This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and subtypes.
Testing the association between copy number variation 32 arm-level results and 10 molecular subtypes across 197 patients, 23 significant findings detected with Q value < 0.25.
-
7p gain cnv correlated to 'CN_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
7q gain cnv correlated to 'CN_CNMF' and 'MIRSEQ_MATURE_CNMF'.
-
8p gain cnv correlated to 'CN_CNMF' and 'RPPA_CHIERARCHICAL'.
-
8q gain cnv correlated to 'CN_CNMF', 'RPPA_CNMF', 'RPPA_CHIERARCHICAL', 'MIRSEQ_CNMF', 'MIRSEQ_CHIERARCHICAL', and 'MIRSEQ_MATURE_CNMF'.
-
6q loss cnv correlated to 'CN_CNMF'.
-
8p loss cnv correlated to 'METHLYATION_CNMF', 'MRNASEQ_CNMF', 'MRNASEQ_CHIERARCHICAL', and 'MIRSEQ_CNMF'.
-
13q loss cnv correlated to 'CN_CNMF'.
-
16q loss cnv correlated to 'CN_CNMF' and 'MIRSEQ_CHIERARCHICAL'.
-
17p loss cnv correlated to 'CN_CNMF'.
-
18q loss cnv correlated to 'CN_CNMF'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 32 arm-level results and 10 molecular subtypes. Shown in the table are P values (Q values). Thresholded by Q value < 0.25, 23 significant findings detected.
|
Molecular subtypes |
CN CNMF |
METHLYATION CNMF |
RPPA CNMF |
RPPA CHIERARCHICAL |
MRNASEQ CNMF |
MRNASEQ CHIERARCHICAL |
MIRSEQ CNMF |
MIRSEQ CHIERARCHICAL |
MIRSEQ MATURE CNMF |
MIRSEQ MATURE CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Fisher's exact test | Chi-square test | Fisher's exact test | Chi-square test | Fisher's exact test | |
| 8q gain | 0 (0%) | 173 |
2.28e-08 (7.14e-06) |
0.129 (1.00) |
0.00025 (0.0747) |
0.000107 (0.0323) |
0.00976 (1.00) |
0.00615 (1.00) |
0.000351 (0.104) |
1.01e-05 (0.00308) |
4.53e-06 (0.00139) |
0.061 (1.00) |
| 8p loss | 0 (0%) | 149 |
0.00345 (0.98) |
4.24e-06 (0.0013) |
0.556 (1.00) |
0.824 (1.00) |
0.000407 (0.12) |
0.000413 (0.122) |
1.66e-05 (0.00504) |
0.482 (1.00) |
0.0092 (1.00) |
0.0842 (1.00) |
| 7p gain | 0 (0%) | 180 |
9.28e-08 (2.9e-05) |
0.0299 (1.00) |
0.0455 (1.00) |
0.115 (1.00) |
