This pipeline computes the correlation between significant arm-level copy number variations (cnvs) and molecular subtypes.
Testing the association between copy number variation 95 arm-level events and 2 molecular subtypes across 110 patients, 54 significant findings detected with P value < 0.05 and Q value < 0.25.
-
1p gain cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
2p gain cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
2q gain cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
5q gain cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
7p gain cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
7q gain cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
10q gain cnv correlated to 'LINCRNA_CHIERARCHICAL'.
-
12p gain cnv correlated to 'MRNA_CHIERARCHICAL'.
-
16p gain cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
16q gain cnv correlated to 'MRNA_CHIERARCHICAL'.
-
18p gain cnv correlated to 'MRNA_CHIERARCHICAL'.
-
18q gain cnv correlated to 'MRNA_CHIERARCHICAL'.
-
20p gain cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
21q gain cnv correlated to 'LINCRNA_CHIERARCHICAL'.
-
xp gain cnv correlated to 'MRNA_CHIERARCHICAL'.
-
1p loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
3p loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
3q loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
5q loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
10q loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
11p loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
12p loss cnv correlated to 'LINCRNA_CHIERARCHICAL'.
-
12q loss cnv correlated to 'LINCRNA_CHIERARCHICAL'.
-
15q loss cnv correlated to 'LINCRNA_CHIERARCHICAL'.
-
16p loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
18p loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
18q loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
