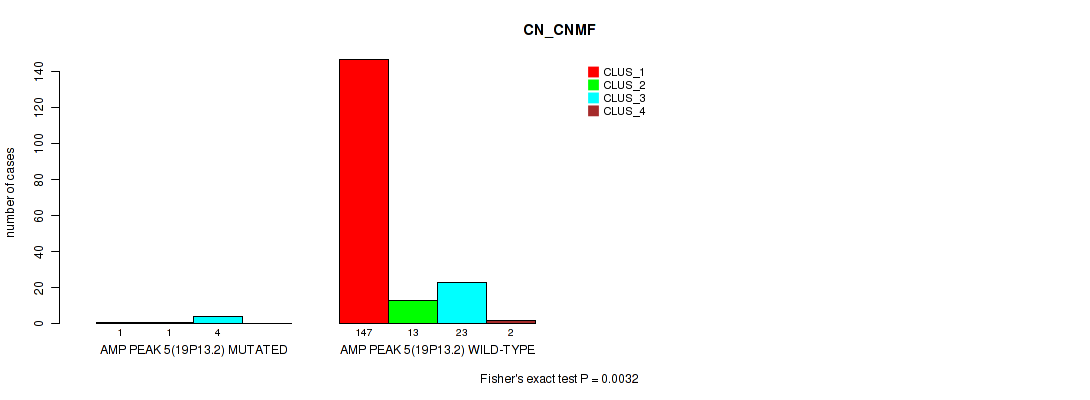
P value = 0.00656 (Fisher's exact test), Q value = 0.016
Table S14. Gene #6: 'amp_20q11.21' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| AMP PEAK 6(20Q11.21) MUTATED | 0 | 0 | 3 | 0 |
| AMP PEAK 6(20Q11.21) WILD-TYPE | 148 | 14 | 24 | 2 |
Figure S14. Get High-res Image Gene #6: 'amp_20q11.21' versus Molecular Subtype #1: 'CN_CNMF'
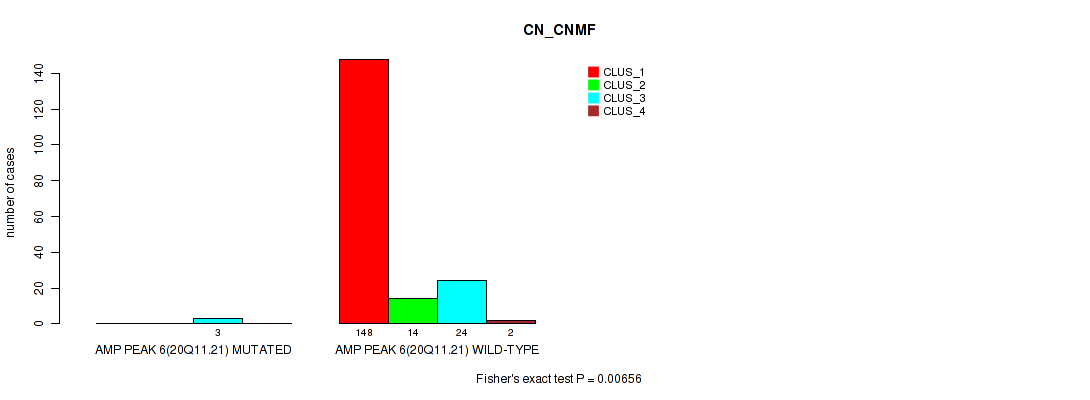
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S15. Gene #7: 'amp_21q22.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| AMP PEAK 7(21Q22.2) MUTATED | 0 | 0 | 14 | 0 |
| AMP PEAK 7(21Q22.2) WILD-TYPE | 148 | 14 | 13 | 2 |
Figure S15. Get High-res Image Gene #7: 'amp_21q22.2' versus Molecular Subtype #1: 'CN_CNMF'
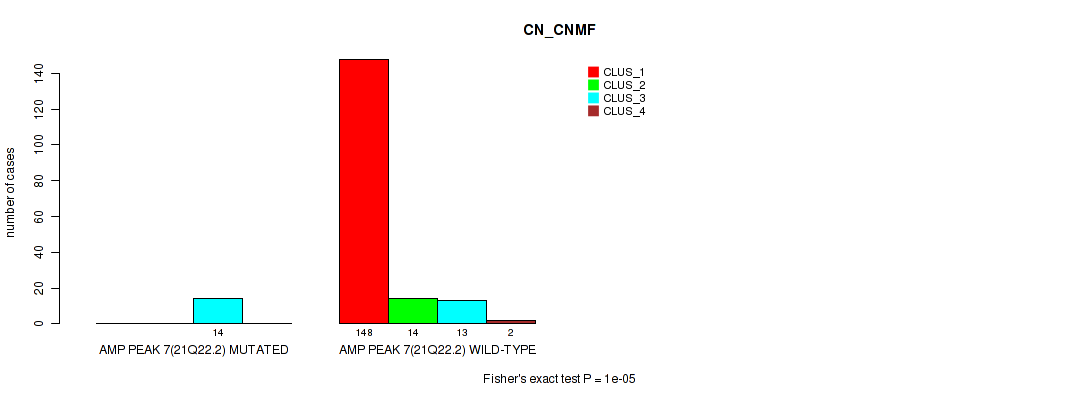
P value = 0.00054 (Fisher's exact test), Q value = 0.0031
Table S16. Gene #7: 'amp_21q22.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| AMP PEAK 7(21Q22.2) MUTATED | 0 | 11 | 2 | 0 |
| AMP PEAK 7(21Q22.2) WILD-TYPE | 33 | 50 | 45 | 44 |
Figure S16. Get High-res Image Gene #7: 'amp_21q22.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
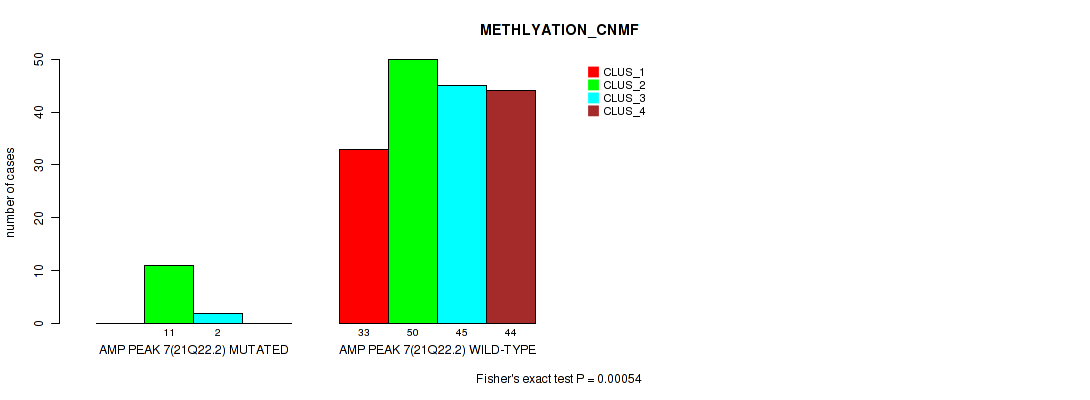
P value = 0.00335 (Fisher's exact test), Q value = 0.0097
Table S17. Gene #7: 'amp_21q22.2' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 52 | 44 |
| AMP PEAK 7(21Q22.2) MUTATED | 10 | 1 | 0 |
| AMP PEAK 7(21Q22.2) WILD-TYPE | 60 | 51 | 44 |
Figure S17. Get High-res Image Gene #7: 'amp_21q22.2' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
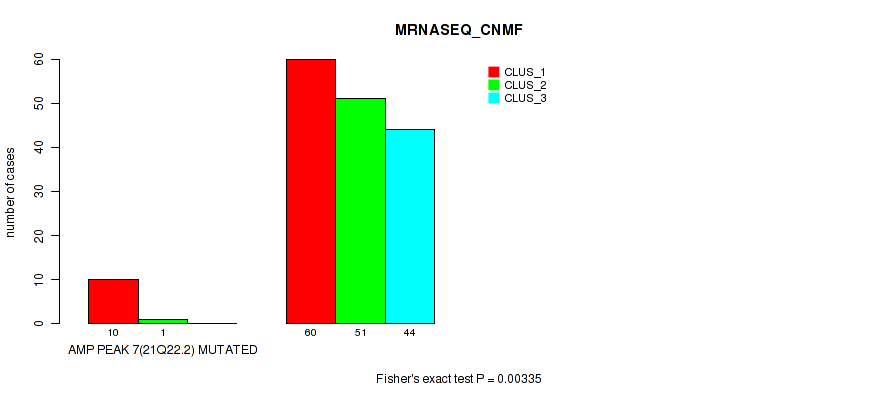
P value = 0.0365 (Fisher's exact test), Q value = 0.068
Table S18. Gene #7: 'amp_21q22.2' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| AMP PEAK 7(21Q22.2) MUTATED | 0 | 0 | 1 | 9 | 0 | 1 |
| AMP PEAK 7(21Q22.2) WILD-TYPE | 16 | 13 | 13 | 46 | 26 | 41 |
Figure S18. Get High-res Image Gene #7: 'amp_21q22.2' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
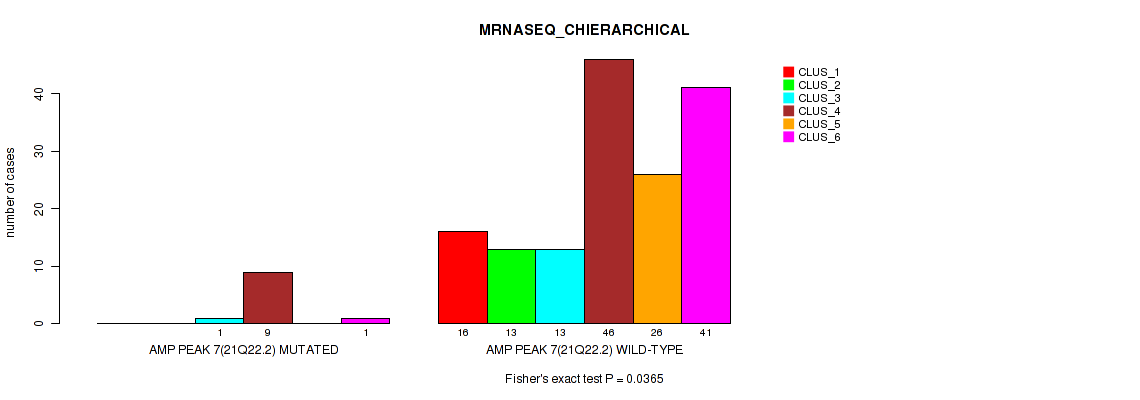
P value = 0.00164 (Fisher's exact test), Q value = 0.0058
Table S19. Gene #7: 'amp_21q22.2' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| AMP PEAK 7(21Q22.2) MUTATED | 1 | 4 | 7 | 0 |
| AMP PEAK 7(21Q22.2) WILD-TYPE | 57 | 33 | 34 | 43 |
Figure S19. Get High-res Image Gene #7: 'amp_21q22.2' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
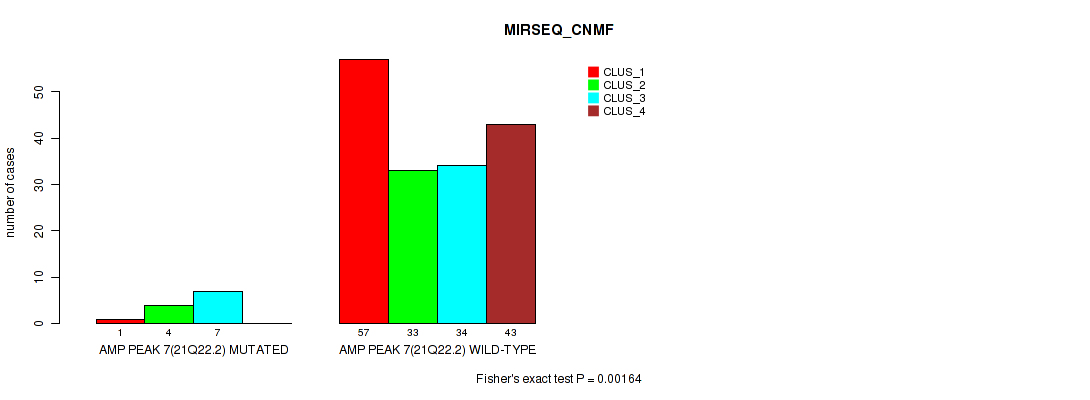
P value = 0.00062 (Fisher's exact test), Q value = 0.0034
Table S20. Gene #8: 'del_3p13' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 2(3P13) MUTATED | 2 | 0 | 6 | 0 |
| DEL PEAK 2(3P13) WILD-TYPE | 146 | 14 | 21 | 2 |
Figure S20. Get High-res Image Gene #8: 'del_3p13' versus Molecular Subtype #1: 'CN_CNMF'
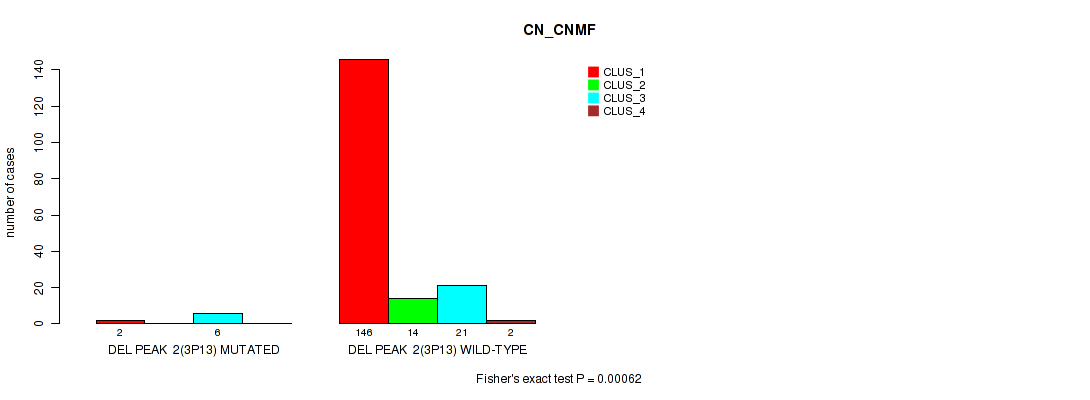
P value = 0.0457 (Fisher's exact test), Q value = 0.082