0.041 (1.00) |
0.0349 (1.00) |
0.0141 (1.00) |
0.000276 (0.0821) |
0.000205 (0.0614) |
0.139 (1.00) |
| 7q gain | 0 (0%) | 181 |
4.16e-07 (0.000129) |
0.0247 (1.00) |
0.0866 (1.00) |
0.219 (1.00) |
0.0626 (1.00) |
0.124 (1.00) |
0.0107 (1.00) |
0.00177 (0.511) |
0.000104 (0.0313) |
0.316 (1.00) |
| 8p gain | 0 (0%) | 186 |
1.93e-06 (0.000596) |
0.109 (1.00) |
0.000875 (0.255) |
0.000425 (0.125) |
0.0403 (1.00) |
0.0688 (1.00) |
0.00748 (1.00) |
0.00609 (1.00) |
0.00366 (1.00) |
0.32 (1.00) |
| 16q loss | 0 (0%) | 169 |
8.64e-09 (2.71e-06) |
0.00343 (0.976) |
0.415 (1.00) |
0.109 (1.00) |
0.0304 (1.00) |
0.00252 (0.721) |
0.00917 (1.00) |
0.000463 (0.136) |
0.0357 (1.00) |
0.243 (1.00) |
| 6q loss | 0 (0%) | 188 |
0.000736 (0.215) |
0.0656 (1.00) |
0.488 (1.00) |
0.629 (1.00) |
0.0824 (1.00) |
0.0149 (1.00) |
0.306 (1.00) |
0.15 (1.00) |
0.386 (1.00) |
0.722 (1.00) |
| 13q loss | 0 (0%) | 182 |
3.11e-06 (0.000957) |
0.0541 (1.00) |
0.00606 (1.00) |
0.0194 (1.00) |
0.0098 (1.00) |
0.0211 (1.00) |
0.243 (1.00) |
0.144 (1.00) |
0.262 (1.00) |
0.543 (1.00) |
| 17p loss | 0 (0%) | 173 |
6.49e-07 (0.000201) |
0.00999 (1.00) |
0.0175 (1.00) |
0.0282 (1.00) |
0.0187 (1.00) |
0.00155 (0.449) |
0.0383 (1.00) |
0.043 (1.00) |
0.0242 (1.00) |
0.322 (1.00) |
| 18q loss | 0 (0%) | 171 |
7.98e-05 (0.0242) |
0.233 (1.00) |
0.0059 (1.00) |
0.00786 (1.00) |
0.113 (1.00) |
0.0341 (1.00) |
0.383 (1.00) |
0.0304 (1.00) |
0.133 (1.00) |
0.334 (1.00) |
| 1p gain | 0 (0%) | 194 |
0.0497 (1.00) |
0.0233 (1.00) |
0.781 (1.00) |
0.636 (1.00) |
0.109 (1.00) |
0.185 (1.00) |
0.566 (1.00) |
0.267 (1.00) |
0.00368 (1.00) |
0.736 (1.00) |
| 1q gain | 0 (0%) | 192 |
0.00405 (1.00) |
0.227 (1.00) |
0.189 (1.00) |
0.129 (1.00) |
0.376 (1.00) |
0.118 (1.00) |
0.405 (1.00) |
0.272 (1.00) |
0.0807 (1.00) |
0.693 (1.00) |
| 3p gain | 0 (0%) | 192 |
0.553 (1.00) |
0.445 (1.00) |
0.189 (1.00) |
0.129 (1.00) |
0.131 (1.00) |
0.118 (1.00) |
0.0404 (1.00) |
0.0242 (1.00) |
0.459 (1.00) |
0.693 (1.00) |
