This pipeline computes the correlation between significantly recurrent gene mutations and molecular subtypes.
Testing the association between mutation status of 23 genes and 6 molecular subtypes across 193 patients, 36 significant findings detected with P value < 0.05 and Q value < 0.25.
-
FLT3 mutation correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
DNMT3A mutation correlated to 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
NPM1 mutation correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
IDH1 mutation correlated to 'CN_CNMF' and 'METHLYATION_CNMF'.
-
WT1 mutation correlated to 'METHLYATION_CNMF'.
-
U2AF1 mutation correlated to 'MIRSEQ_CNMF'.
-
RUNX1 mutation correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CHIERARCHICAL'.
-
CEBPA mutation correlated to 'METHLYATION_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
TP53 mutation correlated to 'CN_CNMF', 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', 'MIRSEQ_CNMF', and 'MIRSEQ_CHIERARCHICAL'.
-
STAG2 mutation correlated to 'MRNASEQ_CHIERARCHICAL' and 'MIRSEQ_CHIERARCHICAL'.
-
KIT mutation correlated to 'METHLYATION_CNMF' and 'MRNASEQ_CHIERARCHICAL'.
-
PTPN11 mutation correlated to 'MRNASEQ_CNMF'.
Table 1. Get Full Table Overview of the association between mutation status of 23 genes and 6 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 36 significant findings detected.
|
Clinical Features |
CN CNMF |
METHLYATION CNMF |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
||
| nMutated (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | |
| TP53 | 15 (8%) | 178 |
0.0174 (0.0799) |
1e-05 (0.000197) |
0.00105 (0.00805) |
0.00237 (0.0149) |
1e-05 (0.000197) |
9e-05 (0.00113) |
| NPM1 | 33 (17%) | 160 |
0.278 (0.547) |
1e-05 (0.000197) |
1e-05 (0.000197) |
1e-05 (0.000197) |
2e-05 (0.000307) |
1e-05 (0.000197) |
| FLT3 | 52 (27%) | 141 |
0.681 (0.805) |
4e-05 (0.000552) |
1e-05 (0.000197) |
0.00082 (0.00742) |
0.0805 (0.258) |
0.00071 (0.007) |
| DNMT3A | 48 (25%) | 145 |
0.341 (0.601) |
2e-05 (0.000307) |
0.0507 (0.189) |
0.00218 (0.0147) |
0.00086 (0.00742) |
0.00203 (0.0147) |
| RUNX1 | 16 (8%) | 177 |
0.459 (0.667) |
0.00871 (0.0462) |
0.00992 (0.0507) |
0.0003 (0.00318) |
0.0565 (0.195) |
0.00224 (0.0147) |
| CEBPA | 13 (7%) | 180 |
1 (1.00) |
0.0348 (0.145) |
0.226 (0.518) |
0.00021 (0.00241) |
0.0124 (0.0609) |
0.0133 (0.0631) |
| IDH1 | 18 (9%) | 175 |
0.0234 (0.104) |
0.00096 (0.00779) |
0.0983 (0.302) |
0.262 (0.525) |
0.447 (0.667) |
0.804 (0.881) |
| STAG2 | 6 (3%) | 187 |
1 (1.00) |
0.337 (0.601) |
0.13 (0.365) |
0.0273 (0.118) |
0.0788 (0.258) |
0.049 (0.188) |
| KIT | 8 (4%) | 185 |
0.744 (0.839) |
0.00361 (0.0208) |
0.325 (0.601) |
0.00604 (0.0333) |
0.676 (0.805) |
0.222 (0.518) |
| WT1 | 12 (6%) | 181 |
0.822 (0.894) |
0.00275 (0.0165) |
0.0598 (0.201) |
0.372 (0.603) |
0.0553 (0.195) |
0.252 (0.525) |
| U2AF1 | 8 (4%) | 185 |
0.242 (0.525) |
0.129 (0.365) |
0.262 (0.525) |
0.263 (0.525) |
0.0487 (0.188) |
0.471 (0.677) |
| PTPN11 | 9 (5%) | 184 |
0.128 (0.365) |
0.164 (0.436) |
0.0477 (0.188) |
0.513 (0.699) |
0.459 (0.667) |
0.185 (0.46) |
| IDH2 | 20 (10%) | 173 |
0.391 (0.627) |
0.215 (0.511) |
0.646 (0.783) |
0.344 (0.601) |
0.753 (0.839) |
0.182 (0.46) |
| TET2 | 17 (9%) | 176 |
0.699 (0.817) |
0.513 (0.699) |
0.35 (0.601) |
0.582 (0.746) |
0.366 (0.601) |
0.753 (0.839) |
| NRAS | 15 (8%) | 178 |
0.149 (0.404) |
0.353 (0.601) |
0.515 (0.699) |
0.931 (0.953) |
0.928 (0.953) |
0.588 (0.746) |
| KRAS | 8 (4%) | 185 |
0.864 (0.91) |
0.576 (0.746) |
0.324 (0.601) |
0.637 (0.778) |
0.301 (0.584) |
0.683 (0.805) |
| PHF6 | 6 (3%) | 187 |
0.517 (0.699) |
0.422 (0.648) |
0.727 (0.836) |
0.476 (0.677) |
0.897 (0.938) |
0.35 (0.601) |
| RAD21 | 5 (3%) | 188 |
0.932 (0.953) |
0.437 (0.663) |
0.605 (0.746) |
0.182 (0.46) |
0.363 (0.601) |
0.603 (0.746) |
| EZH2 | 3 (2%) | 190 |
0.0903 (0.283) |
0.256 (0.525) |
0.593 (0.746) |
0.19 (0.46) |
0.42 (0.648) |
|
| SMC3 | 7 (4%) | 186 |
0.839 (0.899) |
0.418 (0.648) |
0.485 (0.683) |
0.453 (0.667) |
0.744 (0.839) |
0.362 (0.601) |
| ASXL1 | 5 (3%) | 188 |
0.724 (0.836) |
0.187 (0.46) |
0.255 (0.525) |
0.593 (0.746) |
0.11 (0.331) |
0.416 (0.648) |
| SMC1A | 6 (3%) | 187 |
0.852 (0.904) |
0.0544 (0.195) |
0.568 (0.746) |
0.229 (0.518) |
0.335 (0.601) |
0.576 (0.746) |
| SUZ12 | 3 (2%) | 190 |
0.791 (0.873) |
0.84 (0.899) |
0.257 (0.525) |
0.596 (0.746) |
0.326 (0.601) |
0.139 (0.385) |
P value = 0.681 (Fisher's exact test), Q value = 0.81
Table S1. Gene #1: 'FLT3 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| FLT3 MUTATED | 12 | 11 | 18 | 9 |
| FLT3 WILD-TYPE | 37 | 31 | 36 | 30 |
P value = 4e-05 (Fisher's exact test), Q value = 0.00055