21p loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
21q loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
22p loss cnv correlated to 'MRNA_CHIERARCHICAL'.
-
22q loss cnv correlated to 'MRNA_CHIERARCHICAL'.
-
yp loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
-
yq loss cnv correlated to 'MRNA_CHIERARCHICAL' and 'LINCRNA_CHIERARCHICAL'.
Table 1. Get Full Table Overview of the association between significant copy number variation of 95 arm-level events and 2 molecular subtypes. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, 54 significant findings detected.
|
Clinical Features |
MRNA CHIERARCHICAL |
LINCRNA CHIERARCHICAL |
||
| nCNV (%) | nWild-Type | Fisher's exact test | Fisher's exact test | |
| 1p gain | 32 (29%) | 78 |
0.0229 (0.124) |
0.0286 (0.139) |
| 2p gain | 23 (21%) | 87 |
0.00555 (0.0662) |
0.0186 (0.114) |
| 2q gain | 21 (19%) | 89 |
0.0167 (0.11) |
0.0353 (0.156) |
| 5q gain | 26 (24%) | 84 |
0.0156 (0.11) |
0.00038 (0.0361) |
| 7p gain | 58 (53%) | 52 |
0.0301 (0.139) |
0.00974 (0.0839) |
| 7q gain | 48 (44%) | 62 |
0.0205 (0.117) |
0.0323 (0.146) |
| 16p gain | 37 (34%) | 73 |
0.00023 (0.0361) |
0.00248 (0.0581) |
| 20p gain | 24 (22%) | 86 |
0.0472 (0.175) |
0.04 (0.161) |
| 1p loss | 16 (15%) | 94 |
0.0266 (0.136) |
0.0123 (0.0897) |
| 3p loss | 36 (33%) | 74 |
0.00372 (0.0622) |
0.0299 (0.139) |
| 3q loss | 27 (25%) | 83 |
0.00628 (0.0662) |
0.00301 (0.0581) |
| 5q loss | 19 (17%) | 91 |
0.00163 (0.0581) |
0.00243 (0.0581) |
| 10q loss | 28 (25%) | 82 |
0.0496 (0.175) |
0.0271 (0.136) |
| 11p loss | 27 (25%) | 83 |
0.0102 (0.0839) |
0.011 (0.087) |
| 16p loss | 15 (14%) | 95 |
0.00668 (0.0662) |
0.0414 (0.161) |
| 18p loss | 40 (36%) | 70 |
0.0209 (0.117) |
0.00536 (0.0662) |
| 18q loss | 46 (42%) | 64 |
0.0369 (0.156) |
0.012 (0.0897) |
| 21p loss | 42 (38%) | 68 |