| 3q gain | 0 (0%) | 190 |
0.2 (1.00) |
0.299 (1.00) |
0.0748 (1.00) |
0.0499 (1.00) |
0.0186 (1.00) |
0.0482 (1.00) |
0.162 (1.00) |
0.00242 (0.695) |
0.0164 (1.00) |
0.413 (1.00) |
| 9p gain | 0 (0%) | 194 |
0.0497 (1.00) |
0.384 (1.00) |
0.775 (1.00) |
0.0572 (1.00) |
0.26 (1.00) |
0.501 (1.00) |
0.867 (1.00) |
1 (1.00) |
||
| 9q gain | 0 (0%) | 189 |
0.00404 (1.00) |
0.0854 (1.00) |
0.0479 (1.00) |
0.0638 (1.00) |
0.134 (1.00) |
0.176 (1.00) |
0.389 (1.00) |
0.0915 (1.00) |
0.0231 (1.00) |
0.535 (1.00) |
| 10q gain | 0 (0%) | 193 |
0.187 (1.00) |
0.462 (1.00) |
0.113 (1.00) |
0.11 (1.00) |
0.328 (1.00) |
0.131 (1.00) |
0.65 (1.00) |
0.0572 (1.00) |
0.874 (1.00) |
0.625 (1.00) |
| 12q gain | 0 (0%) | 194 |
0.556 (1.00) |
0.195 (1.00) |
0.484 (1.00) |
1 (1.00) |
0.528 (1.00) |
1 (1.00) |
0.935 (1.00) |
0.585 (1.00) |
0.535 (1.00) |
0.736 (1.00) |
| 16p gain | 0 (0%) | 194 |
0.0497 (1.00) |
0.384 (1.00) |
0.781 (1.00) |
0.636 (1.00) |
0.775 (1.00) |
0.0572 (1.00) |
0.566 (1.00) |
0.585 (1.00) |
0.837 (1.00) |
0.736 (1.00) |
| 16q gain | 0 (0%) | 194 |
0.0497 (1.00) |
0.384 (1.00) |
0.781 (1.00) |
0.636 (1.00) |
0.775 (1.00) |
0.0572 (1.00) |
0.566 (1.00) |
0.585 (1.00) |
0.837 (1.00) |
0.736 (1.00) |
| 5q loss | 0 (0%) | 192 |
0.00405 (1.00) |
0.0362 (1.00) |
0.452 (1.00) |
0.318 (1.00) |
0.131 (1.00) |
0.0472 (1.00) |
0.405 (1.00) |
0.0104 (1.00) |
0.602 (1.00) |
1 (1.00) |
| 8q loss | 0 (0%) | 193 |
0.187 (1.00) |
0.317 (1.00) |
0.561 (1.00) |
1 (1.00) |
0.328 (1.00) |
0.131 (1.00) |
0.263 (1.00) |
0.448 (1.00) |
0.605 (1.00) |
0.362 (1.00) |
| 10p loss | 0 (0%) | 190 |
0.0109 (1.00) |
0.0897 (1.00) |
0.0748 (1.00) |
0.0499 (1.00) |
0.181 (1.00) |
0.0401 (1.00) |
0.22 (1.00) |
0.0268 (1.00) |
0.246 (1.00) |
0.413 (1.00) |
| 10q loss | 0 (0%) | 190 |
0.0109 (1.00) |
0.562 (1.00) |
0.189 (1.00) |
0.129 (1.00) |
0.511 (1.00) |
0.15 (1.00) |
0.254 (1.00) |
0.446 (1.00) |
0.246 (1.00) |
0.413 (1.00) |
| 12p loss | 0 (0%) | 186 |
0.011 (1.00) |
0.0644 (1.00) |
0.231 (1.00) |