Table S2. Gene #1: 'FLT3 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| FLT3 MUTATED | 11 | 5 | 7 | 7 | 21 |
| FLT3 WILD-TYPE | 11 | 20 | 55 | 30 | 20 |
Figure S1. Get High-res Image Gene #1: 'FLT3 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 2e-04
Table S3. Gene #1: 'FLT3 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| FLT3 MUTATED | 6 | 17 | 22 |
| FLT3 WILD-TYPE | 68 | 31 | 23 |
Figure S2. Get High-res Image Gene #1: 'FLT3 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.00082 (Fisher's exact test), Q value = 0.0074
Table S4. Gene #1: 'FLT3 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| FLT3 MUTATED | 5 | 3 | 2 | 6 | 13 | 16 |
| FLT3 WILD-TYPE | 11 | 10 | 13 | 51 | 13 | 24 |
Figure S3. Get High-res Image Gene #1: 'FLT3 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.0805 (Fisher's exact test), Q value = 0.26
Table S5. Gene #1: 'FLT3 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| FLT3 MUTATED | 21 | 4 | 14 | 7 | 4 |
| FLT3 WILD-TYPE | 34 | 27 | 28 | 25 | 17 |
P value = 0.00071 (Fisher's exact test), Q value = 0.007
Table S6. Gene #1: 'FLT3 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| FLT3 MUTATED | 29 | 5 | 16 |
| FLT3 WILD-TYPE | 36 | 30 | 65 |
Figure S4. Get High-res Image Gene #1: 'FLT3 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.341 (Fisher's exact test), Q value = 0.6
Table S7. Gene #2: 'DNMT3A MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| DNMT3A MUTATED | 13 | 6 | 16 | 9 |
| DNMT3A WILD-TYPE | 36 | 36 | 38 | 30 |
P value = 2e-05 (Fisher's exact test), Q value = 0.00031
Table S8. Gene #2: 'DNMT3A MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| DNMT3A MUTATED | 16 | 2 | 13 | 6 | 9 |
| DNMT3A WILD-TYPE | 6 | 23 | 49 | 31 | 32 |
Figure S5. Get High-res Image Gene #2: 'DNMT3A MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.0507 (Fisher's exact test), Q value = 0.19
Table S9. Gene #2: 'DNMT3A MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| DNMT3A MUTATED | 12 | 17 | 12 |
| DNMT3A WILD-TYPE | 62 | 31 | 33 |
P value = 0.00218 (Fisher's exact test), Q value = 0.015
Table S10. Gene #2: 'DNMT3A MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| DNMT3A MUTATED | 0 | 3 | 0 | 14 | 11 | 13 |
| DNMT3A WILD-TYPE | 16 | 10 | 15 | 43 | 15 | 27 |
Figure S6. Get High-res Image Gene #2: 'DNMT3A MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.00086 (Fisher's exact test), Q value = 0.0074
Table S11. Gene #2: 'DNMT3A MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| DNMT3A MUTATED | 23 | 2 | 11 | 3 | 5 |
| DNMT3A WILD-TYPE | 32 | 29 | 31 | 29 | 16 |
Figure S7. Get High-res Image Gene #2: 'DNMT3A MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 0.00203 (Fisher's exact test), Q value = 0.015
Table S12. Gene #2: 'DNMT3A MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| DNMT3A MUTATED | 25 | 3 | 16 |
| DNMT3A WILD-TYPE | 40 | 32 | 65 |
Figure S8. Get High-res Image Gene #2: 'DNMT3A MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.278 (Fisher's exact test), Q value = 0.55
Table S13. Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| NPM1 MUTATED | 12 | 8 | 7 | 4 |
| NPM1 WILD-TYPE | 37 | 34 | 47 | 35 |
P value = 1e-05 (Fisher's exact test), Q value = 2e-04
Table S14. Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| NPM1 MUTATED | 9 | 3 | 2 | 0 | 18 |
| NPM1 WILD-TYPE | 13 | 22 | 60 | 37 | 23 |
Figure S9. Get High-res Image Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 2e-04
Table S15. Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| NPM1 MUTATED | 1 | 10 | 17 |
| NPM1 WILD-TYPE | 73 | 38 | 28 |
Figure S10. Get High-res Image Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 2e-04
Table S16. Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| NPM1 MUTATED | 0 | 0 | 0 | 1 | 15 | 12 |
| NPM1 WILD-TYPE | 16 | 13 | 15 | 56 | 11 | 28 |
Figure S11. Get High-res Image Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
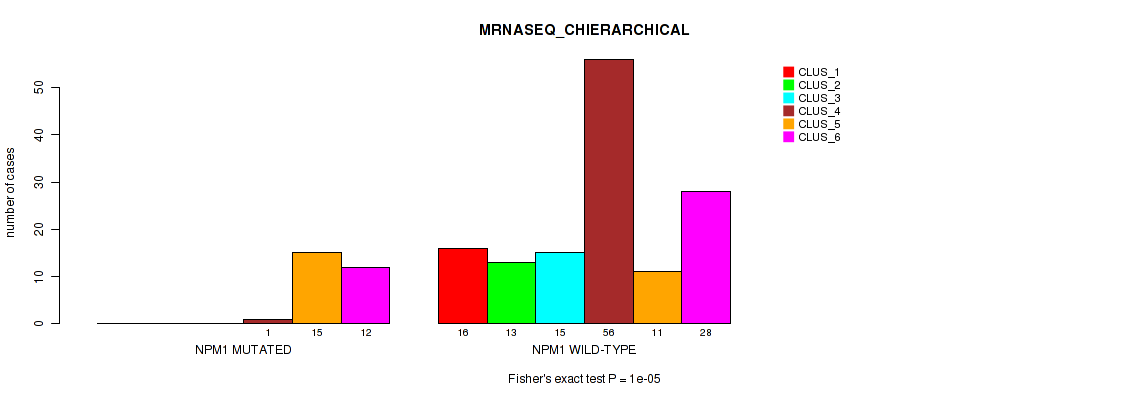
P value = 2e-05 (Fisher's exact test), Q value = 0.00031
Table S17. Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| NPM1 MUTATED | 21 | 0 | 6 | 1 | 4 |
| NPM1 WILD-TYPE | 34 | 31 | 36 | 31 | 17 |
Figure S12. Get High-res Image Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 2e-04
Table S18. Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| NPM1 MUTATED | 30 | 1 | 1 |
| NPM1 WILD-TYPE | 35 | 34 | 80 |
Figure S13. Get High-res Image Gene #3: 'NPM1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.391 (Fisher's exact test), Q value = 0.63
Table S19. Gene #4: 'IDH2 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| IDH2 MUTATED | 6 | 2 | 8 | 3 |