0.0174 (0.11) |
0.0065 (0.0662) |
| 21q loss | 38 (35%) | 72 |
0.0201 (0.117) |
0.00306 (0.0581) |
| yp loss | 63 (57%) | 47 |
0.00668 (0.0662) |
0.00219 (0.0581) |
| yq loss | 64 (58%) | 46 |
0.0497 (0.175) |
0.00393 (0.0622) |
| 10q gain | 7 (6%) | 103 |
0.355 (0.564) |
0.0408 (0.161) |
| 12p gain | 23 (21%) | 87 |
0.0398 (0.161) |
0.113 (0.27) |
| 16q gain | 28 (25%) | 82 |
0.026 (0.136) |
0.0909 (0.24) |
| 18p gain | 8 (7%) | 102 |
0.0169 (0.11) |
0.0765 (0.229) |
| 18q gain | 7 (6%) | 103 |
0.0433 (0.164) |
0.226 (0.413) |
| 21q gain | 16 (15%) | 94 |
0.396 (0.593) |
0.0495 (0.175) |
| xp gain | 46 (42%) | 64 |
0.0365 (0.156) |
0.884 (0.933) |
| 12p loss | 24 (22%) | 86 |
0.0948 (0.244) |
0.00201 (0.0581) |
| 12q loss | 23 (21%) | 87 |
0.109 (0.27) |
0.00172 (0.0581) |
| 15q loss | 52 (47%) | 58 |
0.086 (0.234) |
0.00975 (0.0839) |
| 22p loss | 47 (43%) | 63 |
0.00553 (0.0662) |
0.521 (0.704) |
| 22q loss | 49 (45%) | 61 |
0.00697 (0.0662) |
0.627 (0.759) |
| 1q gain | 66 (60%) | 44 |
0.299 (0.502) |
0.455 (0.654) |
| 3p gain | 10 (9%) | 100 |
0.765 (0.855) |
0.839 (0.901) |
| 3q gain | 18 (16%) | 92 |
0.607 (0.744) |
0.487 (0.67) |
| 4p gain | 8 (7%) | 102 |
1 (1.00) |
0.808 (0.884) |
| 4q gain | 7 (6%) | 103 |
0.416 (0.608) |
0.13 (0.291) |
| 5p gain | 57 (52%) | 53 |
0.409 (0.602) |
0.21 (0.398) |
| 6p gain | 20 (18%) | 90 |
0.777 (0.863) |
0.326 (0.534) |
| 6q gain | 10 (9%) | 100 |
1 (1.00) |
0.644 (0.774) |
| 8p gain | 20 (18%) | 90 |
0.201 (0.389) |
0.861 (0.919) |
| 8q gain | 34 (31%) | 76 |
0.604 (0.744) |
0.965 (0.996) |
| 9p gain | 7 (6%) | 103 |
0.534 (0.704) |
0.289 (0.494) |
| 9q gain | 5 (5%) | 105 |
1 (1.00) |
0.73 (0.834) |
| 10p gain | 15 (14%) | 95 |
0.086 (0.234) |
0.276 (0.485) |
| 11p gain | 21 (19%) | 89 |
0.283 (0.492) |
0.16 (0.337) |
| 11q gain | 24 (22%) | 86 |
0.837 (0.901) |
0.561 (0.725) |