0.629 (1.00) |
0.034 (1.00) |
0.0208 (1.00) |
0.241 (1.00) |
0.271 (1.00) |
0.348 (1.00) |
0.691 (1.00) |
| 15q loss | 0 (0%) | 194 |
0.0497 (1.00) |
0.0233 (1.00) |
0.781 (1.00) |
0.636 (1.00) |
0.109 (1.00) |
0.185 (1.00) |
0.553 (1.00) |
0.0452 (1.00) |
0.00377 (1.00) |
0.512 (1.00) |
| 16p loss | 0 (0%) | 193 |
0.018 (1.00) |
0.00649 (1.00) |
0.0353 (1.00) |
0.178 (1.00) |
0.00923 (1.00) |
0.0452 (1.00) |
0.00377 (1.00) |
0.0468 (1.00) |
||
| 18p loss | 0 (0%) | 178 |
0.00197 (0.568) |
0.177 (1.00) |
0.0263 (1.00) |
0.0279 (1.00) |
0.47 (1.00) |
0.117 (1.00) |
0.166 (1.00) |
0.129 (1.00) |
0.0274 (1.00) |
0.0638 (1.00) |
| 20p loss | 0 (0%) | 192 |
0.553 (1.00) |
0.0362 (1.00) |
0.537 (1.00) |
0.736 (1.00) |
0.131 (1.00) |
0.304 (1.00) |
0.263 (1.00) |
0.0283 (1.00) |
0.459 (1.00) |
0.268 (1.00) |
| 21q loss | 0 (0%) | 192 |
0.0598 (1.00) |
0.273 (1.00) |
0.484 (1.00) |
0.777 (1.00) |
0.131 (1.00) |
0.147 (1.00) |
0.753 (1.00) |
0.187 (1.00) |
0.769 (1.00) |
0.835 (1.00) |
| 22q loss | 0 (0%) | 192 |
0.0186 (1.00) |
0.273 (1.00) |
0.837 (1.00) |
0.684 (1.00) |
0.376 (1.00) |
0.118 (1.00) |
0.787 (1.00) |
1 (1.00) |
0.491 (1.00) |
1 (1.00) |
| Xq loss | 0 (0%) | 194 |
0.123 (1.00) |
0.0233 (1.00) |
0.109 (1.00) |
0.185 (1.00) |
0.349 (1.00) |
0.585 (1.00) |
0.311 (1.00) |
0.26 (1.00) |
P value = 9.28e-08 (Fisher's exact test), Q value = 2.9e-05
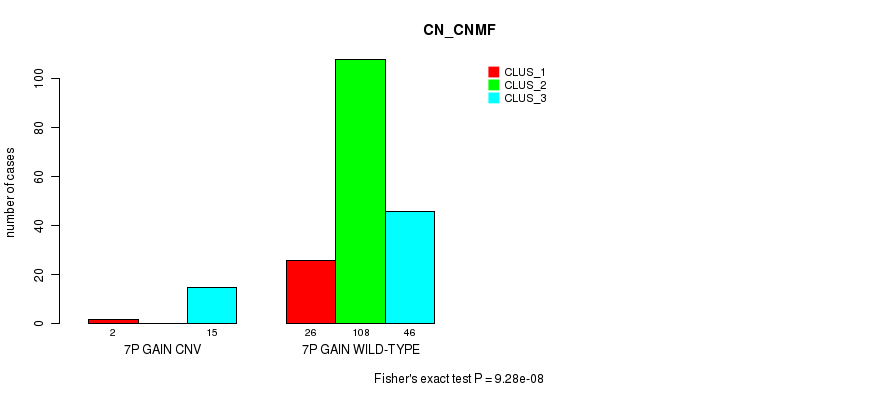
Table S1. Gene #5: '7p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 28 | 108 | 61 |
| 7P GAIN CNV | 2 | 0 | 15 |
| 7P GAIN WILD-TYPE | 26 | 108 | 46 |
Figure S1. Get High-res Image Gene #5: '7p gain' versus Molecular Subtype #1: 'CN_CNMF'
P value = 0.000276 (Fisher's exact test), Q value = 0.082
Table S2. Gene #5: '7p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 34 | 21 | 74 | 67 |
| 7P GAIN CNV | 1 | 8 | 5 | 3 |
| 7P GAIN WILD-TYPE | 33 | 13 | 69 | 64 |
Figure S2. Get High-res Image Gene #5: '7p gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'