| IDH2 WILD-TYPE | 43 | 40 | 46 | 36 |
P value = 0.215 (Fisher's exact test), Q value = 0.51
Table S20. Gene #4: 'IDH2 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| IDH2 MUTATED | 0 | 2 | 10 | 2 | 4 |
| IDH2 WILD-TYPE | 22 | 23 | 52 | 35 | 37 |
P value = 0.646 (Fisher's exact test), Q value = 0.78
Table S21. Gene #4: 'IDH2 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| IDH2 MUTATED | 8 | 6 | 3 |
| IDH2 WILD-TYPE | 66 | 42 | 42 |
P value = 0.344 (Fisher's exact test), Q value = 0.6
Table S22. Gene #4: 'IDH2 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| IDH2 MUTATED | 0 | 2 | 0 | 9 | 2 | 4 |
| IDH2 WILD-TYPE | 16 | 11 | 15 | 48 | 24 | 36 |
P value = 0.753 (Fisher's exact test), Q value = 0.84
Table S23. Gene #4: 'IDH2 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| IDH2 MUTATED | 6 | 1 | 4 | 4 | 2 |
| IDH2 WILD-TYPE | 49 | 30 | 38 | 28 | 19 |
P value = 0.182 (Fisher's exact test), Q value = 0.46
Table S24. Gene #4: 'IDH2 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| IDH2 MUTATED | 5 | 1 | 11 |
| IDH2 WILD-TYPE | 60 | 34 | 70 |
P value = 0.0234 (Fisher's exact test), Q value = 0.1
Table S25. Gene #5: 'IDH1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| IDH1 MUTATED | 7 | 1 | 2 | 7 |
| IDH1 WILD-TYPE | 42 | 41 | 52 | 32 |
Figure S14. Get High-res Image Gene #5: 'IDH1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.00096 (Fisher's exact test), Q value = 0.0078
Table S26. Gene #5: 'IDH1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| IDH1 MUTATED | 3 | 5 | 10 | 0 | 0 |
| IDH1 WILD-TYPE | 19 | 20 | 52 | 37 | 41 |
Figure S15. Get High-res Image Gene #5: 'IDH1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.0983 (Fisher's exact test), Q value = 0.3
Table S27. Gene #5: 'IDH1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| IDH1 MUTATED | 8 | 1 | 6 |
| IDH1 WILD-TYPE | 66 | 47 | 39 |
P value = 0.262 (Fisher's exact test), Q value = 0.53
Table S28. Gene #5: 'IDH1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| IDH1 MUTATED | 0 | 0 | 0 | 8 | 4 | 3 |
| IDH1 WILD-TYPE | 16 | 13 | 15 | 49 | 22 | 37 |
P value = 0.447 (Fisher's exact test), Q value = 0.67
Table S29. Gene #5: 'IDH1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| IDH1 MUTATED | 5 | 3 | 7 | 1 | 2 |
| IDH1 WILD-TYPE | 50 | 28 | 35 | 31 | 19 |
P value = 0.804 (Fisher's exact test), Q value = 0.88
Table S30. Gene #5: 'IDH1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| IDH1 MUTATED | 8 | 3 | 7 |
| IDH1 WILD-TYPE | 57 | 32 | 74 |
P value = 0.699 (Fisher's exact test), Q value = 0.82
Table S31. Gene #6: 'TET2 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| TET2 MUTATED | 3 | 4 | 4 | 5 |
| TET2 WILD-TYPE | 46 | 38 | 50 | 34 |
P value = 0.513 (Fisher's exact test), Q value = 0.7
Table S32. Gene #6: 'TET2 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| TET2 MUTATED | 1 | 2 | 8 | 1 | 4 |
| TET2 WILD-TYPE | 21 | 23 | 54 | 36 | 37 |
P value = 0.35 (Fisher's exact test), Q value = 0.6
Table S33. Gene #6: 'TET2 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| TET2 MUTATED | 9 | 2 | 4 |
| TET2 WILD-TYPE | 65 | 46 | 41 |
P value = 0.582 (Fisher's exact test), Q value = 0.75
Table S34. Gene #6: 'TET2 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| TET2 MUTATED | 0 | 0 | 2 | 7 | 3 | 3 |
| TET2 WILD-TYPE | 16 | 13 | 13 | 50 | 23 | 37 |
P value = 0.366 (Fisher's exact test), Q value = 0.6
Table S35. Gene #6: 'TET2 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| TET2 MUTATED | 5 | 2 | 5 | 1 | 4 |
| TET2 WILD-TYPE | 50 | 29 | 37 | 31 | 17 |
P value = 0.753 (Fisher's exact test), Q value = 0.84
Table S36. Gene #6: 'TET2 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| TET2 MUTATED | 7 | 2 | 8 |
| TET2 WILD-TYPE | 58 | 33 | 73 |
P value = 0.149 (Fisher's exact test), Q value = 0.4
Table S37. Gene #7: 'NRAS MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| NRAS MUTATED | 3 | 2 | 3 | 7 |
| NRAS WILD-TYPE | 46 | 40 | 51 | 32 |
P value = 0.353 (Fisher's exact test), Q value = 0.6
Table S38. Gene #7: 'NRAS MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| NRAS MUTATED | 0 | 1 | 5 | 3 | 6 |
| NRAS WILD-TYPE | 22 | 24 | 57 | 34 | 35 |
P value = 0.515 (Fisher's exact test), Q value = 0.7
Table S39. Gene #7: 'NRAS MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| NRAS MUTATED | 5 | 5 | 2 |
| NRAS WILD-TYPE | 69 | 43 | 43 |
P value = 0.931 (Fisher's exact test), Q value = 0.95
Table S40. Gene #7: 'NRAS MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| NRAS MUTATED | 0 | 1 | 1 | 4 | 2 | 4 |
| NRAS WILD-TYPE | 16 | 12 | 14 | 53 | 24 | 36 |
P value = 0.928 (Fisher's exact test), Q value = 0.95