| 12q gain | 22 (20%) | 88 |
0.0873 (0.234) |
0.232 (0.42) |
| 13p gain | 7 (6%) | 103 |
0.537 (0.704) |
0.477 (0.661) |
| 13q gain | 4 (4%) | 106 |
0.687 (0.796) |
0.469 (0.659) |
| 14p gain | 26 (24%) | 84 |
0.805 (0.884) |
0.717 (0.825) |
| 14q gain | 27 (25%) | 83 |
0.96 (0.996) |
0.142 (0.311) |
| 15p gain | 9 (8%) | 101 |
0.109 (0.27) |
0.189 (0.382) |
| 17p gain | 22 (20%) | 88 |
0.593 (0.744) |
0.684 (0.796) |
| 17q gain | 30 (27%) | 80 |
0.891 (0.935) |
0.156 (0.334) |
| 19p gain | 14 (13%) | 96 |
0.196 (0.384) |
0.314 (0.524) |
| 19q gain | 15 (14%) | 95 |
0.266 (0.473) |
0.384 (0.579) |
| 20q gain | 29 (26%) | 81 |
0.0546 (0.189) |
0.17 (0.35) |
| 21p gain | 4 (4%) | 106 |
0.363 (0.564) |
0.377 (0.574) |
| 22p gain | 4 (4%) | 106 |
0.687 (0.796) |
0.472 (0.659) |
| 22q gain | 3 (3%) | 107 |
0.333 (0.54) |
0.259 (0.465) |
| xq gain | 49 (45%) | 61 |
0.103 (0.262) |
0.97 (0.996) |
| yp gain | 24 (22%) | 86 |
0.0657 (0.208) |
0.22 (0.406) |
| yq gain | 17 (15%) | 93 |
0.365 (0.564) |
0.75 (0.843) |
| 1q loss | 6 (5%) | 104 |
0.559 (0.725) |
0.653 (0.775) |
| 2p loss | 8 (7%) | 102 |
0.574 (0.733) |
0.472 (0.659) |
| 2q loss | 10 (9%) | 100 |
0.362 (0.564) |
0.369 (0.565) |
| 4p loss | 19 (17%) | 91 |
0.064 (0.207) |
0.191 (0.382) |
| 4q loss | 19 (17%) | 91 |
0.0643 (0.207) |
0.0692 (0.216) |
| 5p loss | 7 (6%) | 103 |
0.215 (0.401) |
0.15 (0.325) |
| 6p loss | 19 (17%) | 91 |
0.113 (0.27) |
0.123 (0.281) |
| 6q loss | 31 (28%) | 79 |
0.136 (0.301) |
0.119 (0.279) |
| 7p loss | 8 (7%) | 102 |
0.46 (0.657) |
0.167 (0.348) |
| 7q loss | 9 (8%) | 101 |
0.675 (0.796) |
0.127 (0.287) |
| 8p loss | 33 (30%) | 77 |
0.196 (0.384) |
0.215 (0.401) |
| 8q loss | 15 (14%) | 95 |
0.285 (0.492) |
0.295 (0.5) |
| 9p loss | 53 (48%) | 57 |
0.538 (0.704) |
0.365 (0.564) |
| 9q loss | 54 (49%) | 56 |
0.535 (0.704) |
0.527 (0.704) |