Table S21. Gene #8: 'del_3p13' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| DEL PEAK 2(3P13) MUTATED | 0 | 6 | 2 | 0 |
| DEL PEAK 2(3P13) WILD-TYPE | 33 | 55 | 45 | 44 |
Figure S21. Get High-res Image Gene #8: 'del_3p13' versus Molecular Subtype #2: 'METHLYATION_CNMF'
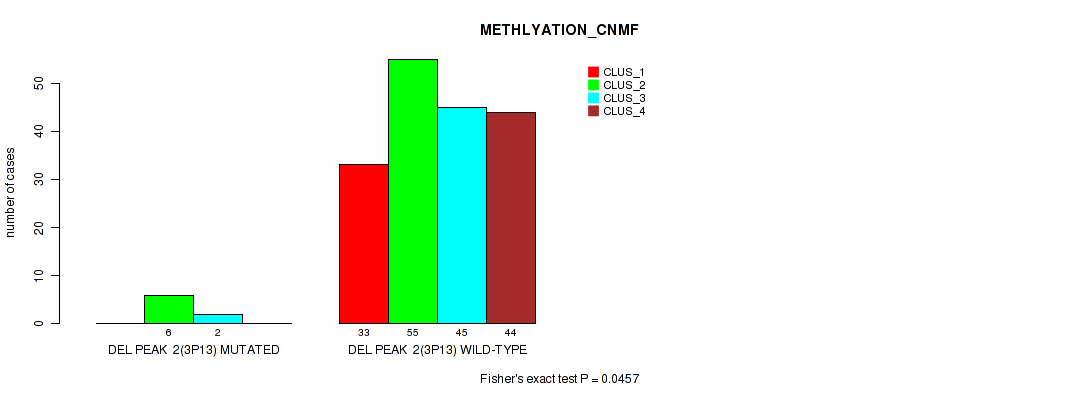
P value = 0.0368 (Fisher's exact test), Q value = 0.068
Table S22. Gene #8: 'del_3p13' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| DEL PEAK 2(3P13) MUTATED | 0 | 4 | 2 | 1 |
| DEL PEAK 2(3P13) WILD-TYPE | 58 | 33 | 39 | 42 |
Figure S22. Get High-res Image Gene #8: 'del_3p13' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
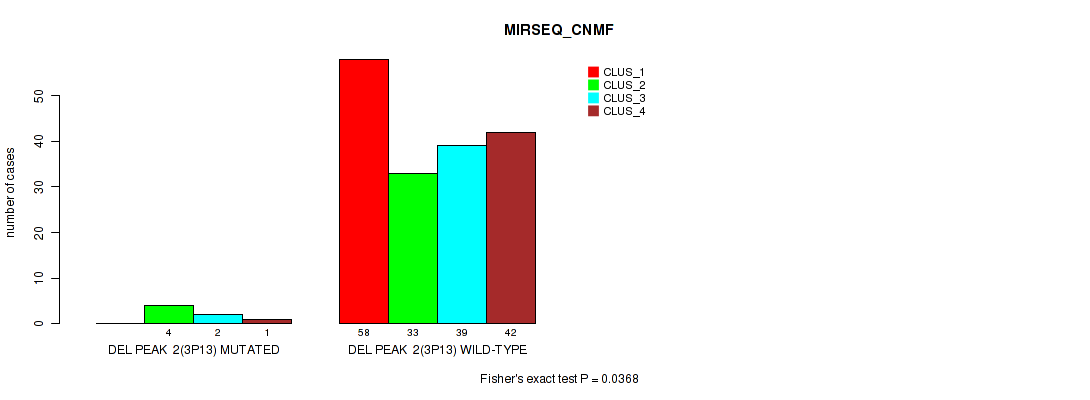
P value = 0.0484 (Fisher's exact test), Q value = 0.084
Table S23. Gene #8: 'del_3p13' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| DEL PEAK 2(3P13) MUTATED | 0 | 3 | 4 |
| DEL PEAK 2(3P13) WILD-TYPE | 67 | 31 | 74 |
Figure S23. Get High-res Image Gene #8: 'del_3p13' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
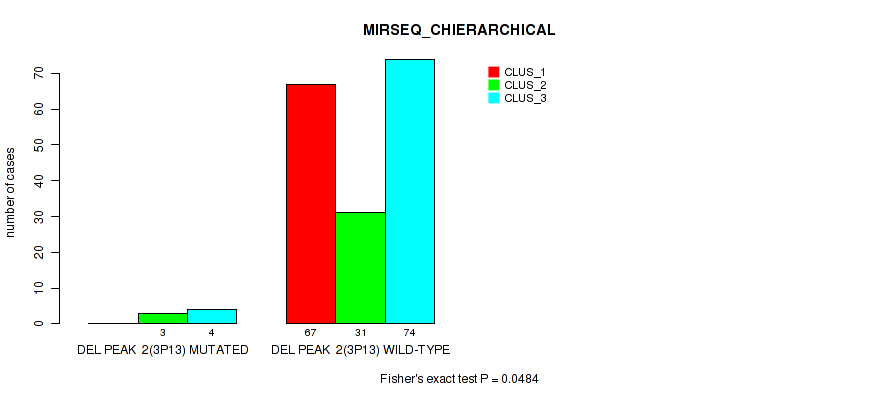
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
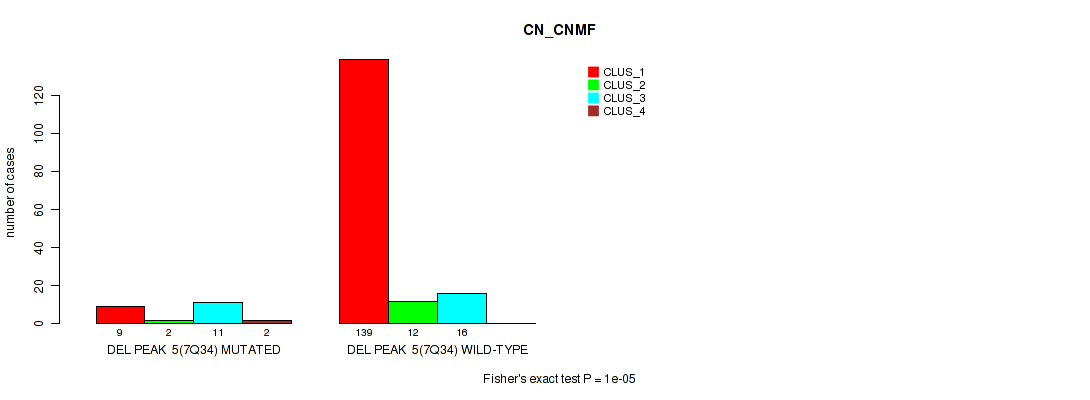
Table S24. Gene #9: 'del_5q31.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 3(5Q31.2) MUTATED | 1 | 1 | 15 | 1 |
| DEL PEAK 3(5Q31.2) WILD-TYPE | 147 | 13 | 12 | 1 |
Figure S24. Get High-res Image Gene #9: 'del_5q31.2' versus Molecular Subtype #1: 'CN_CNMF'
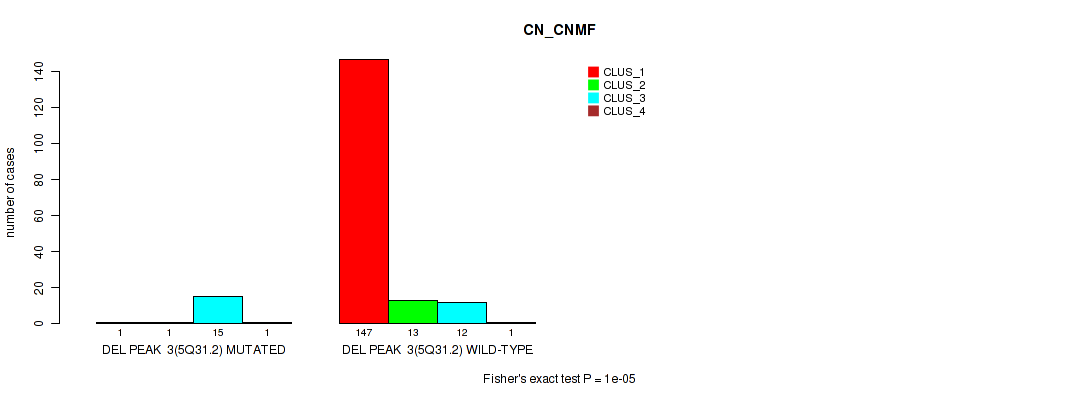
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S25. Gene #9: 'del_5q31.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| DEL PEAK 3(5Q31.2) MUTATED | 0 | 15 | 3 | 0 |
| DEL PEAK 3(5Q31.2) WILD-TYPE | 33 | 46 | 44 | 44 |
Figure S25. Get High-res Image Gene #9: 'del_5q31.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
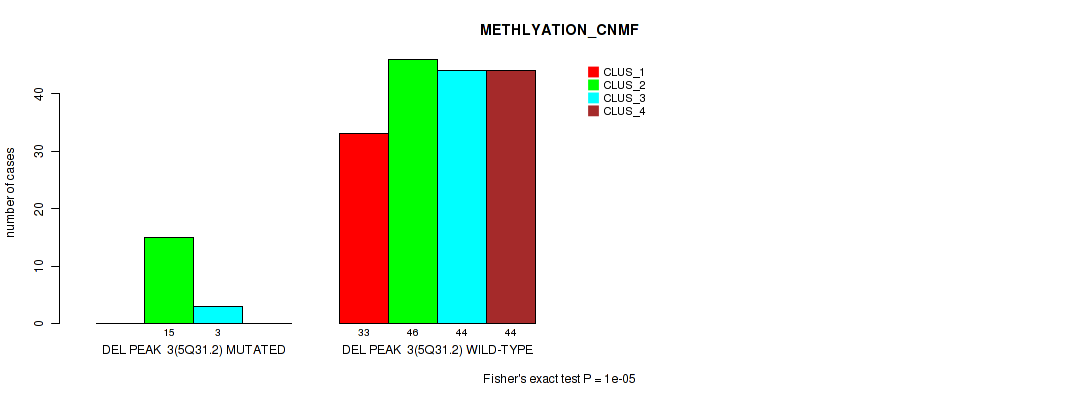
P value = 0.00535 (Fisher's exact test), Q value = 0.014
Table S26. Gene #9: 'del_5q31.2' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 52 | 44 |
| DEL PEAK 3(5Q31.2) MUTATED | 13 | 2 | 1 |
| DEL PEAK 3(5Q31.2) WILD-TYPE | 57 | 50 | 43 |
Figure S26. Get High-res Image Gene #9: 'del_5q31.2' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
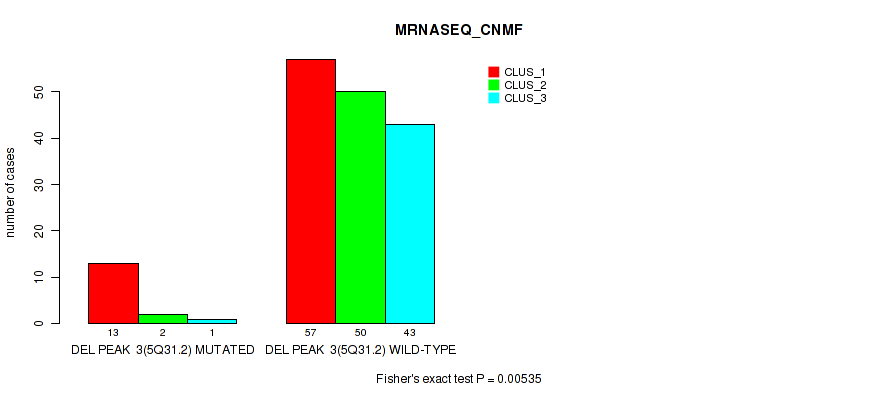
P value = 0.00083 (Fisher's exact test), Q value = 0.0038
Table S27. Gene #9: 'del_5q31.2' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| DEL PEAK 3(5Q31.2) MUTATED | 0 | 0 | 0 | 14 | 0 | 2 |
| DEL PEAK 3(5Q31.2) WILD-TYPE | 16 | 13 | 14 | 41 | 26 | 40 |
Figure S27. Get High-res Image Gene #9: 'del_5q31.2' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
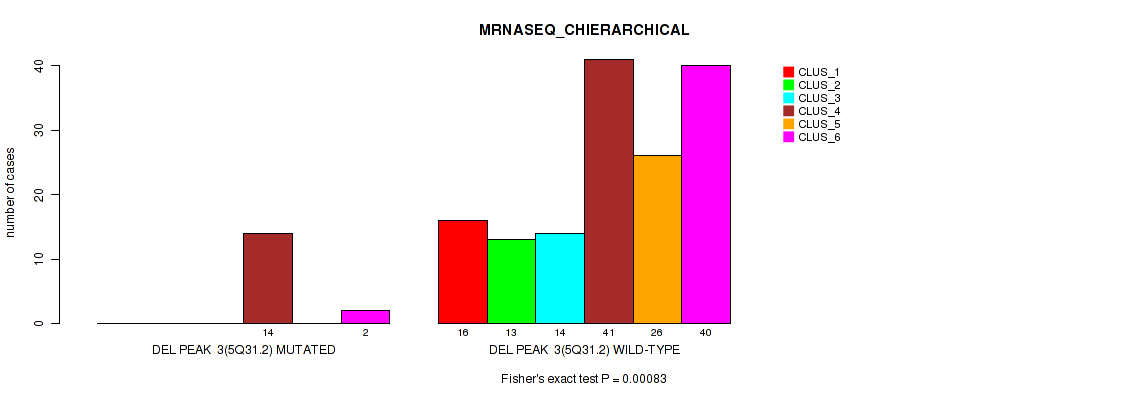
P value = 0.00069 (Fisher's exact test), Q value = 0.0034