Table S41. Gene #7: 'NRAS MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| NRAS MUTATED | 5 | 2 | 2 | 3 | 2 |
| NRAS WILD-TYPE | 50 | 29 | 40 | 29 | 19 |
P value = 0.588 (Fisher's exact test), Q value = 0.75
Table S42. Gene #7: 'NRAS MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| NRAS MUTATED | 7 | 2 | 5 |
| NRAS WILD-TYPE | 58 | 33 | 76 |
P value = 0.822 (Fisher's exact test), Q value = 0.89
Table S43. Gene #8: 'WT1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| WT1 MUTATED | 3 | 2 | 3 | 4 |
| WT1 WILD-TYPE | 46 | 40 | 51 | 35 |
P value = 0.00275 (Fisher's exact test), Q value = 0.016
Table S44. Gene #8: 'WT1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| WT1 MUTATED | 0 | 2 | 0 | 2 | 7 |
| WT1 WILD-TYPE | 22 | 23 | 62 | 35 | 34 |
Figure S16. Get High-res Image Gene #8: 'WT1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.0598 (Fisher's exact test), Q value = 0.2
Table S45. Gene #8: 'WT1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| WT1 MUTATED | 2 | 2 | 6 |
| WT1 WILD-TYPE | 72 | 46 | 39 |
P value = 0.372 (Fisher's exact test), Q value = 0.6
Table S46. Gene #8: 'WT1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| WT1 MUTATED | 0 | 1 | 0 | 3 | 4 | 2 |
| WT1 WILD-TYPE | 16 | 12 | 15 | 54 | 22 | 38 |
P value = 0.0553 (Fisher's exact test), Q value = 0.19
Table S47. Gene #8: 'WT1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| WT1 MUTATED | 2 | 0 | 6 | 3 | 0 |
| WT1 WILD-TYPE | 53 | 31 | 36 | 29 | 21 |
P value = 0.252 (Fisher's exact test), Q value = 0.53
Table S48. Gene #8: 'WT1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| WT1 MUTATED | 5 | 0 | 6 |
| WT1 WILD-TYPE | 60 | 35 | 75 |
P value = 0.242 (Fisher's exact test), Q value = 0.53
Table S49. Gene #9: 'U2AF1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| U2AF1 MUTATED | 2 | 0 | 1 | 3 |
| U2AF1 WILD-TYPE | 47 | 42 | 53 | 36 |
P value = 0.129 (Fisher's exact test), Q value = 0.37
Table S50. Gene #9: 'U2AF1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| U2AF1 MUTATED | 1 | 0 | 6 | 1 | 0 |
| U2AF1 WILD-TYPE | 21 | 25 | 56 | 36 | 41 |
P value = 0.262 (Fisher's exact test), Q value = 0.53
Table S51. Gene #9: 'U2AF1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| U2AF1 MUTATED | 5 | 2 | 0 |
| U2AF1 WILD-TYPE | 69 | 46 | 45 |
P value = 0.263 (Fisher's exact test), Q value = 0.53
Table S52. Gene #9: 'U2AF1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| U2AF1 MUTATED | 0 | 0 | 0 | 6 | 0 | 1 |
| U2AF1 WILD-TYPE | 16 | 13 | 15 | 51 | 26 | 39 |
P value = 0.0487 (Fisher's exact test), Q value = 0.19
Table S53. Gene #9: 'U2AF1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| U2AF1 MUTATED | 1 | 1 | 0 | 2 | 3 |
| U2AF1 WILD-TYPE | 54 | 30 | 42 | 30 | 18 |
Figure S17. Get High-res Image Gene #9: 'U2AF1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 0.471 (Fisher's exact test), Q value = 0.68
Table S54. Gene #9: 'U2AF1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| U2AF1 MUTATED | 1 | 2 | 4 |
| U2AF1 WILD-TYPE | 64 | 33 | 77 |
P value = 0.459 (Fisher's exact test), Q value = 0.67
Table S55. Gene #10: 'RUNX1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| RUNX1 MUTATED | 2 | 4 | 7 | 3 |
| RUNX1 WILD-TYPE | 47 | 38 | 47 | 36 |
P value = 0.00871 (Fisher's exact test), Q value = 0.046
Table S56. Gene #10: 'RUNX1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| RUNX1 MUTATED | 0 | 0 | 11 | 2 | 1 |
| RUNX1 WILD-TYPE | 22 | 25 | 51 | 35 | 40 |
Figure S18. Get High-res Image Gene #10: 'RUNX1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.00992 (Fisher's exact test), Q value = 0.051
Table S57. Gene #10: 'RUNX1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| RUNX1 MUTATED | 11 | 3 | 0 |
| RUNX1 WILD-TYPE | 63 | 45 | 45 |
Figure S19. Get High-res Image Gene #10: 'RUNX1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 3e-04 (Fisher's exact test), Q value = 0.0032
Table S58. Gene #10: 'RUNX1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| RUNX1 MUTATED | 0 | 1 | 0 | 13 | 0 | 0 |
| RUNX1 WILD-TYPE | 16 | 12 | 15 | 44 | 26 | 40 |
Figure S20. Get High-res Image Gene #10: 'RUNX1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.0565 (Fisher's exact test), Q value = 0.19
Table S59. Gene #10: 'RUNX1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| RUNX1 MUTATED | 1 | 3 | 2 | 4 | 4 |
| RUNX1 WILD-TYPE | 54 | 28 | 40 | 28 | 17 |
P value = 0.00224 (Fisher's exact test), Q value = 0.015
Table S60. Gene #10: 'RUNX1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| RUNX1 MUTATED | 0 | 3 | 11 |
| RUNX1 WILD-TYPE | 65 | 32 | 70 |
Figure S21. Get High-res Image Gene #10: 'RUNX1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 1 (Fisher's exact test), Q value = 1