| 10p loss | 26 (24%) | 84 |
0.0815 (0.231) |
0.114 (0.27) |
| 11q loss | 22 (20%) | 88 |
0.121 (0.281) |
0.175 (0.358) |
| 13p loss | 50 (45%) | 60 |
0.354 (0.564) |
0.832 (0.901) |
| 13q loss | 48 (44%) | 62 |
0.586 (0.743) |
0.0726 (0.222) |
| 14p loss | 34 (31%) | 76 |
0.931 (0.972) |
0.32 (0.529) |
| 14q loss | 10 (9%) | 100 |
0.0772 (0.229) |
0.407 (0.602) |
| 15p loss | 58 (53%) | 52 |
0.0942 (0.244) |
0.0558 (0.189) |
| 16q loss | 24 (22%) | 86 |
0.0786 (0.229) |
0.084 (0.234) |
| 17p loss | 24 (22%) | 86 |
0.511 (0.698) |
1 (1.00) |
| 17q loss | 18 (16%) | 92 |
1 (1.00) |
0.575 (0.733) |
| 19p loss | 45 (41%) | 65 |
0.0638 (0.207) |
0.0794 (0.229) |
| 19q loss | 35 (32%) | 75 |
0.81 (0.884) |
0.733 (0.834) |
| 20p loss | 26 (24%) | 84 |
0.597 (0.744) |
0.206 (0.396) |
| 20q loss | 21 (19%) | 89 |
0.744 (0.842) |
0.648 (0.774) |
| xp loss | 13 (12%) | 97 |
0.867 (0.92) |
0.612 (0.745) |
| xq loss | 11 (10%) | 99 |
0.602 (0.744) |
0.435 (0.631) |
P value = 0.0229 (Fisher's exact test), Q value = 0.12
Table S1. Gene #1: '1p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 1P GAIN MUTATED | 5 | 6 | 21 |
| 1P GAIN WILD-TYPE | 28 | 21 | 29 |
Figure S1. Get High-res Image Gene #1: '1p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
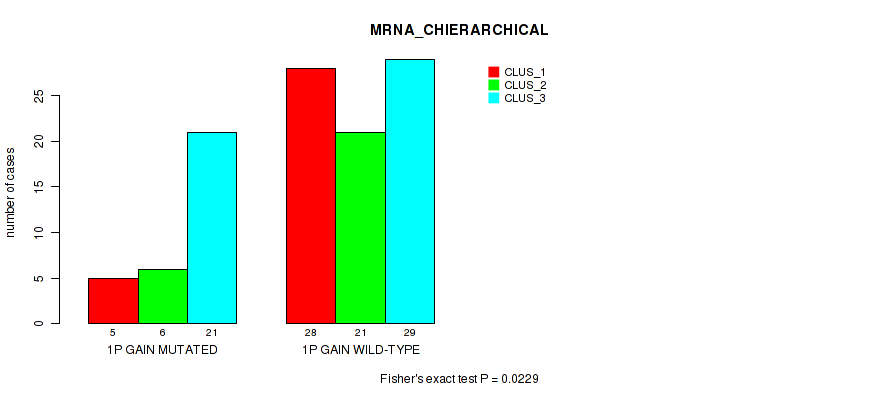
P value = 0.0286 (Fisher's exact test), Q value = 0.14
Table S2. Gene #1: '1p gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 1P GAIN MUTATED | 7 | 5 | 20 |
| 1P GAIN WILD-TYPE | 26 | 25 | 27 |
Figure S2. Get High-res Image Gene #1: '1p gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
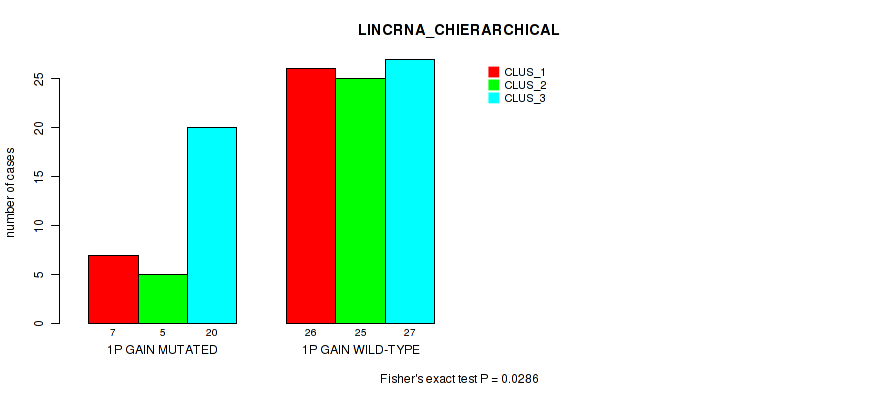
P value = 0.00555 (Fisher's exact test), Q value = 0.066
Table S3. Gene #3: '2p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 2P GAIN MUTATED | 12 | 7 | 4 |
| 2P GAIN WILD-TYPE | 21 | 20 | 46 |