P value = 0.000205 (Chi-square test), Q value = 0.061
Table S3. Gene #5: '7p gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 73 | 18 | 12 | 15 | 78 |
| 7P GAIN CNV | 6 | 0 | 0 | 6 | 5 |
| 7P GAIN WILD-TYPE | 67 | 18 | 12 | 9 | 73 |
Figure S3. Get High-res Image Gene #5: '7p gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'

P value = 4.16e-07 (Fisher's exact test), Q value = 0.00013
Table S4. Gene #6: '7q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 28 | 108 | 61 |
| 7Q GAIN CNV | 2 | 0 | 14 |
| 7Q GAIN WILD-TYPE | 26 | 108 | 47 |
Figure S4. Get High-res Image Gene #6: '7q gain' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.000104 (Chi-square test), Q value = 0.031
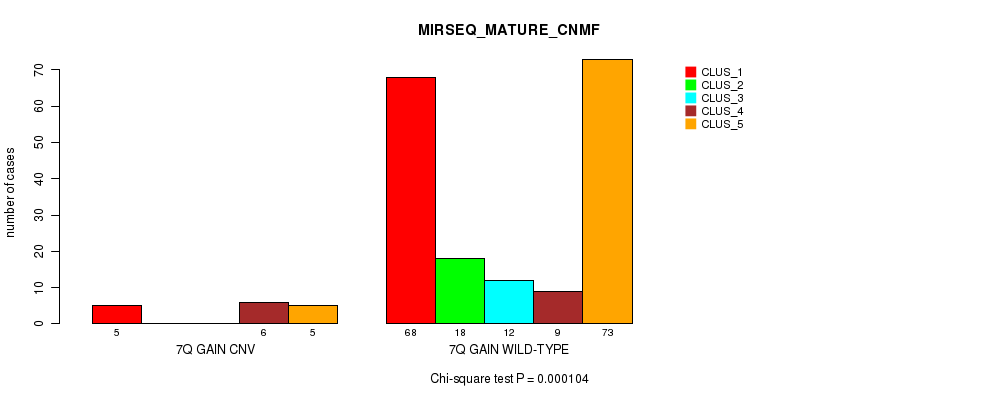
Table S5. Gene #6: '7q gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 73 | 18 | 12 | 15 | 78 |
| 7Q GAIN CNV | 5 | 0 | 0 | 6 | 5 |
| 7Q GAIN WILD-TYPE | 68 | 18 | 12 | 9 | 73 |
Figure S5. Get High-res Image Gene #6: '7q gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
P value = 1.93e-06 (Fisher's exact test), Q value = 6e-04
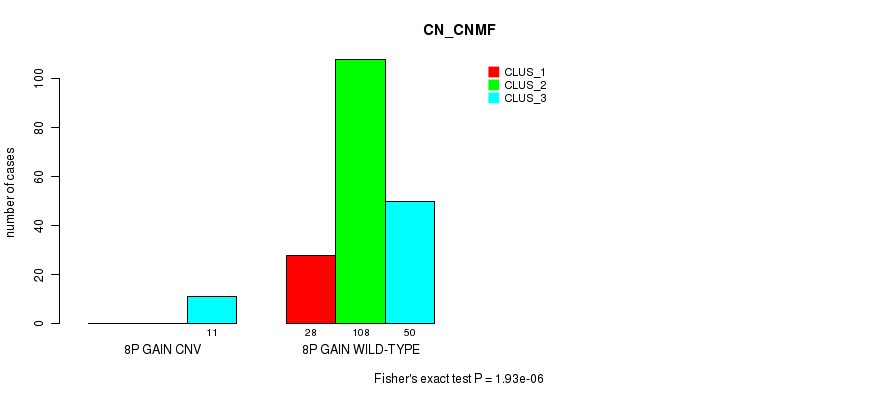
Table S6. Gene #7: '8p gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 28 | 108 | 61 |
| 8P GAIN CNV | 0 | 0 | 11 |
| 8P GAIN WILD-TYPE | 28 | 108 | 50 |
Figure S6. Get High-res Image Gene #7: '8p gain' versus Molecular Subtype #1: 'CN_CNMF'
P value = 0.000425 (Fisher's exact test), Q value = 0.12
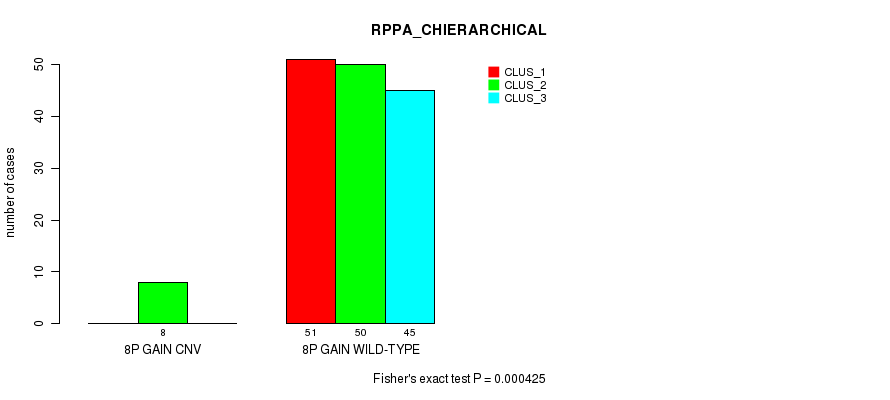
Table S7. Gene #7: '8p gain' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 51 | 58 | 45 |
| 8P GAIN CNV | 0 | 8 | 0 |
| 8P GAIN WILD-TYPE | 51 | 50 | 45 |
Figure S7. Get High-res Image Gene #7: '8p gain' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
P value = 2.28e-08 (Fisher's exact test), Q value = 7.1e-06
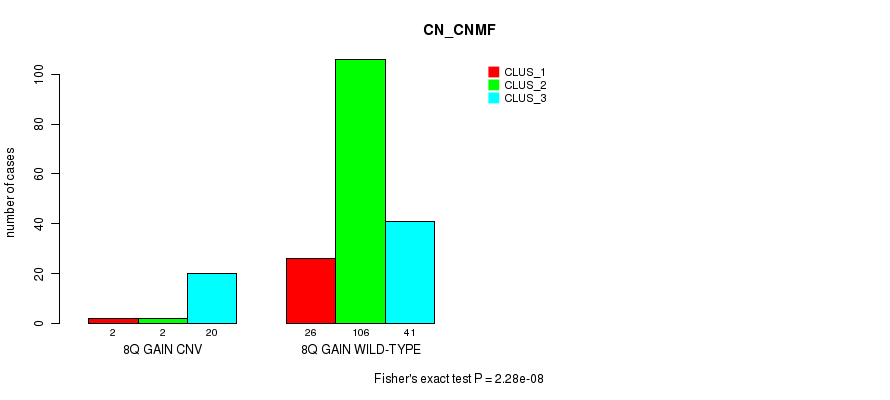
Table S8. Gene #8: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 28 | 108 | 61 |
| 8Q GAIN CNV | 2 | 2 | 20 |
| 8Q GAIN WILD-TYPE | 26 | 106 | 41 |