Table S28. Gene #9: 'del_5q31.2' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| DEL PEAK 3(5Q31.2) MUTATED | 1 | 10 | 3 | 3 |
| DEL PEAK 3(5Q31.2) WILD-TYPE | 57 | 27 | 38 | 40 |
Figure S28. Get High-res Image Gene #9: 'del_5q31.2' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
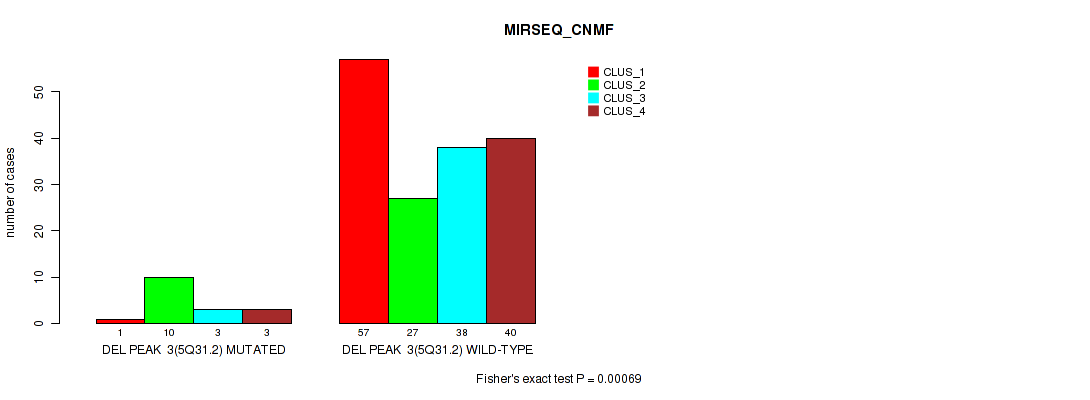
P value = 0.00119 (Fisher's exact test), Q value = 0.0047
Table S29. Gene #9: 'del_5q31.2' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| DEL PEAK 3(5Q31.2) MUTATED | 1 | 8 | 8 |
| DEL PEAK 3(5Q31.2) WILD-TYPE | 66 | 26 | 70 |
Figure S29. Get High-res Image Gene #9: 'del_5q31.2' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
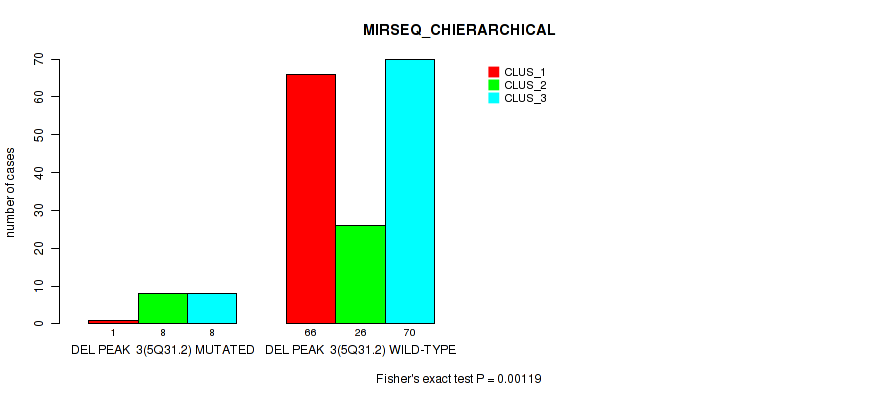
P value = 0.00021 (Fisher's exact test), Q value = 0.0016
Table S30. Gene #10: 'del_7p12.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 4(7P12.1) MUTATED | 7 | 0 | 8 | 1 |
| DEL PEAK 4(7P12.1) WILD-TYPE | 141 | 14 | 19 | 1 |
Figure S30. Get High-res Image Gene #10: 'del_7p12.1' versus Molecular Subtype #1: 'CN_CNMF'
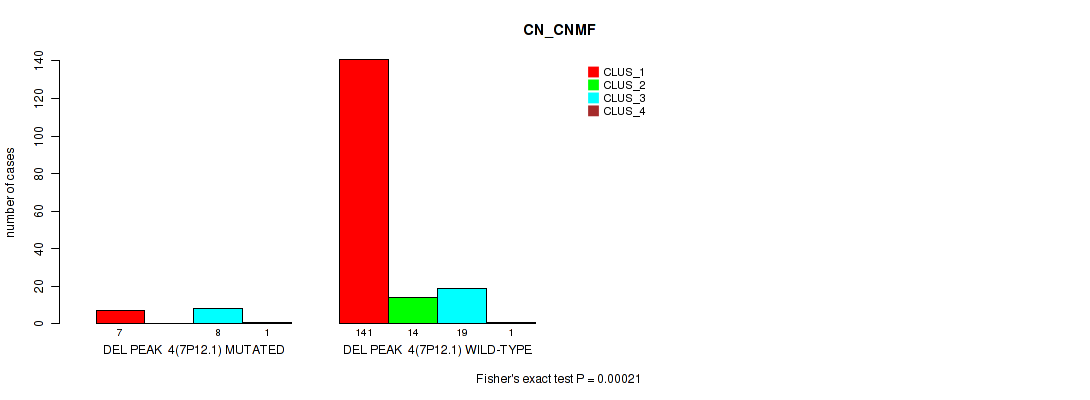
P value = 0.00037 (Fisher's exact test), Q value = 0.0023
Table S31. Gene #10: 'del_7p12.1' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| DEL PEAK 4(7P12.1) MUTATED | 0 | 13 | 1 | 2 |
| DEL PEAK 4(7P12.1) WILD-TYPE | 33 | 48 | 46 | 42 |
Figure S31. Get High-res Image Gene #10: 'del_7p12.1' versus Molecular Subtype #2: 'METHLYATION_CNMF'
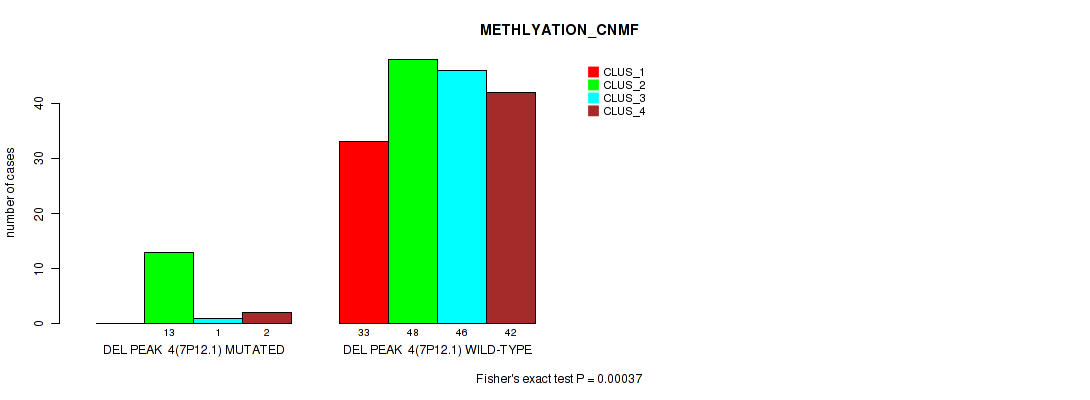
P value = 0.00183 (Fisher's exact test), Q value = 0.0063
Table S32. Gene #10: 'del_7p12.1' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 52 | 44 |
| DEL PEAK 4(7P12.1) MUTATED | 12 | 0 | 3 |
| DEL PEAK 4(7P12.1) WILD-TYPE | 58 | 52 | 41 |
Figure S32. Get High-res Image Gene #10: 'del_7p12.1' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
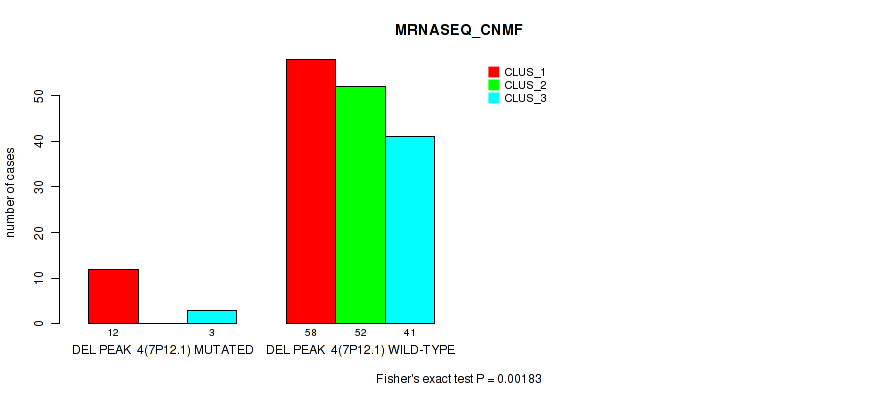
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S33. Gene #10: 'del_7p12.1' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| DEL PEAK 4(7P12.1) MUTATED | 0 | 0 | 0 | 15 | 0 | 0 |
| DEL PEAK 4(7P12.1) WILD-TYPE | 16 | 13 | 14 | 40 | 26 | 42 |
Figure S33. Get High-res Image Gene #10: 'del_7p12.1' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
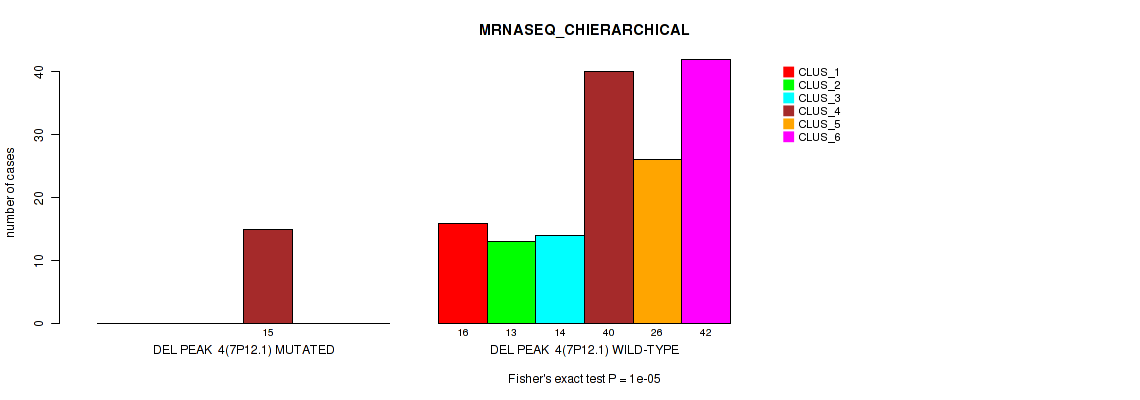
P value = 0.00568 (Fisher's exact test), Q value = 0.015
Table S34. Gene #10: 'del_7p12.1' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| DEL PEAK 4(7P12.1) MUTATED | 1 | 8 | 2 | 5 |
| DEL PEAK 4(7P12.1) WILD-TYPE | 57 | 29 | 39 | 38 |
Figure S34. Get High-res Image Gene #10: 'del_7p12.1' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
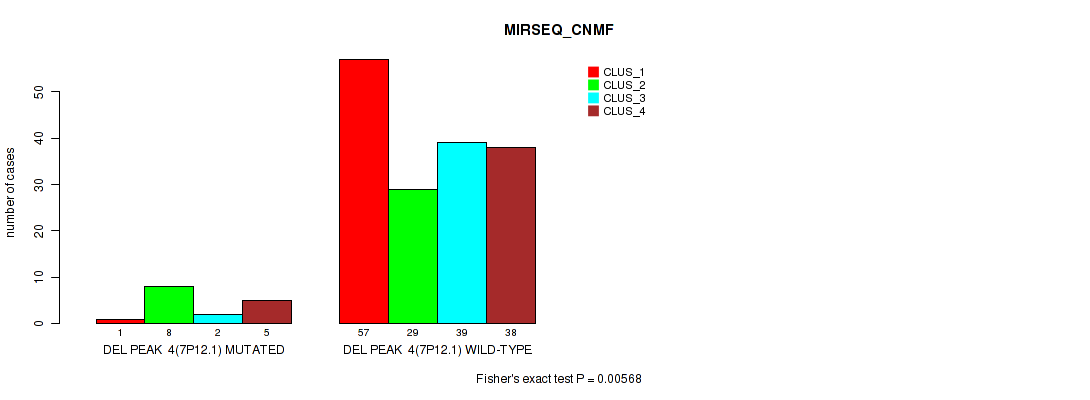
P value = 0.00684 (Fisher's exact test), Q value = 0.016
Table S35. Gene #10: 'del_7p12.1' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| DEL PEAK 4(7P12.1) MUTATED | 1 | 6 | 9 |