Table S61. Gene #11: 'CEBPA MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| CEBPA MUTATED | 3 | 3 | 4 | 2 |
| CEBPA WILD-TYPE | 46 | 39 | 50 | 37 |
P value = 0.0348 (Fisher's exact test), Q value = 0.15
Table S62. Gene #11: 'CEBPA MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| CEBPA MUTATED | 0 | 2 | 2 | 7 | 2 |
| CEBPA WILD-TYPE | 22 | 23 | 60 | 30 | 39 |
Figure S22. Get High-res Image Gene #11: 'CEBPA MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.226 (Fisher's exact test), Q value = 0.52
Table S63. Gene #11: 'CEBPA MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| CEBPA MUTATED | 9 | 2 | 2 |
| CEBPA WILD-TYPE | 65 | 46 | 43 |
P value = 0.00021 (Fisher's exact test), Q value = 0.0024
Table S64. Gene #11: 'CEBPA MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| CEBPA MUTATED | 0 | 0 | 7 | 4 | 1 | 1 |
| CEBPA WILD-TYPE | 16 | 13 | 8 | 53 | 25 | 39 |
Figure S23. Get High-res Image Gene #11: 'CEBPA MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
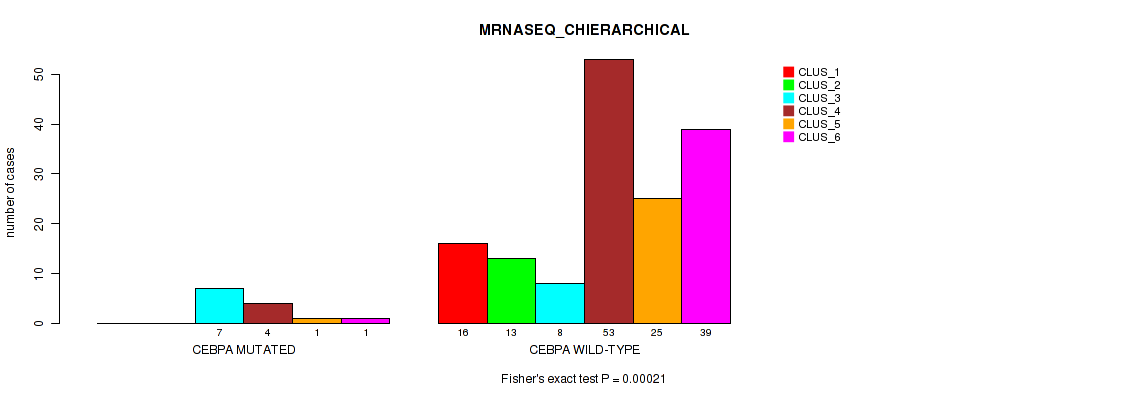
P value = 0.0124 (Fisher's exact test), Q value = 0.061
Table S65. Gene #11: 'CEBPA MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| CEBPA MUTATED | 1 | 0 | 7 | 4 | 1 |
| CEBPA WILD-TYPE | 54 | 31 | 35 | 28 | 20 |
Figure S24. Get High-res Image Gene #11: 'CEBPA MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 0.0133 (Fisher's exact test), Q value = 0.063
Table S66. Gene #11: 'CEBPA MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| CEBPA MUTATED | 2 | 0 | 11 |
| CEBPA WILD-TYPE | 63 | 35 | 70 |
Figure S25. Get High-res Image Gene #11: 'CEBPA MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
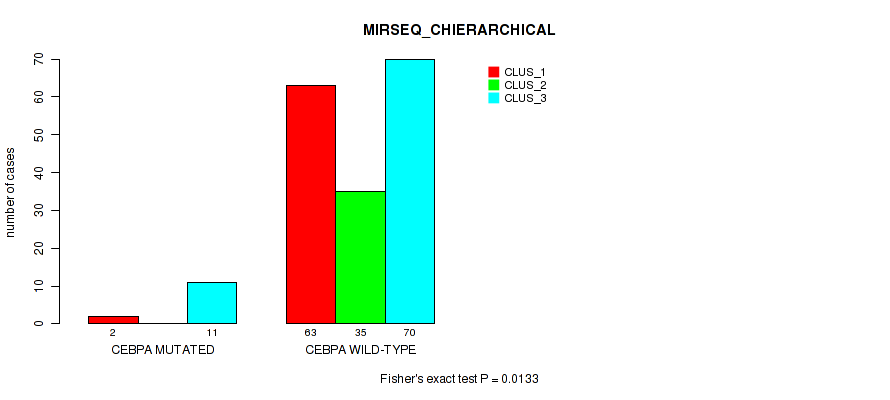
P value = 0.0174 (Fisher's exact test), Q value = 0.08
Table S67. Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| TP53 MUTATED | 0 | 7 | 5 | 3 |
| TP53 WILD-TYPE | 49 | 35 | 49 | 36 |
Figure S26. Get High-res Image Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'

P value = 1e-05 (Fisher's exact test), Q value = 2e-04
Table S68. Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| TP53 MUTATED | 0 | 0 | 15 | 0 | 0 |
| TP53 WILD-TYPE | 22 | 25 | 47 | 37 | 41 |
Figure S27. Get High-res Image Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
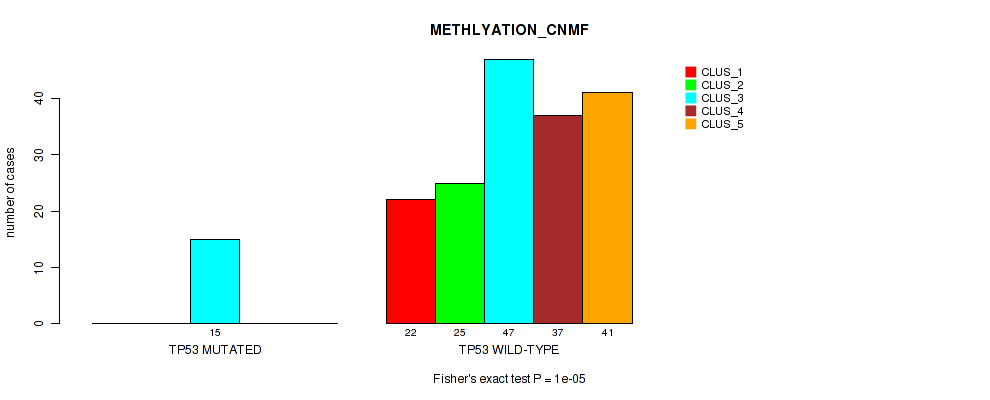
P value = 0.00105 (Fisher's exact test), Q value = 0.008
Table S69. Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| TP53 MUTATED | 12 | 1 | 0 |
| TP53 WILD-TYPE | 62 | 47 | 45 |
Figure S28. Get High-res Image Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.00237 (Fisher's exact test), Q value = 0.015
Table S70. Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| TP53 MUTATED | 0 | 0 | 0 | 12 | 0 | 1 |
| TP53 WILD-TYPE | 16 | 13 | 15 | 45 | 26 | 39 |
Figure S29. Get High-res Image Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 1e-05 (Fisher's exact test), Q value = 2e-04
Table S71. Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| TP53 MUTATED | 0 | 6 | 0 | 2 | 6 |
| TP53 WILD-TYPE | 55 | 25 | 42 | 30 | 15 |
Figure S30. Get High-res Image Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'

P value = 9e-05 (Fisher's exact test), Q value = 0.0011
Table S72. Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| TP53 MUTATED | 0 | 8 | 6 |
| TP53 WILD-TYPE | 65 | 27 | 75 |
Figure S31. Get High-res Image Gene #12: 'TP53 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
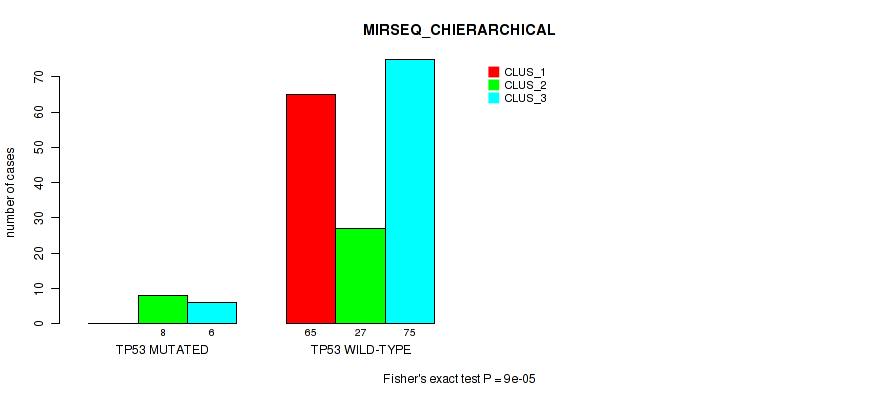
P value = 0.864 (Fisher's exact test), Q value = 0.91
Table S73. Gene #13: 'KRAS MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| KRAS MUTATED | 3 | 1 | 2 | 2 |
| KRAS WILD-TYPE | 46 | 41 | 52 | 37 |
P value = 0.576 (Fisher's exact test), Q value = 0.75
Table S74. Gene #13: 'KRAS MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| KRAS MUTATED | 2 | 0 | 2 | 1 | 1 |
| KRAS WILD-TYPE | 20 | 25 | 60 | 36 | 40 |
P value = 0.324 (Fisher's exact test), Q value = 0.6
Table S75. Gene #13: 'KRAS MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| KRAS MUTATED | 4 | 3 | 0 |
| KRAS WILD-TYPE | 70 | 45 | 45 |
P value = 0.637 (Fisher's exact test), Q value = 0.78
Table S76. Gene #13: 'KRAS MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| KRAS MUTATED | 0 | 0 | 0 | 4 | 0 | 3 |
| KRAS WILD-TYPE | 16 | 13 | 15 | 53 | 26 | 37 |
P value = 0.301 (Fisher's exact test), Q value = 0.58
Table S77. Gene #13: 'KRAS MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| KRAS MUTATED | 3 | 3 | 1 | 0 | 0 |
| KRAS WILD-TYPE | 52 | 28 | 41 | 32 | 21 |
P value = 0.683 (Fisher's exact test), Q value = 0.81
Table S78. Gene #13: 'KRAS MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| KRAS MUTATED | 3 | 2 | 2 |