Figure S8. Get High-res Image Gene #8: '8q gain' versus Molecular Subtype #1: 'CN_CNMF'
P value = 0.00025 (Fisher's exact test), Q value = 0.075
Table S9. Gene #8: '8q gain' versus Molecular Subtype #3: 'RPPA_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 41 | 56 | 57 |
| 8Q GAIN CNV | 0 | 4 | 14 |
| 8Q GAIN WILD-TYPE | 41 | 52 | 43 |
Figure S9. Get High-res Image Gene #8: '8q gain' versus Molecular Subtype #3: 'RPPA_CNMF'

P value = 0.000107 (Fisher's exact test), Q value = 0.032
Table S10. Gene #8: '8q gain' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 51 | 58 | 45 |
| 8Q GAIN CNV | 0 | 14 | 4 |
| 8Q GAIN WILD-TYPE | 51 | 44 | 41 |
Figure S10. Get High-res Image Gene #8: '8q gain' versus Molecular Subtype #4: 'RPPA_CHIERARCHICAL'

P value = 0.000351 (Chi-square test), Q value = 0.1
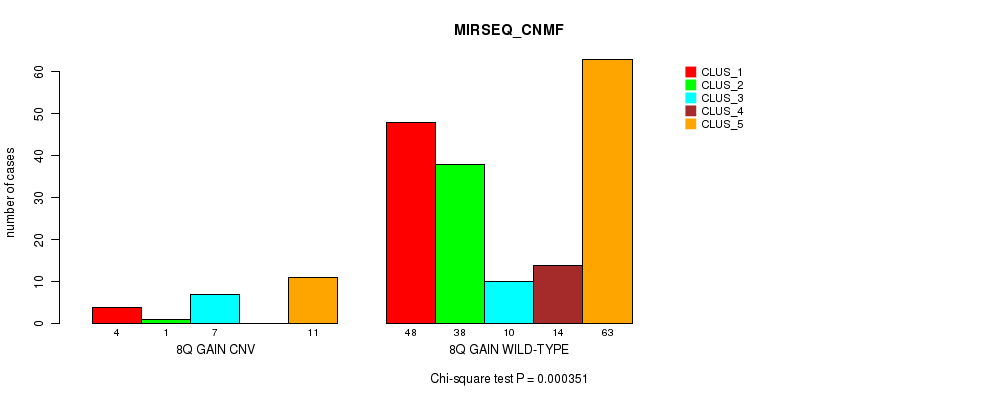
Table S11. Gene #8: '8q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 52 | 39 | 17 | 14 | 74 |
| 8Q GAIN CNV | 4 | 1 | 7 | 0 | 11 |
| 8Q GAIN WILD-TYPE | 48 | 38 | 10 | 14 | 63 |
Figure S11. Get High-res Image Gene #8: '8q gain' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
P value = 1.01e-05 (Fisher's exact test), Q value = 0.0031
Table S12. Gene #8: '8q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 34 | 21 | 74 | 67 |
| 8Q GAIN CNV | 2 | 9 | 11 | 1 |
| 8Q GAIN WILD-TYPE | 32 | 12 | 63 | 66 |
Figure S12. Get High-res Image Gene #8: '8q gain' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'

P value = 4.53e-06 (Chi-square test), Q value = 0.0014
Table S13. Gene #8: '8q gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 73 | 18 | 12 | 15 | 78 |
| 8Q GAIN CNV | 4 | 1 | 0 | 8 | 10 |
| 8Q GAIN WILD-TYPE | 69 | 17 | 12 | 7 | 68 |
Figure S13. Get High-res Image Gene #8: '8q gain' versus Molecular Subtype #9: 'MIRSEQ_MATURE_CNMF'

P value = 0.000736 (Fisher's exact test), Q value = 0.21
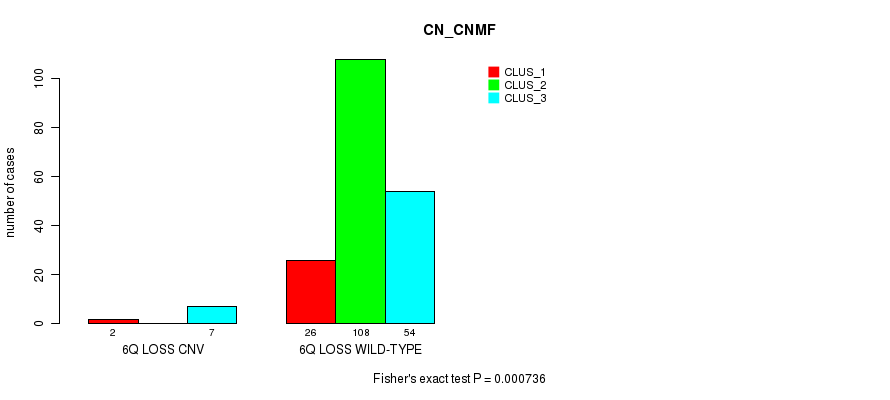
Table S14. Gene #16: '6q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 28 | 108 | 61 |
| 6Q LOSS CNV | 2 | 0 | 7 |