| DEL PEAK 4(7P12.1) WILD-TYPE | 66 | 28 | 69 |
Figure S35. Get High-res Image Gene #10: 'del_7p12.1' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
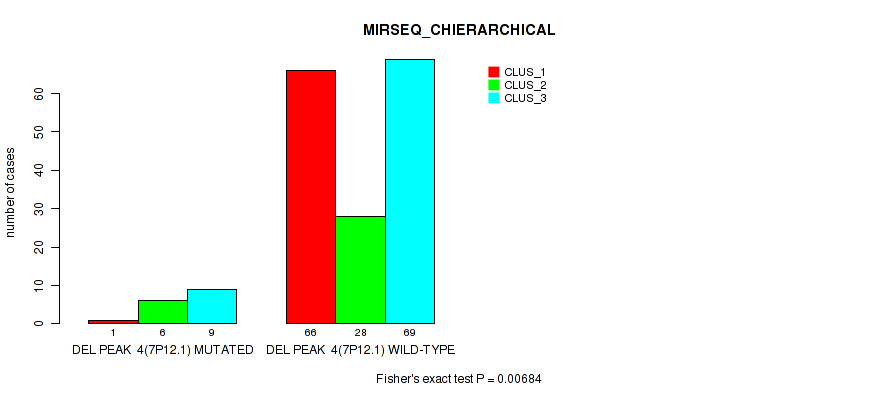
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S36. Gene #11: 'del_7q34' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 5(7Q34) MUTATED | 9 | 2 | 11 | 2 |
| DEL PEAK 5(7Q34) WILD-TYPE | 139 | 12 | 16 | 0 |
Figure S36. Get High-res Image Gene #11: 'del_7q34' versus Molecular Subtype #1: 'CN_CNMF'
P value = 0.00011 (Fisher's exact test), Q value = 0.00094
Table S37. Gene #11: 'del_7q34' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| DEL PEAK 5(7Q34) MUTATED | 1 | 18 | 2 | 3 |
| DEL PEAK 5(7Q34) WILD-TYPE | 32 | 43 | 45 | 41 |
Figure S37. Get High-res Image Gene #11: 'del_7q34' versus Molecular Subtype #2: 'METHLYATION_CNMF'
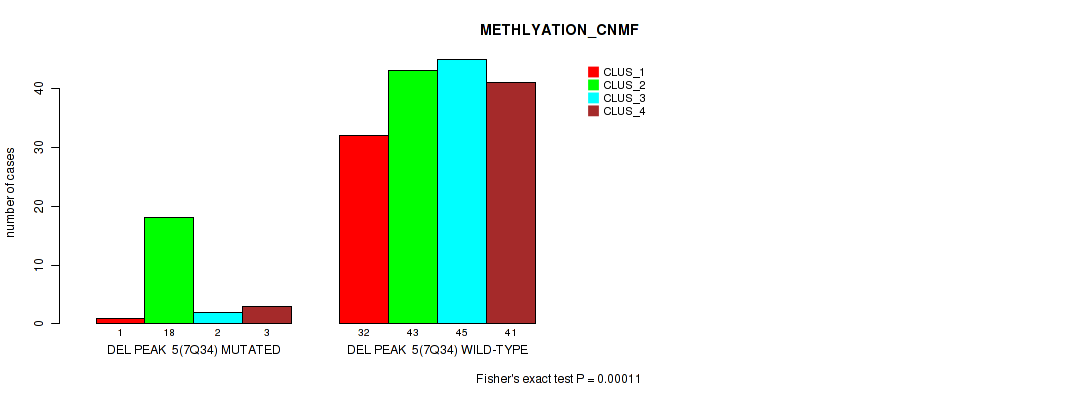
P value = 0.023 (Fisher's exact test), Q value = 0.047
Table S38. Gene #11: 'del_7q34' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 52 | 44 |
| DEL PEAK 5(7Q34) MUTATED | 14 | 2 | 4 |
| DEL PEAK 5(7Q34) WILD-TYPE | 56 | 50 | 40 |
Figure S38. Get High-res Image Gene #11: 'del_7q34' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
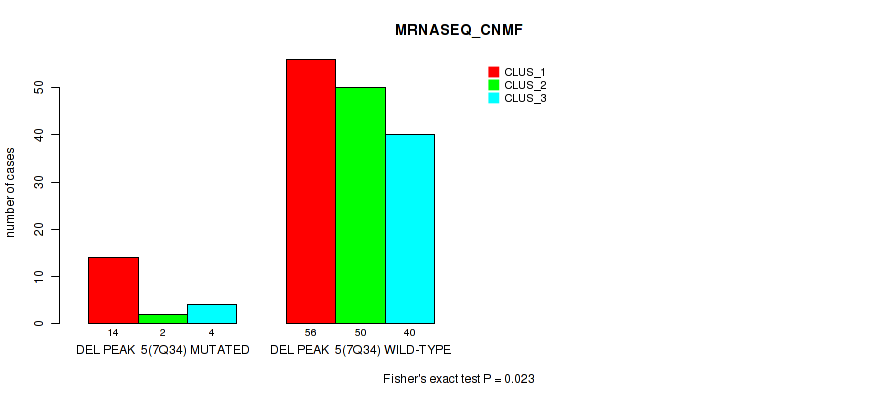
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S39. Gene #11: 'del_7q34' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| DEL PEAK 5(7Q34) MUTATED | 1 | 1 | 0 | 18 | 0 | 0 |
| DEL PEAK 5(7Q34) WILD-TYPE | 15 | 12 | 14 | 37 | 26 | 42 |
Figure S39. Get High-res Image Gene #11: 'del_7q34' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
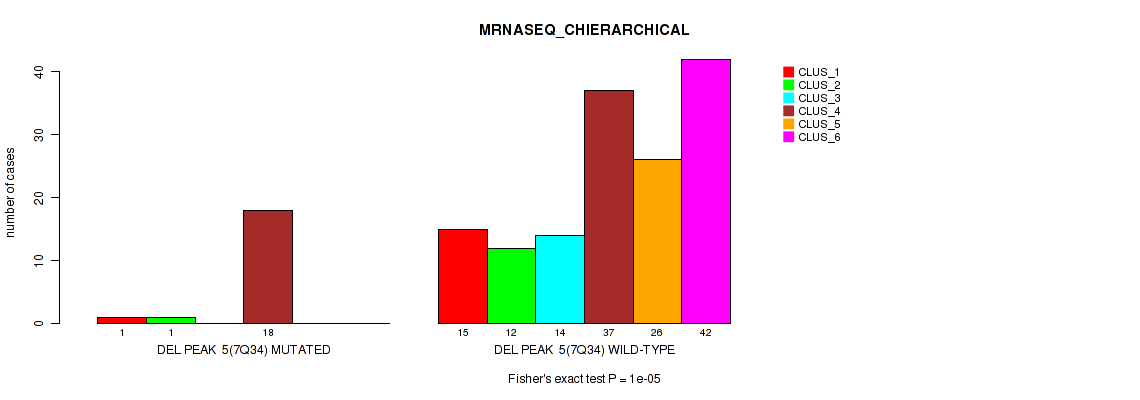
P value = 0.00087 (Fisher's exact test), Q value = 0.0039
Table S40. Gene #11: 'del_7q34' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| DEL PEAK 5(7Q34) MUTATED | 2 | 12 | 5 | 4 |
| DEL PEAK 5(7Q34) WILD-TYPE | 56 | 25 | 36 | 39 |
Figure S40. Get High-res Image Gene #11: 'del_7q34' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
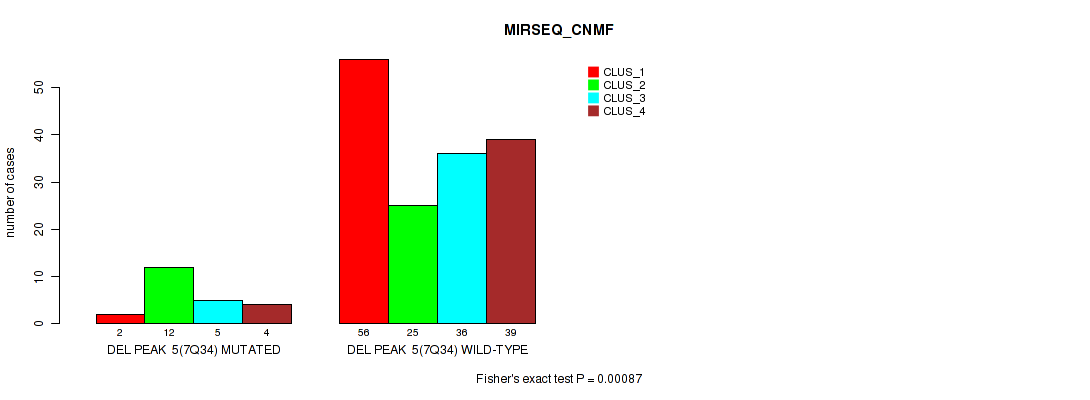
P value = 0.00068 (Fisher's exact test), Q value = 0.0034
Table S41. Gene #11: 'del_7q34' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| DEL PEAK 5(7Q34) MUTATED | 2 | 10 | 11 |
| DEL PEAK 5(7Q34) WILD-TYPE | 65 | 24 | 67 |
Figure S41. Get High-res Image Gene #11: 'del_7q34' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
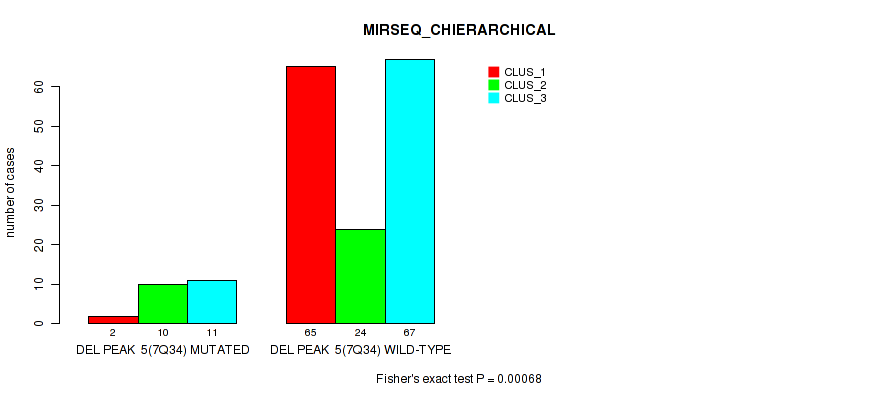
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S42. Gene #12: 'del_7q34' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 6(7Q34) MUTATED | 9 | 2 | 11 | 2 |
| DEL PEAK 6(7Q34) WILD-TYPE | 139 | 12 | 16 | 0 |
Figure S42. Get High-res Image Gene #12: 'del_7q34' versus Molecular Subtype #1: 'CN_CNMF'
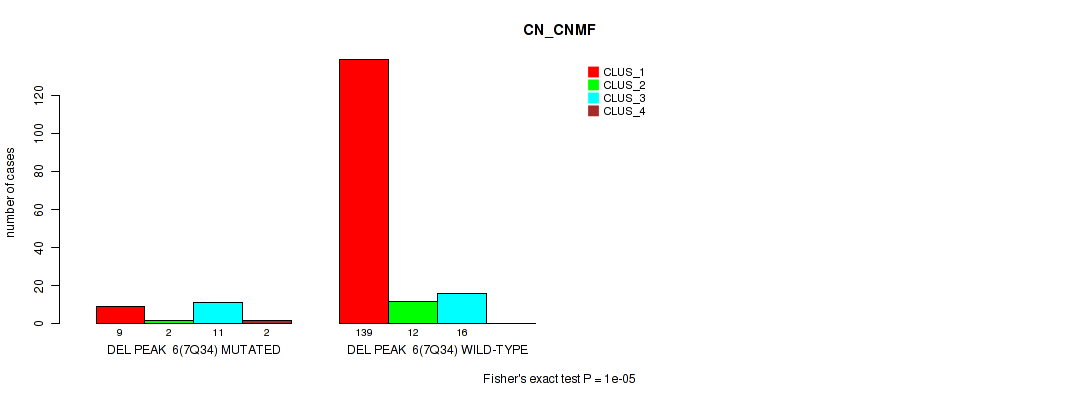