| KRAS WILD-TYPE | 62 | 33 | 79 |
P value = 0.517 (Fisher's exact test), Q value = 0.7
Table S79. Gene #14: 'PHF6 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| PHF6 MUTATED | 1 | 2 | 3 | 0 |
| PHF6 WILD-TYPE | 48 | 40 | 51 | 39 |
P value = 0.422 (Fisher's exact test), Q value = 0.65
Table S80. Gene #14: 'PHF6 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| PHF6 MUTATED | 0 | 1 | 1 | 3 | 1 |
| PHF6 WILD-TYPE | 22 | 24 | 61 | 34 | 40 |
P value = 0.727 (Fisher's exact test), Q value = 0.84
Table S81. Gene #14: 'PHF6 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| PHF6 MUTATED | 2 | 1 | 2 |
| PHF6 WILD-TYPE | 72 | 47 | 43 |
P value = 0.476 (Fisher's exact test), Q value = 0.68
Table S82. Gene #14: 'PHF6 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| PHF6 MUTATED | 0 | 0 | 0 | 4 | 1 | 0 |
| PHF6 WILD-TYPE | 16 | 13 | 15 | 53 | 25 | 40 |
P value = 0.897 (Fisher's exact test), Q value = 0.94
Table S83. Gene #14: 'PHF6 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| PHF6 MUTATED | 2 | 0 | 1 | 1 | 1 |
| PHF6 WILD-TYPE | 53 | 31 | 41 | 31 | 20 |
P value = 0.35 (Fisher's exact test), Q value = 0.6
Table S84. Gene #14: 'PHF6 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| PHF6 MUTATED | 1 | 0 | 4 |
| PHF6 WILD-TYPE | 64 | 35 | 77 |
P value = 1 (Fisher's exact test), Q value = 1
Table S85. Gene #15: 'STAG2 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| STAG2 MUTATED | 2 | 1 | 2 | 1 |
| STAG2 WILD-TYPE | 47 | 41 | 52 | 38 |
P value = 0.337 (Fisher's exact test), Q value = 0.6
Table S86. Gene #15: 'STAG2 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| STAG2 MUTATED | 0 | 0 | 2 | 0 | 3 |
| STAG2 WILD-TYPE | 22 | 25 | 60 | 37 | 38 |
P value = 0.13 (Fisher's exact test), Q value = 0.37
Table S87. Gene #15: 'STAG2 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| STAG2 MUTATED | 2 | 0 | 3 |
| STAG2 WILD-TYPE | 72 | 48 | 42 |
P value = 0.0273 (Fisher's exact test), Q value = 0.12
Table S88. Gene #15: 'STAG2 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| STAG2 MUTATED | 0 | 0 | 0 | 1 | 4 | 0 |
| STAG2 WILD-TYPE | 16 | 13 | 15 | 56 | 22 | 40 |
Figure S32. Get High-res Image Gene #15: 'STAG2 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.0788 (Fisher's exact test), Q value = 0.26
Table S89. Gene #15: 'STAG2 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| STAG2 MUTATED | 0 | 1 | 2 | 0 | 2 |
| STAG2 WILD-TYPE | 55 | 30 | 40 | 32 | 19 |
P value = 0.049 (Fisher's exact test), Q value = 0.19
Table S90. Gene #15: 'STAG2 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| STAG2 MUTATED | 4 | 1 | 0 |
| STAG2 WILD-TYPE | 61 | 34 | 81 |
Figure S33. Get High-res Image Gene #15: 'STAG2 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'

P value = 0.744 (Fisher's exact test), Q value = 0.84
Table S91. Gene #16: 'KIT MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| KIT MUTATED | 1 | 1 | 3 | 2 |
| KIT WILD-TYPE | 48 | 41 | 51 | 37 |
P value = 0.00361 (Fisher's exact test), Q value = 0.021
Table S92. Gene #16: 'KIT MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| KIT MUTATED | 1 | 0 | 1 | 6 | 0 |
| KIT WILD-TYPE | 21 | 25 | 61 | 31 | 41 |
Figure S34. Get High-res Image Gene #16: 'KIT MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.325 (Fisher's exact test), Q value = 0.6
Table S93. Gene #16: 'KIT MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| KIT MUTATED | 4 | 3 | 0 |
| KIT WILD-TYPE | 70 | 45 | 45 |
P value = 0.00604 (Fisher's exact test), Q value = 0.033
Table S94. Gene #16: 'KIT MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| KIT MUTATED | 0 | 3 | 2 | 2 | 0 | 0 |
| KIT WILD-TYPE | 16 | 10 | 13 | 55 | 26 | 40 |
Figure S35. Get High-res Image Gene #16: 'KIT MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'

P value = 0.676 (Fisher's exact test), Q value = 0.81
Table S95. Gene #16: 'KIT MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| KIT MUTATED | 1 | 2 | 3 | 1 | 1 |
| KIT WILD-TYPE | 54 | 29 | 39 | 31 | 20 |
P value = 0.222 (Fisher's exact test), Q value = 0.52
Table S96. Gene #16: 'KIT MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| KIT MUTATED | 1 | 3 | 4 |
| KIT WILD-TYPE | 64 | 32 | 77 |
P value = 0.932 (Fisher's exact test), Q value = 0.95
Table S97. Gene #17: 'RAD21 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| RAD21 MUTATED | 2 | 1 | 1 | 1 |
| RAD21 WILD-TYPE | 47 | 41 | 53 | 38 |
P value = 0.437 (Fisher's exact test), Q value = 0.66
Table S98. Gene #17: 'RAD21 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| RAD21 MUTATED | 0 | 0 | 1 | 1 | 3 |
| RAD21 WILD-TYPE | 22 | 25 | 61 | 36 | 38 |