| 6Q LOSS WILD-TYPE | 26 | 108 | 54 |
Figure S14. Get High-res Image Gene #16: '6q loss' versus Molecular Subtype #1: 'CN_CNMF'
P value = 4.24e-06 (Fisher's exact test), Q value = 0.0013
Table S15. Gene #17: '8p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 57 | 65 | 75 |
| 8P LOSS CNV | 11 | 5 | 32 |
| 8P LOSS WILD-TYPE | 46 | 60 | 43 |
Figure S15. Get High-res Image Gene #17: '8p loss' versus Molecular Subtype #2: 'METHLYATION_CNMF'

P value = 0.000407 (Fisher's exact test), Q value = 0.12
Table S16. Gene #17: '8p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 67 | 60 | 67 |
| 8P LOSS CNV | 12 | 8 | 28 |
| 8P LOSS WILD-TYPE | 55 | 52 | 39 |
Figure S16. Get High-res Image Gene #17: '8p loss' versus Molecular Subtype #5: 'MRNASEQ_CNMF'

P value = 0.000413 (Fisher's exact test), Q value = 0.12
Table S17. Gene #17: '8p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 49 | 68 | 77 |
| 8P LOSS CNV | 10 | 28 | 10 |
| 8P LOSS WILD-TYPE | 39 | 40 | 67 |
Figure S17. Get High-res Image Gene #17: '8p loss' versus Molecular Subtype #6: 'MRNASEQ_CHIERARCHICAL'

P value = 1.66e-05 (Chi-square test), Q value = 0.005
Table S18. Gene #17: '8p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 | CLUS_5 |
|---|---|---|---|---|---|
| ALL | 52 | 39 | 17 | 14 | 74 |
| 8P LOSS CNV | 24 | 2 | 2 | 0 | 20 |
| 8P LOSS WILD-TYPE | 28 | 37 | 15 | 14 | 54 |
Figure S18. Get High-res Image Gene #17: '8p loss' versus Molecular Subtype #7: 'MIRSEQ_CNMF'

P value = 3.11e-06 (Fisher's exact test), Q value = 0.00096
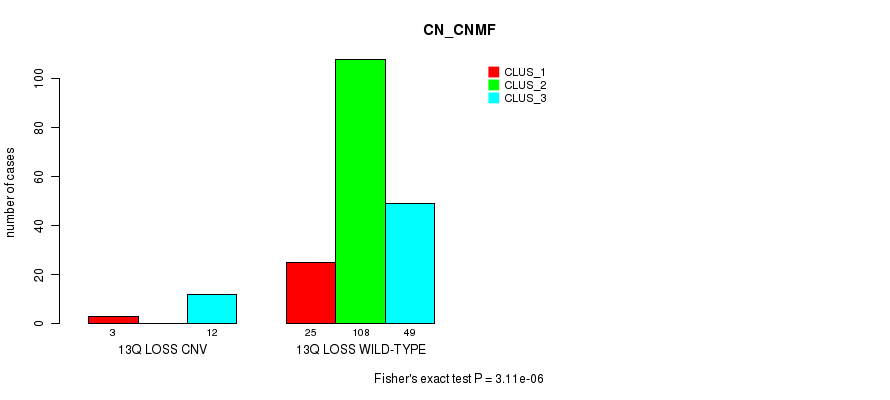
Table S19. Gene #22: '13q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 28 | 108 | 61 |
| 13Q LOSS CNV | 3 | 0 | 12 |
| 13Q LOSS WILD-TYPE | 25 | 108 | 49 |
Figure S19. Get High-res Image Gene #22: '13q loss' versus Molecular Subtype #1: 'CN_CNMF'
P value = 8.64e-09 (Fisher's exact test), Q value = 2.7e-06
Table S20. Gene #25: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 28 | 108 | 61 |
| 16Q LOSS CNV | 1 | 4 | 23 |