P value = 0.00012 (Fisher's exact test), Q value = 0.00096
Table S43. Gene #12: 'del_7q34' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| DEL PEAK 6(7Q34) MUTATED | 1 | 18 | 2 | 3 |
| DEL PEAK 6(7Q34) WILD-TYPE | 32 | 43 | 45 | 41 |
Figure S43. Get High-res Image Gene #12: 'del_7q34' versus Molecular Subtype #2: 'METHLYATION_CNMF'
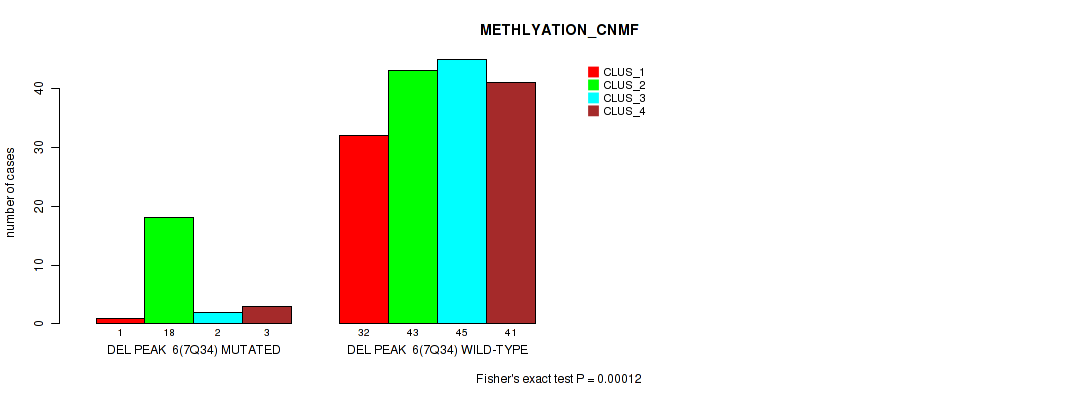
P value = 0.0217 (Fisher's exact test), Q value = 0.046
Table S44. Gene #12: 'del_7q34' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 52 | 44 |
| DEL PEAK 6(7Q34) MUTATED | 14 | 2 | 4 |
| DEL PEAK 6(7Q34) WILD-TYPE | 56 | 50 | 40 |
Figure S44. Get High-res Image Gene #12: 'del_7q34' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
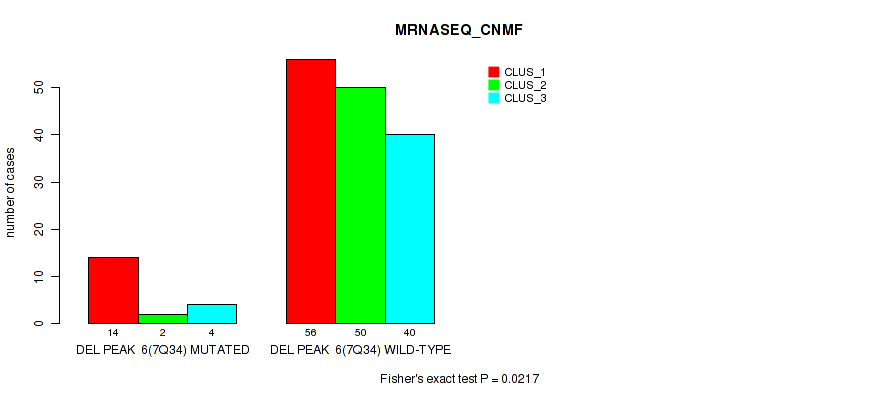
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S45. Gene #12: 'del_7q34' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| DEL PEAK 6(7Q34) MUTATED | 1 | 1 | 0 | 18 | 0 | 0 |
| DEL PEAK 6(7Q34) WILD-TYPE | 15 | 12 | 14 | 37 | 26 | 42 |
Figure S45. Get High-res Image Gene #12: 'del_7q34' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
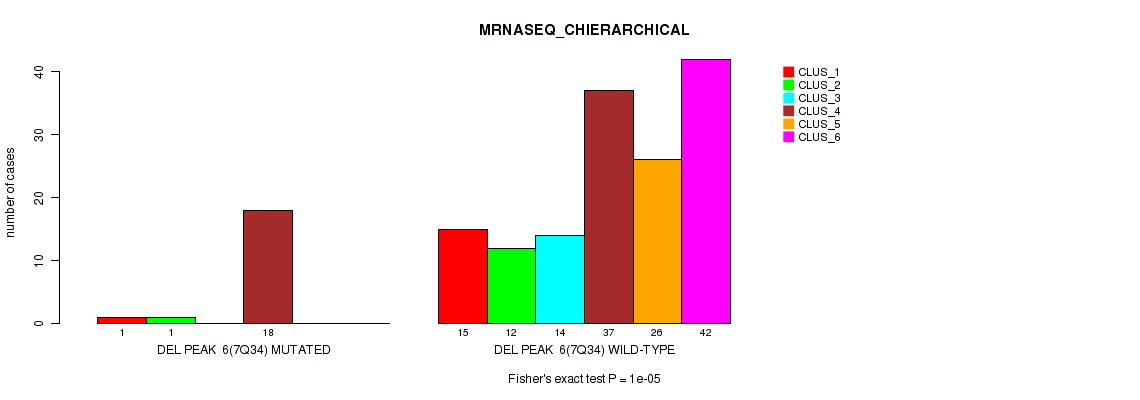
P value = 9e-04 (Fisher's exact test), Q value = 0.0039
Table S46. Gene #12: 'del_7q34' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| DEL PEAK 6(7Q34) MUTATED | 2 | 12 | 5 | 4 |
| DEL PEAK 6(7Q34) WILD-TYPE | 56 | 25 | 36 | 39 |
Figure S46. Get High-res Image Gene #12: 'del_7q34' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
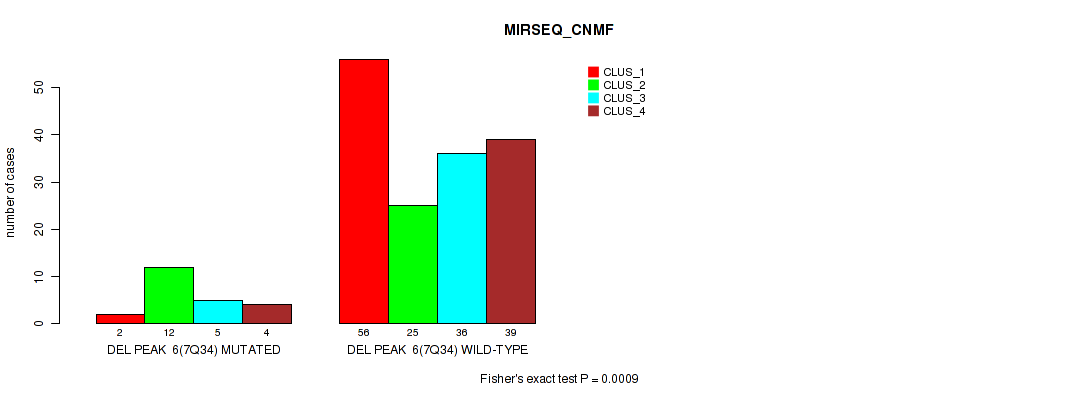
P value = 0.00072 (Fisher's exact test), Q value = 0.0035
Table S47. Gene #12: 'del_7q34' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| DEL PEAK 6(7Q34) MUTATED | 2 | 10 | 11 |
| DEL PEAK 6(7Q34) WILD-TYPE | 65 | 24 | 67 |
Figure S47. Get High-res Image Gene #12: 'del_7q34' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
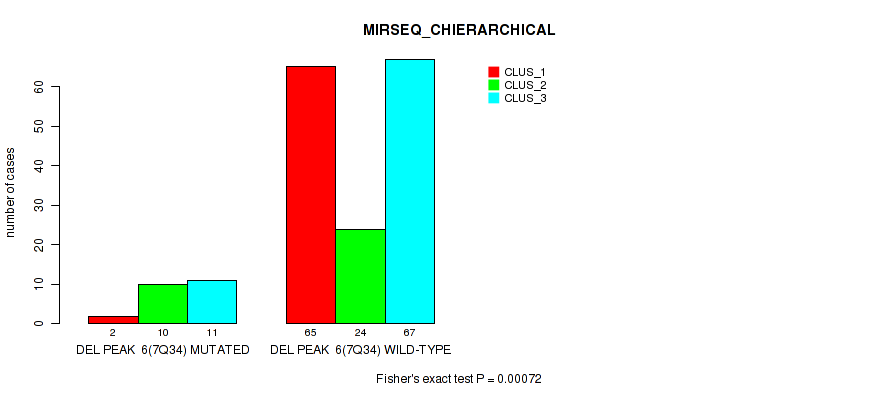
P value = 3e-05 (Fisher's exact test), Q value = 0.00028
Table S48. Gene #14: 'del_12p13.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 10(12P13.2) MUTATED | 2 | 0 | 7 | 1 |
| DEL PEAK 10(12P13.2) WILD-TYPE | 146 | 14 | 20 | 1 |
Figure S48. Get High-res Image Gene #14: 'del_12p13.2' versus Molecular Subtype #1: 'CN_CNMF'
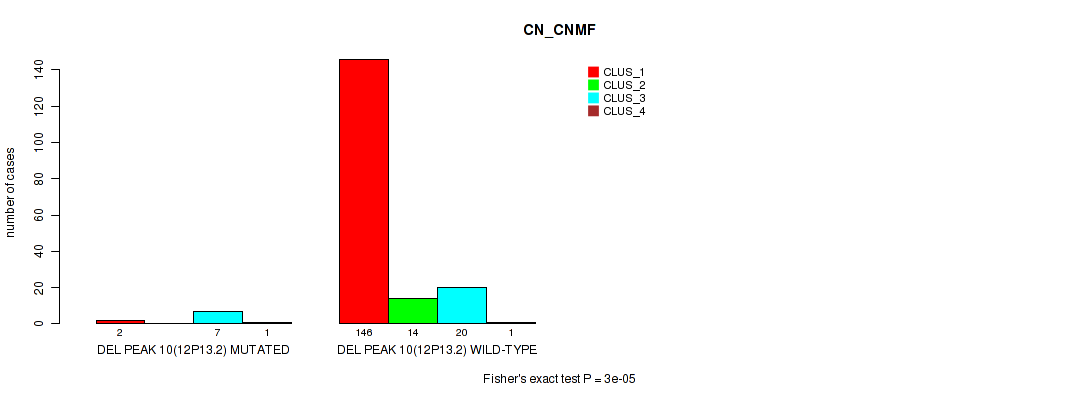
P value = 0.00158 (Fisher's exact test), Q value = 0.0057
Table S49. Gene #14: 'del_12p13.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| DEL PEAK 10(12P13.2) MUTATED | 0 | 9 | 1 | 0 |
| DEL PEAK 10(12P13.2) WILD-TYPE | 33 | 52 | 46 | 44 |
Figure S49. Get High-res Image Gene #14: 'del_12p13.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
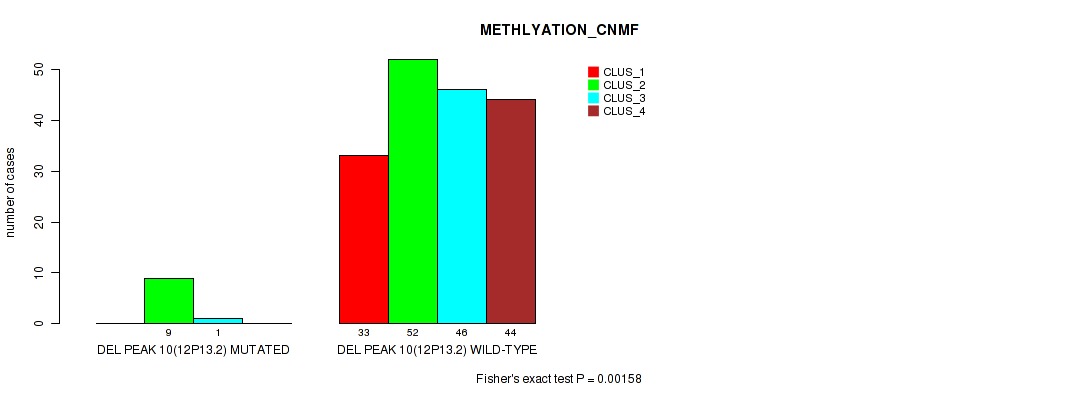
P value = 0.0101 (Fisher's exact test), Q value = 0.022