P value = 0.605 (Fisher's exact test), Q value = 0.75
Table S99. Gene #17: 'RAD21 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| RAD21 MUTATED | 1 | 2 | 2 |
| RAD21 WILD-TYPE | 73 | 46 | 43 |
P value = 0.182 (Fisher's exact test), Q value = 0.46
Table S100. Gene #17: 'RAD21 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| RAD21 MUTATED | 0 | 0 | 1 | 0 | 2 | 2 |
| RAD21 WILD-TYPE | 16 | 13 | 14 | 57 | 24 | 38 |
P value = 0.363 (Fisher's exact test), Q value = 0.6
Table S101. Gene #17: 'RAD21 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| RAD21 MUTATED | 2 | 0 | 3 | 0 | 0 |
| RAD21 WILD-TYPE | 53 | 31 | 39 | 32 | 21 |
P value = 0.603 (Fisher's exact test), Q value = 0.75
Table S102. Gene #17: 'RAD21 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| RAD21 MUTATED | 3 | 0 | 2 |
| RAD21 WILD-TYPE | 62 | 35 | 79 |
P value = 0.0903 (Fisher's exact test), Q value = 0.28
Table S103. Gene #18: 'EZH2 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| EZH2 MUTATED | 0 | 1 | 0 | 2 |
| EZH2 WILD-TYPE | 49 | 41 | 54 | 37 |
P value = 0.256 (Fisher's exact test), Q value = 0.53
Table S104. Gene #18: 'EZH2 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| EZH2 MUTATED | 3 | 0 | 0 |
| EZH2 WILD-TYPE | 71 | 48 | 45 |
P value = 0.593 (Fisher's exact test), Q value = 0.75
Table S105. Gene #18: 'EZH2 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| EZH2 MUTATED | 0 | 0 | 0 | 3 | 0 | 0 |
| EZH2 WILD-TYPE | 16 | 13 | 15 | 54 | 26 | 40 |
P value = 0.19 (Fisher's exact test), Q value = 0.46
Table S106. Gene #18: 'EZH2 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| EZH2 MUTATED | 0 | 1 | 0 | 1 | 1 |
| EZH2 WILD-TYPE | 55 | 30 | 42 | 31 | 20 |
P value = 0.42 (Fisher's exact test), Q value = 0.65
Table S107. Gene #18: 'EZH2 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| EZH2 MUTATED | 0 | 1 | 2 |
| EZH2 WILD-TYPE | 65 | 34 | 79 |
P value = 0.839 (Fisher's exact test), Q value = 0.9
Table S108. Gene #19: 'SMC3 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| SMC3 MUTATED | 3 | 1 | 2 | 1 |
| SMC3 WILD-TYPE | 46 | 41 | 52 | 38 |
P value = 0.418 (Fisher's exact test), Q value = 0.65
Table S109. Gene #19: 'SMC3 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| SMC3 MUTATED | 1 | 0 | 1 | 2 | 3 |
| SMC3 WILD-TYPE | 21 | 25 | 61 | 35 | 38 |
P value = 0.485 (Fisher's exact test), Q value = 0.68
Table S110. Gene #19: 'SMC3 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| SMC3 MUTATED | 2 | 2 | 3 |
| SMC3 WILD-TYPE | 72 | 46 | 42 |
P value = 0.453 (Fisher's exact test), Q value = 0.67
Table S111. Gene #19: 'SMC3 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| SMC3 MUTATED | 0 | 0 | 1 | 2 | 3 | 1 |
| SMC3 WILD-TYPE | 16 | 13 | 14 | 55 | 23 | 39 |
P value = 0.744 (Fisher's exact test), Q value = 0.84
Table S112. Gene #19: 'SMC3 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| SMC3 MUTATED | 2 | 0 | 2 | 2 | 1 |
| SMC3 WILD-TYPE | 53 | 31 | 40 | 30 | 20 |
P value = 0.362 (Fisher's exact test), Q value = 0.6
Table S113. Gene #19: 'SMC3 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| SMC3 MUTATED | 4 | 0 | 3 |
| SMC3 WILD-TYPE | 61 | 35 | 78 |
P value = 0.724 (Fisher's exact test), Q value = 0.84
Table S114. Gene #20: 'ASXL1 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| ASXL1 MUTATED | 2 | 0 | 2 | 1 |
| ASXL1 WILD-TYPE | 47 | 42 | 52 | 38 |
P value = 0.187 (Fisher's exact test), Q value = 0.46
Table S115. Gene #20: 'ASXL1 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| ASXL1 MUTATED | 0 | 0 | 4 | 0 | 0 |
| ASXL1 WILD-TYPE | 22 | 25 | 58 | 37 | 41 |
P value = 0.255 (Fisher's exact test), Q value = 0.53
Table S116. Gene #20: 'ASXL1 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| ASXL1 MUTATED | 3 | 0 | 0 |
| ASXL1 WILD-TYPE | 71 | 48 | 45 |
P value = 0.593 (Fisher's exact test), Q value = 0.75
Table S117. Gene #20: 'ASXL1 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| ASXL1 MUTATED | 0 | 0 | 0 | 3 | 0 | 0 |
| ASXL1 WILD-TYPE | 16 | 13 | 15 | 54 | 26 | 40 |
P value = 0.11 (Fisher's exact test), Q value = 0.33
Table S118. Gene #20: 'ASXL1 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| ASXL1 MUTATED | 0 | 1 | 0 | 2 | 0 |
| ASXL1 WILD-TYPE | 55 | 30 | 42 | 30 | 21 |
P value = 0.416 (Fisher's exact test), Q value = 0.65
Table S119. Gene #20: 'ASXL1 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| ASXL1 MUTATED | 0 | 1 | 2 |
| ASXL1 WILD-TYPE | 65 | 34 | 79 |
P value = 0.852 (Fisher's exact test), Q value = 0.9
Table S120. Gene #21: 'SMC1A MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| SMC1A MUTATED | 2 | 2 | 1 | 1 |
| SMC1A WILD-TYPE | 47 | 40 | 53 | 38 |