| 16Q LOSS WILD-TYPE | 27 | 104 | 38 |
Figure S20. Get High-res Image Gene #25: '16q loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 0.000463 (Fisher's exact test), Q value = 0.14
Table S21. Gene #25: '16q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 | CLUS_4 |
|---|---|---|---|---|
| ALL | 34 | 21 | 74 | 67 |
| 16Q LOSS CNV | 2 | 9 | 12 | 4 |
| 16Q LOSS WILD-TYPE | 32 | 12 | 62 | 63 |
Figure S21. Get High-res Image Gene #25: '16q loss' versus Molecular Subtype #8: 'MIRSEQ_CHIERARCHICAL'
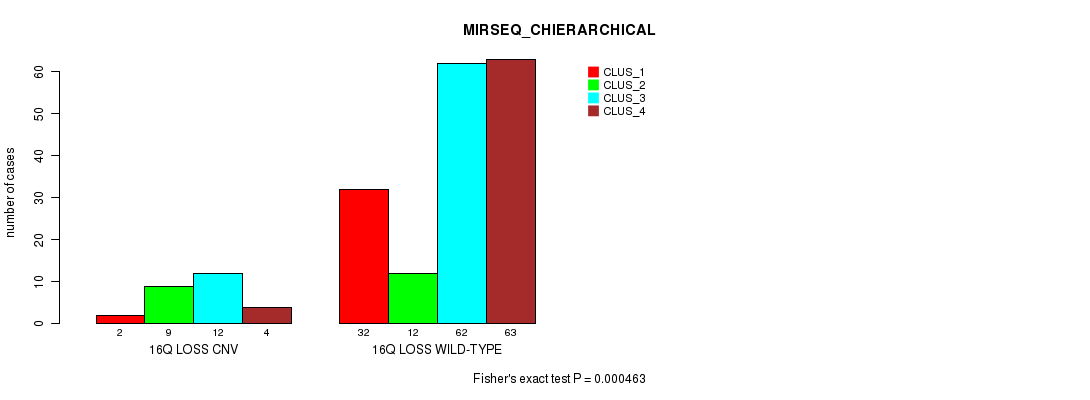
P value = 6.49e-07 (Fisher's exact test), Q value = 2e-04
Table S22. Gene #26: '17p loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 28 | 108 | 61 |
| 17P LOSS CNV | 0 | 5 | 19 |
| 17P LOSS WILD-TYPE | 28 | 103 | 42 |
Figure S22. Get High-res Image Gene #26: '17p loss' versus Molecular Subtype #1: 'CN_CNMF'

P value = 7.98e-05 (Fisher's exact test), Q value = 0.024
Table S23. Gene #28: '18q loss' versus Molecular Subtype #1: 'CN_CNMF'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 28 | 108 | 61 |
| 18Q LOSS CNV | 1 | 7 | 18 |
| 18Q LOSS WILD-TYPE | 27 | 101 | 43 |
Figure S23. Get High-res Image Gene #28: '18q loss' versus Molecular Subtype #1: 'CN_CNMF'
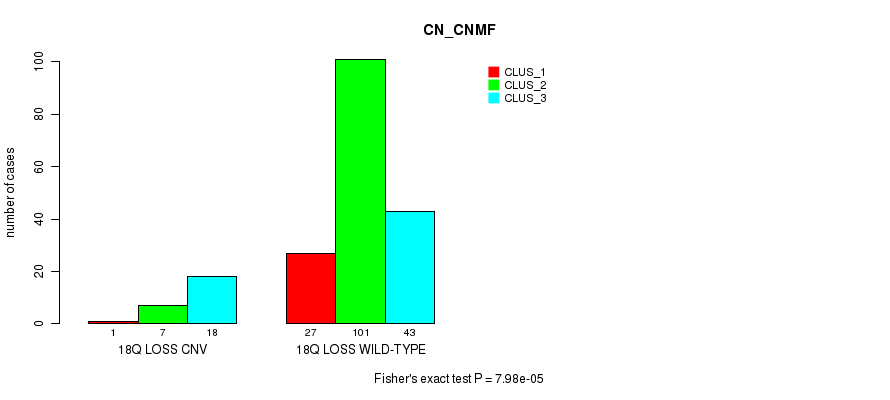
-
Mutation data file = broad_values_by_arm.mutsig.cluster.txt
-
Molecular subtypes file = PRAD-TP.transferedmergedcluster.txt
-
Number of patients = 197
-
Number of significantly arm-level cnvs = 32
-
Number of molecular subtypes = 10
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multi-class clinical features (nominal or ordinal), Chi-square tests (Greenwood and Nikulin 1996) were used to estimate the P values using the 'chisq.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.
In addition to the links below, the full results of the analysis summarized in this report can also be downloaded programmatically using firehose_get, or interactively from either the Broad GDAC website or TCGA Data Coordination Center Portal.