Table S50. Gene #14: 'del_12p13.2' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| DEL PEAK 10(12P13.2) MUTATED | 0 | 4 | 6 |
| DEL PEAK 10(12P13.2) WILD-TYPE | 67 | 30 | 72 |
Figure S50. Get High-res Image Gene #14: 'del_12p13.2' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
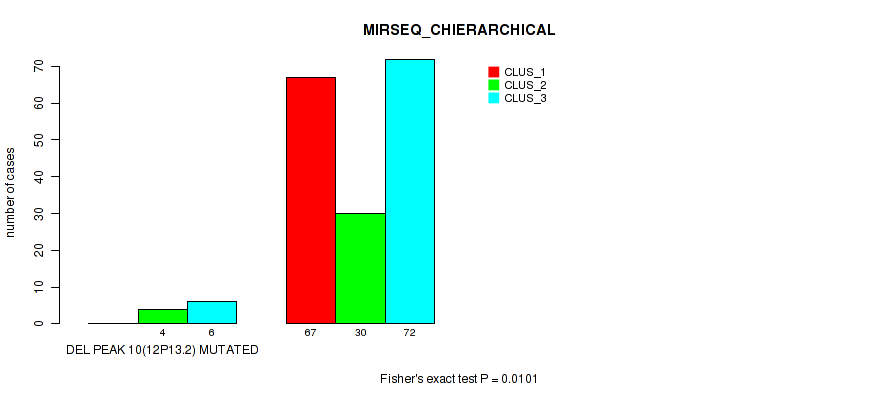
P value = 0.00121 (Fisher's exact test), Q value = 0.0047
Table S51. Gene #15: 'del_12q21.33' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 11(12Q21.33) MUTATED | 0 | 0 | 2 | 1 |
| DEL PEAK 11(12Q21.33) WILD-TYPE | 148 | 14 | 25 | 1 |
Figure S51. Get High-res Image Gene #15: 'del_12q21.33' versus Molecular Subtype #1: 'CN_CNMF'
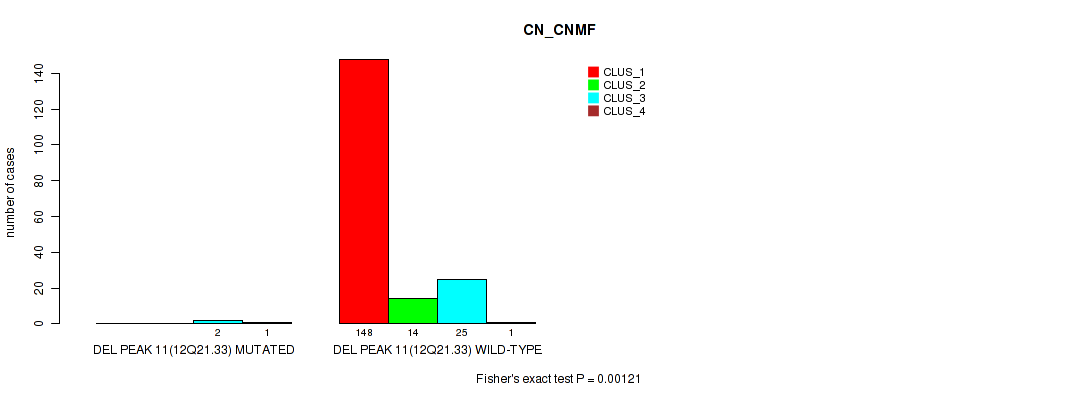
P value = 0.0086 (Fisher's exact test), Q value = 0.02
Table S52. Gene #15: 'del_12q21.33' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| DEL PEAK 11(12Q21.33) MUTATED | 0 | 3 | 0 | 0 |
| DEL PEAK 11(12Q21.33) WILD-TYPE | 58 | 34 | 41 | 43 |
Figure S52. Get High-res Image Gene #15: 'del_12q21.33' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
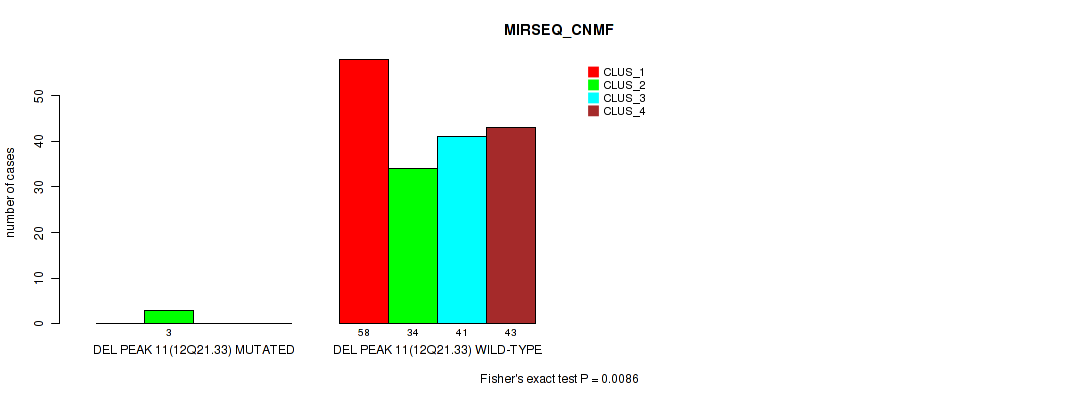
P value = 0.00611 (Fisher's exact test), Q value = 0.015
Table S53. Gene #15: 'del_12q21.33' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| DEL PEAK 11(12Q21.33) MUTATED | 0 | 3 | 0 |
| DEL PEAK 11(12Q21.33) WILD-TYPE | 67 | 31 | 78 |
Figure S53. Get High-res Image Gene #15: 'del_12q21.33' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
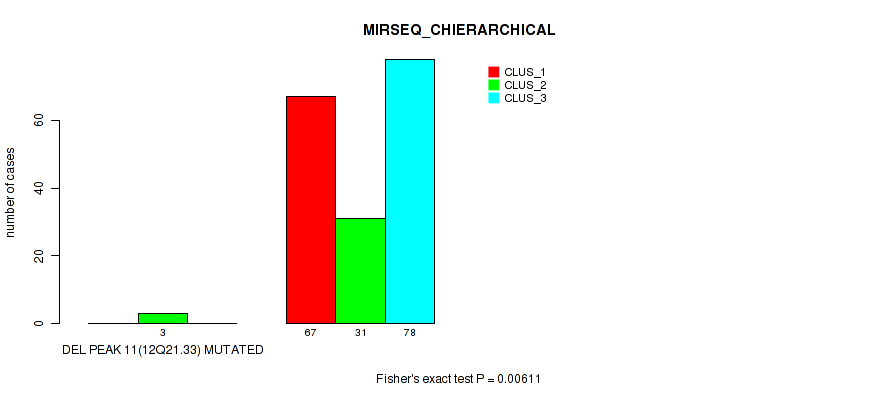
P value = 0.00152 (Fisher's exact test), Q value = 0.0057
Table S54. Gene #16: 'del_16q23.1' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 12(16Q23.1) MUTATED | 3 | 0 | 6 | 0 |
| DEL PEAK 12(16Q23.1) WILD-TYPE | 145 | 14 | 21 | 2 |
Figure S54. Get High-res Image Gene #16: 'del_16q23.1' versus Molecular Subtype #1: 'CN_CNMF'
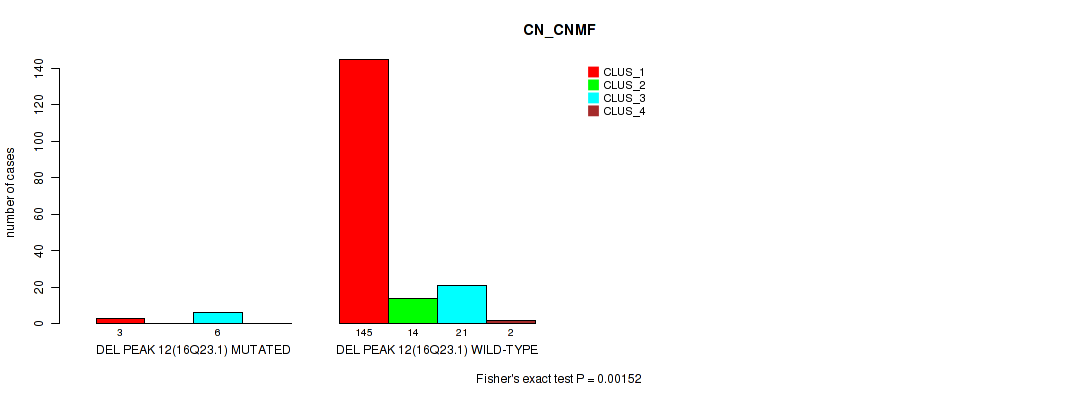
P value = 0.0226 (Fisher's exact test), Q value = 0.047
Table S55. Gene #16: 'del_16q23.1' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| DEL PEAK 12(16Q23.1) MUTATED | 0 | 7 | 2 | 0 |
| DEL PEAK 12(16Q23.1) WILD-TYPE | 33 | 54 | 45 | 44 |
Figure S55. Get High-res Image Gene #16: 'del_16q23.1' versus Molecular Subtype #2: 'METHLYATION_CNMF'
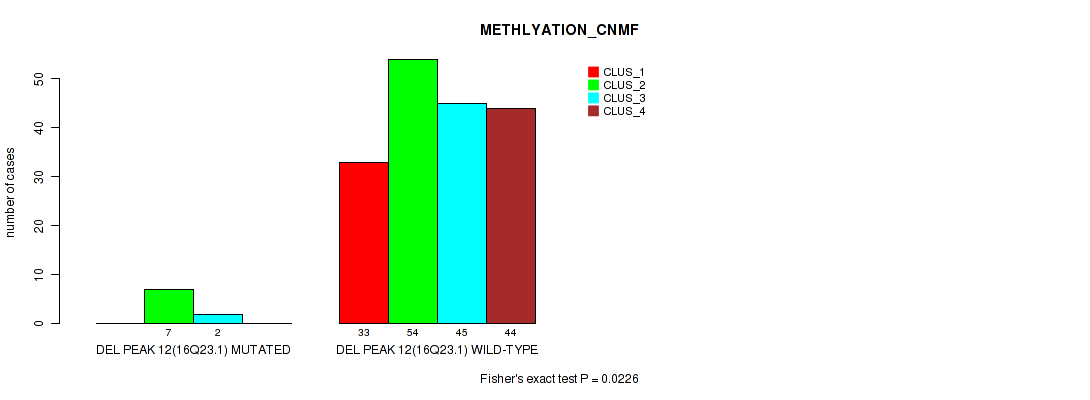
P value = 0.0451 (Fisher's exact test), Q value = 0.082
Table S56. Gene #16: 'del_16q23.1' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 58 | 37 | 41 | 43 |
| DEL PEAK 12(16Q23.1) MUTATED | 1 | 4 | 3 | 0 |
| DEL PEAK 12(16Q23.1) WILD-TYPE | 57 | 33 | 38 | 43 |
Figure S56. Get High-res Image Gene #16: 'del_16q23.1' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
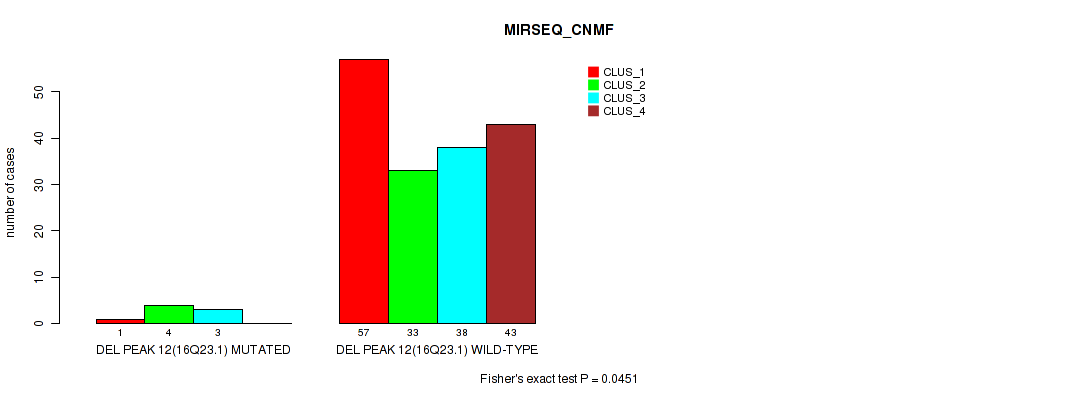
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S57. Gene #17: 'del_17p13.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 13(17P13.2) MUTATED | 0 | 0 | 15 | 0 |