P value = 0.0544 (Fisher's exact test), Q value = 0.19
Table S121. Gene #21: 'SMC1A MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| SMC1A MUTATED | 0 | 1 | 0 | 1 | 4 |
| SMC1A WILD-TYPE | 22 | 24 | 62 | 36 | 37 |
P value = 0.568 (Fisher's exact test), Q value = 0.75
Table S122. Gene #21: 'SMC1A MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| SMC1A MUTATED | 2 | 3 | 1 |
| SMC1A WILD-TYPE | 72 | 45 | 44 |
P value = 0.229 (Fisher's exact test), Q value = 0.52
Table S123. Gene #21: 'SMC1A MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| SMC1A MUTATED | 0 | 0 | 1 | 1 | 0 | 4 |
| SMC1A WILD-TYPE | 16 | 13 | 14 | 56 | 26 | 36 |
P value = 0.335 (Fisher's exact test), Q value = 0.6
Table S124. Gene #21: 'SMC1A MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| SMC1A MUTATED | 3 | 0 | 3 | 0 | 0 |
| SMC1A WILD-TYPE | 52 | 31 | 39 | 32 | 21 |
P value = 0.576 (Fisher's exact test), Q value = 0.75
Table S125. Gene #21: 'SMC1A MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| SMC1A MUTATED | 3 | 0 | 3 |
| SMC1A WILD-TYPE | 62 | 35 | 78 |
P value = 0.128 (Fisher's exact test), Q value = 0.37
Table S126. Gene #22: 'PTPN11 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| PTPN11 MUTATED | 3 | 4 | 1 | 0 |
| PTPN11 WILD-TYPE | 46 | 38 | 53 | 39 |
P value = 0.164 (Fisher's exact test), Q value = 0.44
Table S127. Gene #22: 'PTPN11 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| PTPN11 MUTATED | 2 | 0 | 1 | 2 | 4 |
| PTPN11 WILD-TYPE | 20 | 25 | 61 | 35 | 37 |
P value = 0.0477 (Fisher's exact test), Q value = 0.19
Table S128. Gene #22: 'PTPN11 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| PTPN11 MUTATED | 1 | 2 | 5 |
| PTPN11 WILD-TYPE | 73 | 46 | 40 |
Figure S36. Get High-res Image Gene #22: 'PTPN11 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'

P value = 0.513 (Fisher's exact test), Q value = 0.7
Table S129. Gene #22: 'PTPN11 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| PTPN11 MUTATED | 0 | 1 | 0 | 2 | 3 | 2 |
| PTPN11 WILD-TYPE | 16 | 12 | 15 | 55 | 23 | 38 |
P value = 0.459 (Fisher's exact test), Q value = 0.67
Table S130. Gene #22: 'PTPN11 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| PTPN11 MUTATED | 4 | 1 | 2 | 0 | 2 |
| PTPN11 WILD-TYPE | 51 | 30 | 40 | 32 | 19 |
P value = 0.185 (Fisher's exact test), Q value = 0.46
Table S131. Gene #22: 'PTPN11 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| PTPN11 MUTATED | 6 | 1 | 2 |
| PTPN11 WILD-TYPE | 59 | 34 | 79 |
P value = 0.791 (Fisher's exact test), Q value = 0.87
Table S132. Gene #23: 'SUZ12 MUTATION STATUS' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 49 | 42 | 54 | 39 |
| SUZ12 MUTATED | 0 | 1 | 1 | 1 |
| SUZ12 WILD-TYPE | 49 | 41 | 53 | 38 |
P value = 0.84 (Fisher's exact test), Q value = 0.9
Table S133. Gene #23: 'SUZ12 MUTATION STATUS' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 22 | 25 | 62 | 37 | 41 |
| SUZ12 MUTATED | 0 | 0 | 2 | 1 | 0 |
| SUZ12 WILD-TYPE | 22 | 25 | 60 | 36 | 41 |
P value = 0.257 (Fisher's exact test), Q value = 0.53
Table S134. Gene #23: 'SUZ12 MUTATION STATUS' versus Molecular Subtype #3: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 74 | 48 | 45 |
| SUZ12 MUTATED | 3 | 0 | 0 |
| SUZ12 WILD-TYPE | 71 | 48 | 45 |
P value = 0.596 (Fisher's exact test), Q value = 0.75
Table S135. Gene #23: 'SUZ12 MUTATION STATUS' versus Molecular Subtype #4: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 | CLUS_6 |
|---|---|---|---|---|---|---|
| ALL | 16 | 13 | 15 | 57 | 26 | 40 |
| SUZ12 MUTATED | 0 | 0 | 0 | 3 | 0 | 0 |
| SUZ12 WILD-TYPE | 16 | 13 | 15 | 54 | 26 | 40 |
P value = 0.326 (Fisher's exact test), Q value = 0.6
Table S136. Gene #23: 'SUZ12 MUTATION STATUS' versus Molecular Subtype #5: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 55 | 31 | 42 | 32 | 21 |
| SUZ12 MUTATED | 0 | 1 | 1 | 0 | 1 |
| SUZ12 WILD-TYPE | 55 | 30 | 41 | 32 | 20 |
P value = 0.139 (Fisher's exact test), Q value = 0.38
Table S137. Gene #23: 'SUZ12 MUTATION STATUS' versus Molecular Subtype #6: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 65 | 35 | 81 |
| SUZ12 MUTATED | 0 | 2 | 1 |
| SUZ12 WILD-TYPE | 65 | 33 | 80 |
-
Mutation data file = sample_sig_gene_table.txt from Mutsig_2CV pipeline
-
Processed Mutation data file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_Correlate_Genomic_Events_Preprocess/LAML-TB/22570965/transformed.cor.cli.txt
-
Molecular subtypes file = /xchip/cga/gdac-prod/tcga-gdac/jobResults/GDAC_mergedClustering/LAML-TB/22541002/LAML-TB.transferedmergedcluster.txt
-
Number of patients = 193
-
Number of significantly mutated genes = 23
-
Number of Molecular subtypes = 6
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.