| DEL PEAK 13(17P13.2) WILD-TYPE | 148 | 14 | 12 | 2 |
Figure S57. Get High-res Image Gene #17: 'del_17p13.2' versus Molecular Subtype #1: 'CN_CNMF'
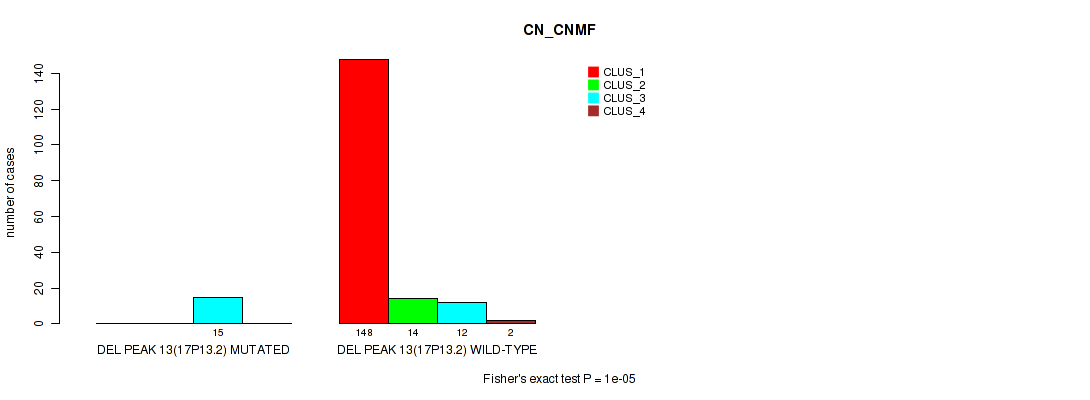
P value = 0.00025 (Fisher's exact test), Q value = 0.0017
Table S58. Gene #17: 'del_17p13.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| DEL PEAK 13(17P13.2) MUTATED | 0 | 12 | 3 | 0 |
| DEL PEAK 13(17P13.2) WILD-TYPE | 33 | 49 | 44 | 44 |
Figure S58. Get High-res Image Gene #17: 'del_17p13.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
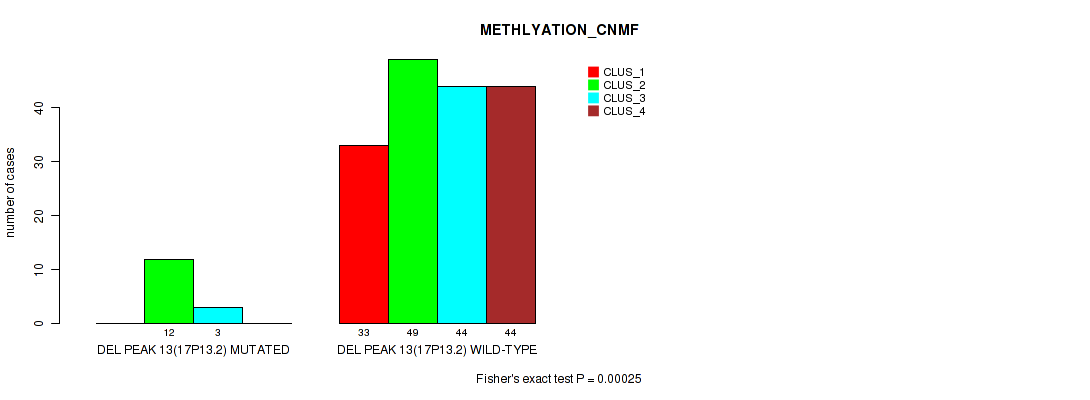
P value = 0.0111 (Fisher's exact test), Q value = 0.024
Table S59. Gene #17: 'del_17p13.2' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 70 | 52 | 44 |
| DEL PEAK 13(17P13.2) MUTATED | 12 | 2 | 1 |
| DEL PEAK 13(17P13.2) WILD-TYPE | 58 | 50 | 43 |
Figure S59. Get High-res Image Gene #17: 'del_17p13.2' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
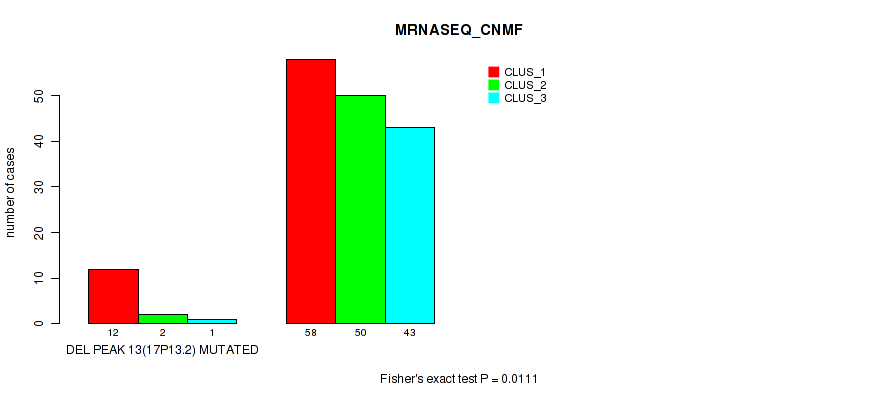
P value = 0.00023 (Fisher's exact test), Q value = 0.0016
Table S60. Gene #17: 'del_17p13.2' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| DEL PEAK 13(17P13.2) MUTATED | 0 | 0 | 0 | 14 | 0 | 1 |
| DEL PEAK 13(17P13.2) WILD-TYPE | 16 | 13 | 14 | 41 | 26 | 41 |
Figure S60. Get High-res Image Gene #17: 'del_17p13.2' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
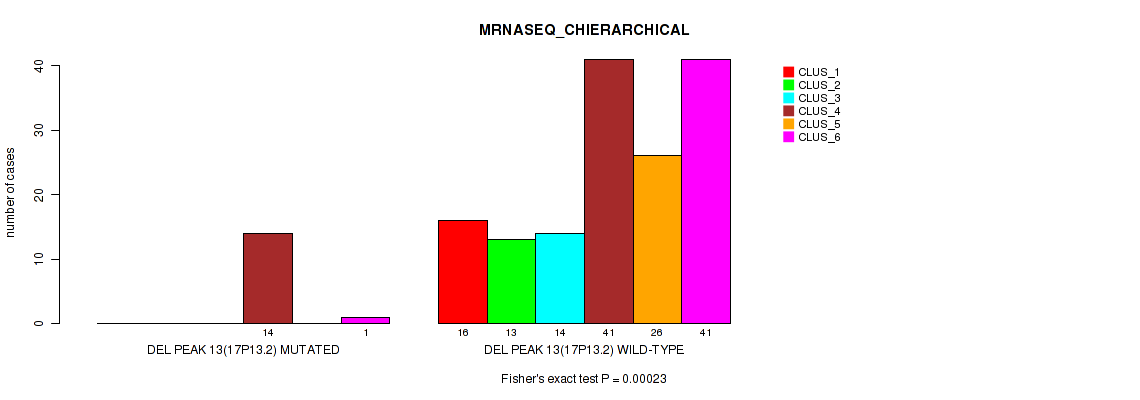
P value = 0.0012 (Fisher's exact test), Q value = 0.0047
Table S61. Gene #17: 'del_17p13.2' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| DEL PEAK 13(17P13.2) MUTATED | 0 | 5 | 10 |
| DEL PEAK 13(17P13.2) WILD-TYPE | 67 | 29 | 68 |
Figure S61. Get High-res Image Gene #17: 'del_17p13.2' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
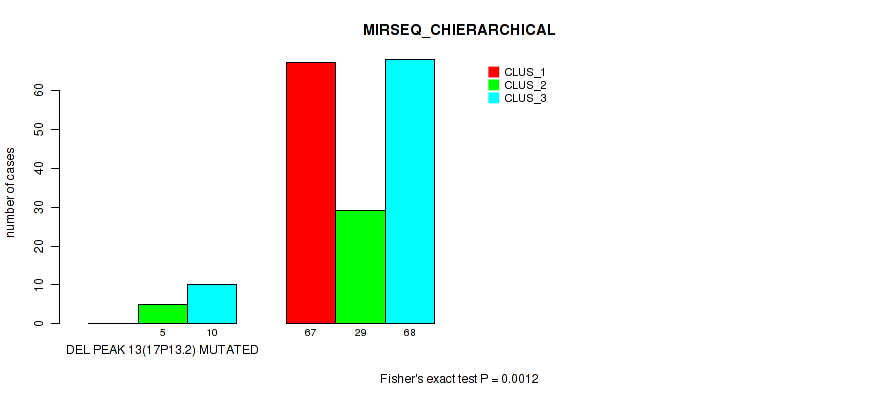
P value = 1e-05 (Fisher's exact test), Q value = 0.00011
Table S62. Gene #18: 'del_17q11.2' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 14(17Q11.2) MUTATED | 2 | 2 | 9 | 0 |
| DEL PEAK 14(17Q11.2) WILD-TYPE | 146 | 12 | 18 | 2 |
Figure S62. Get High-res Image Gene #18: 'del_17q11.2' versus Molecular Subtype #1: 'CN_CNMF'
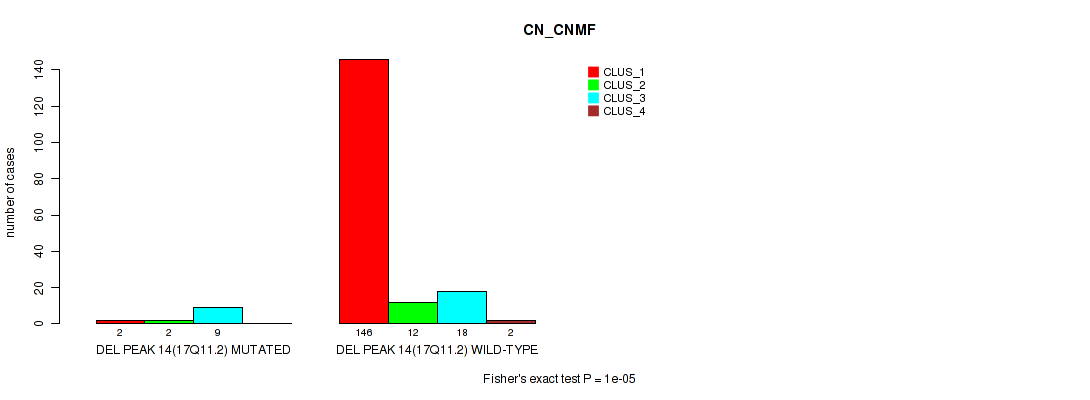
P value = 0.00512 (Fisher's exact test), Q value = 0.014
Table S63. Gene #18: 'del_17q11.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| DEL PEAK 14(17Q11.2) MUTATED | 0 | 9 | 4 | 0 |
| DEL PEAK 14(17Q11.2) WILD-TYPE | 33 | 52 | 43 | 44 |
Figure S63. Get High-res Image Gene #18: 'del_17q11.2' versus Molecular Subtype #2: 'METHLYATION_CNMF'
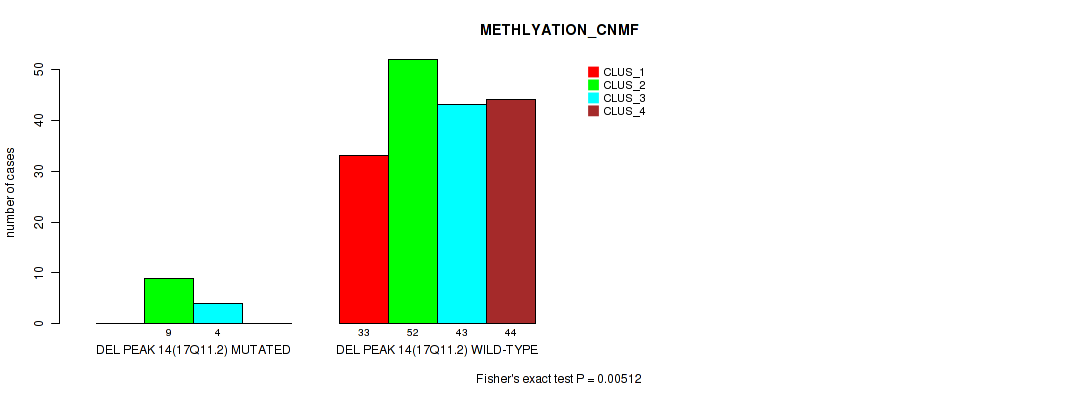
P value = 0.00658 (Fisher's exact test), Q value = 0.016
Table S64. Gene #18: 'del_17q11.2' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 14 | 55 | 26 | 42 |
| DEL PEAK 14(17Q11.2) MUTATED | 0 | 1 | 0 | 11 | 0 | 1 |
| DEL PEAK 14(17Q11.2) WILD-TYPE | 16 | 12 | 14 | 44 | 26 | 41 |
Figure S64. Get High-res Image Gene #18: 'del_17q11.2' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
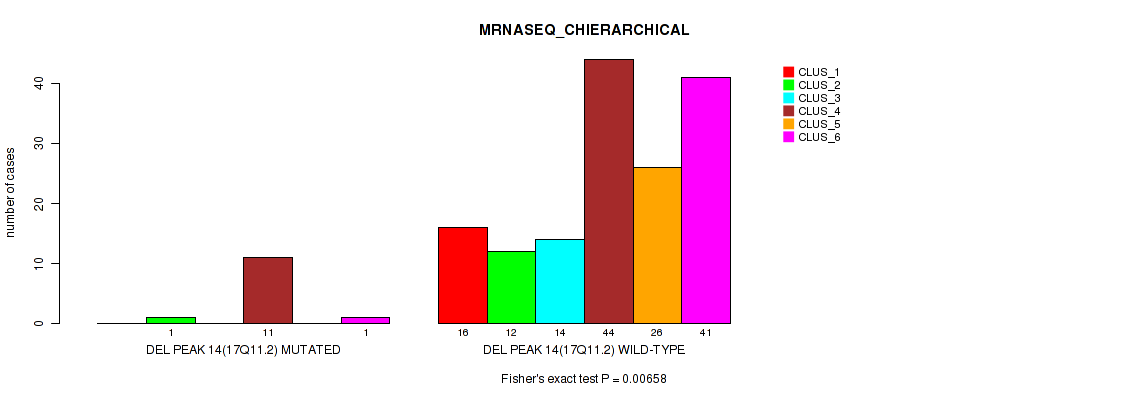
P value = 0.00383 (Fisher's exact test), Q value = 0.011
Table S65. Gene #18: 'del_17q11.2' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| DEL PEAK 14(17Q11.2) MUTATED | 0 | 4 | 9 |
| DEL PEAK 14(17Q11.2) WILD-TYPE | 67 | 30 | 69 |
Figure S65. Get High-res Image Gene #18: 'del_17q11.2' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
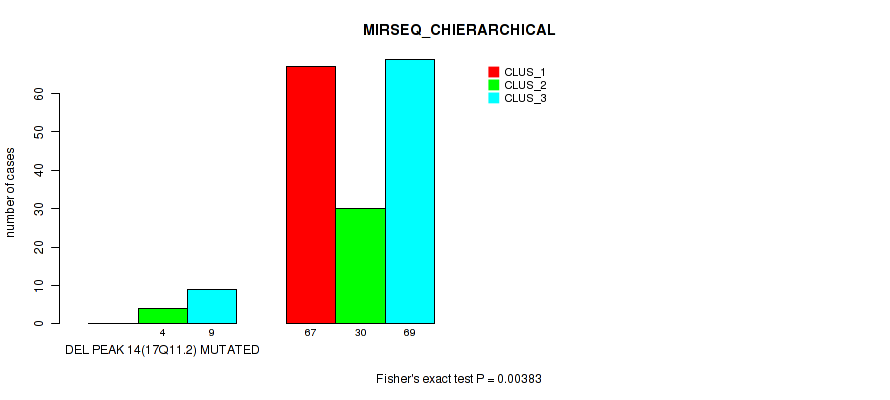
P value = 2e-05 (Fisher's exact test), Q value = 2e-04
Table S66. Gene #19: 'del_18p11.21' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 15(18P11.21) MUTATED | 1 | 0 | 8 | 0 |
| DEL PEAK 15(18P11.21) WILD-TYPE | 147 | 14 | 19 | 2 |
Figure S66. Get High-res Image Gene #19: 'del_18p11.21' versus Molecular Subtype #1: 'CN_CNMF'
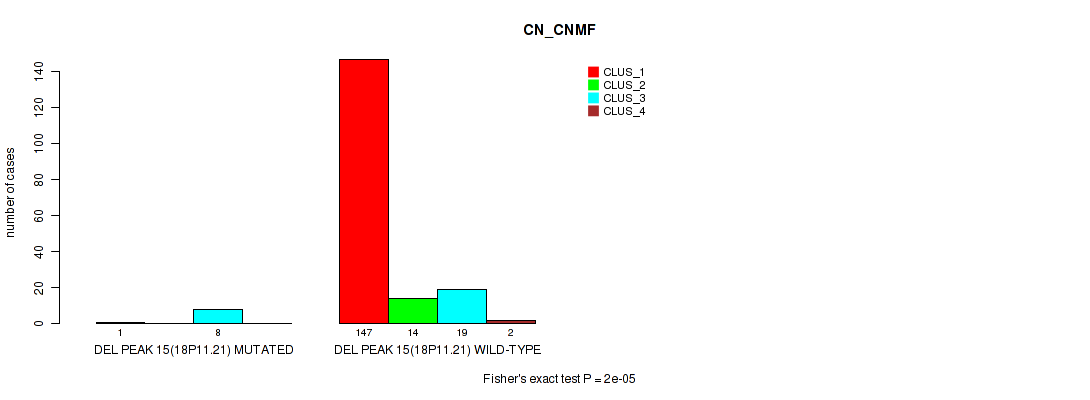
P value = 0.00387 (Fisher's exact test), Q value = 0.011
Table S67. Gene #19: 'del_18p11.21' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 33 | 61 | 47 | 44 |
| DEL PEAK 15(18P11.21) MUTATED | 0 | 8 | 0 | 1 |
| DEL PEAK 15(18P11.21) WILD-TYPE | 33 | 53 | 47 | 43 |
Figure S67. Get High-res Image Gene #19: 'del_18p11.21' versus Molecular Subtype #2: 'METHLYATION_CNMF'
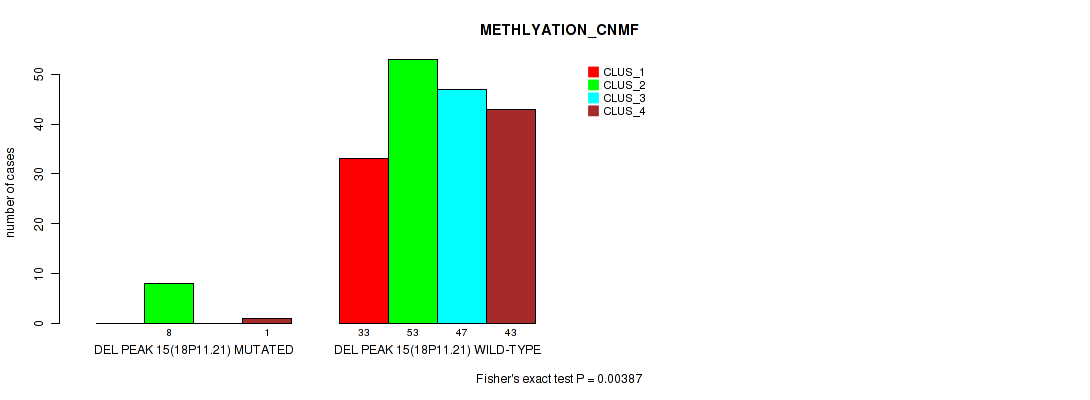
P value = 0.0318 (Fisher's exact test), Q value = 0.063
Table S68. Gene #19: 'del_18p11.21' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 34 | 78 |
| DEL PEAK 15(18P11.21) MUTATED | 0 | 3 | 5 |
| DEL PEAK 15(18P11.21) WILD-TYPE | 67 | 31 | 73 |
Figure S68. Get High-res Image Gene #19: 'del_18p11.21' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
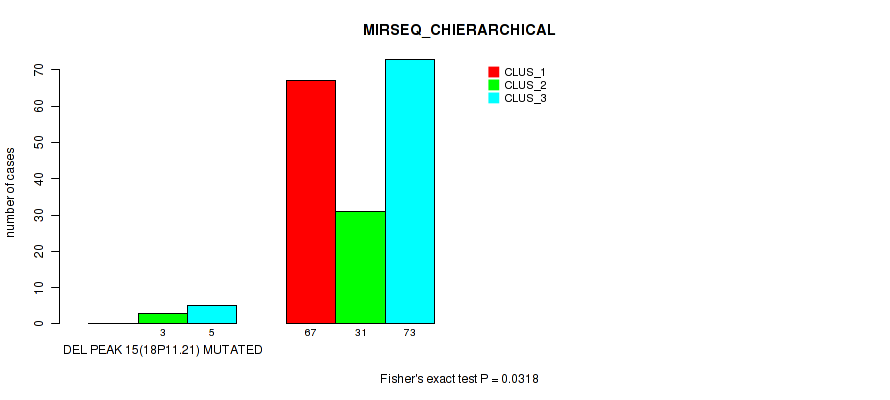
P value = 0.029 (Fisher's exact test), Q value = 0.058
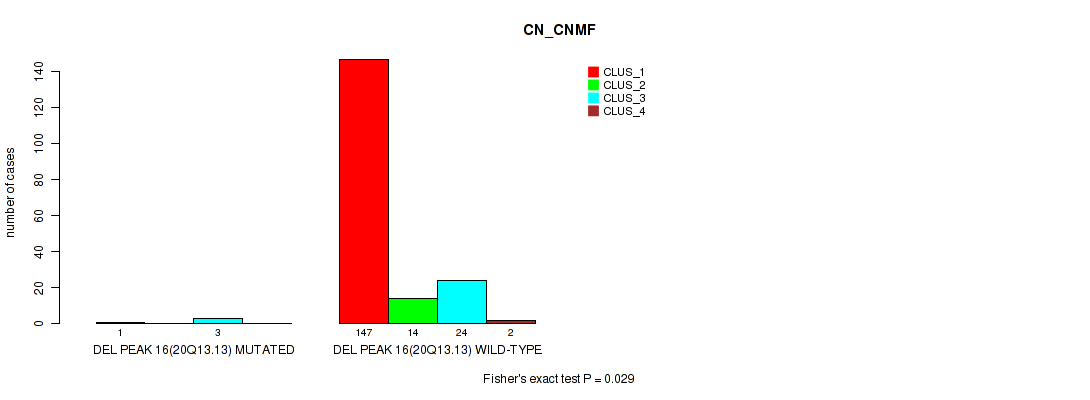
Table S69. Gene #20: 'del_20q13.13' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 148 | 14 | 27 | 2 |
| DEL PEAK 16(20Q13.13) MUTATED | 1 | 0 | 3 | 0 |
| DEL PEAK 16(20Q13.13) WILD-TYPE | 147 | 14 | 24 | 2 |
Figure S69. Get High-res Image Gene #20: 'del_20q13.13' versus Molecular Subtype #1: 'CN_CNMF'
-
Copy number data file = all_lesions.txt from GISTIC pipeline
-
Processed Copy number data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/LAML-TB/19782150/transformed.cor.cli.txt
-
Molecular subtype file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_mergedClustering/LAML-TB/19785823/LAML-TB.transferedmergedcluster.txt
-
Number of patients = 191
-
Number of significantly focal cnvs = 20
-
Number of molecular subtypes = 6
-
Exclude genes that fewer than K tumors have alterations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.