Figure S3. Get High-res Image Gene #3: '2p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
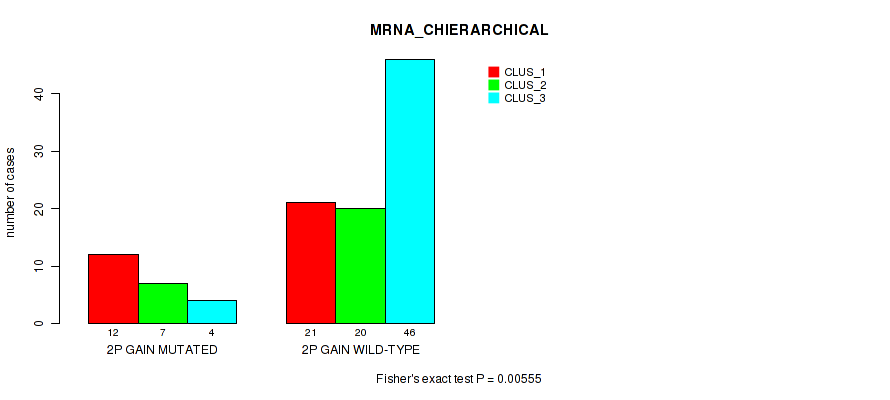
P value = 0.0186 (Fisher's exact test), Q value = 0.11
Table S4. Gene #3: '2p gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 2P GAIN MUTATED | 10 | 9 | 4 |
| 2P GAIN WILD-TYPE | 23 | 21 | 43 |
Figure S4. Get High-res Image Gene #3: '2p gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
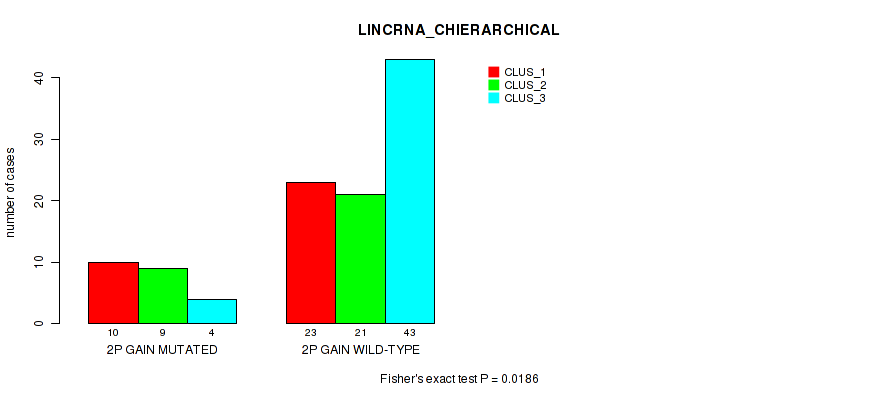
P value = 0.0167 (Fisher's exact test), Q value = 0.11
Table S5. Gene #4: '2q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 2Q GAIN MUTATED | 10 | 7 | 4 |
| 2Q GAIN WILD-TYPE | 23 | 20 | 46 |
Figure S5. Get High-res Image Gene #4: '2q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
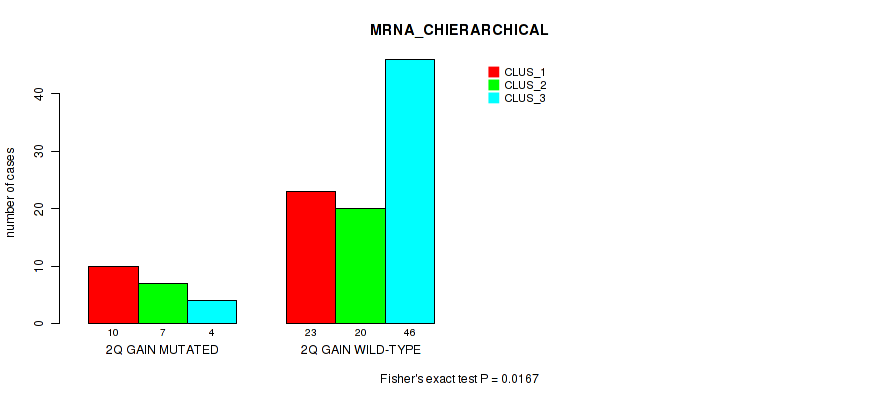
P value = 0.0353 (Fisher's exact test), Q value = 0.16
Table S6. Gene #4: '2q gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 2Q GAIN MUTATED | 8 | 9 | 4 |
| 2Q GAIN WILD-TYPE | 25 | 21 | 43 |
Figure S6. Get High-res Image Gene #4: '2q gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
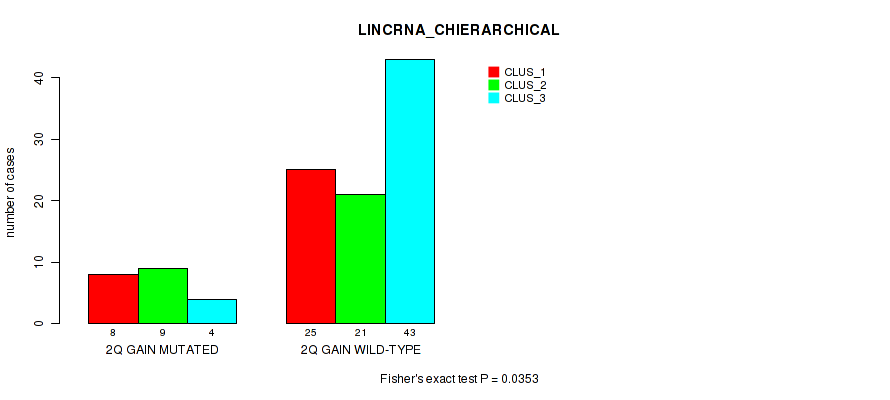
P value = 0.0156 (Fisher's exact test), Q value = 0.11
Table S7. Gene #10: '5q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 5Q GAIN MUTATED | 3 | 5 | 18 |
| 5Q GAIN WILD-TYPE | 30 | 22 | 32 |
Figure S7. Get High-res Image Gene #10: '5q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
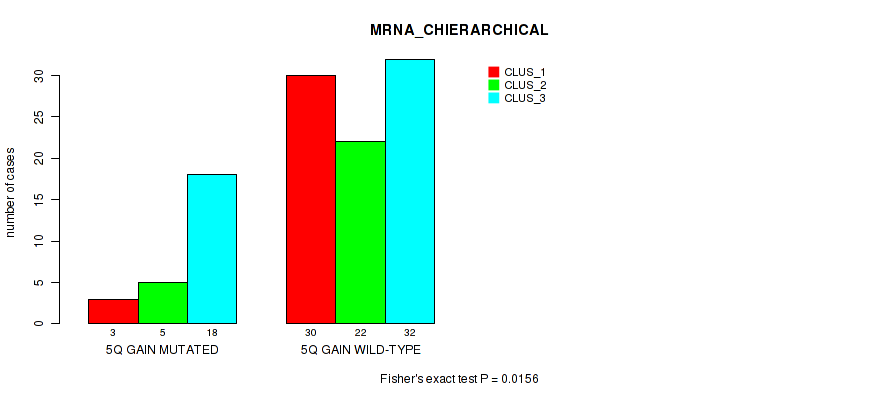
P value = 0.00038 (Fisher's exact test), Q value = 0.036
Table S8. Gene #10: '5q gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 5Q GAIN MUTATED | 3 | 3 | 20 |
| 5Q GAIN WILD-TYPE | 30 | 27 | 27 |
Figure S8. Get High-res Image Gene #10: '5q gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
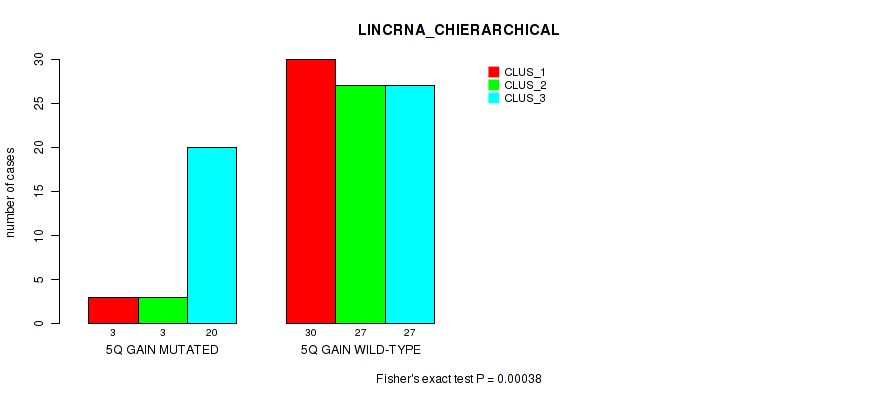
P value = 0.0301 (Fisher's exact test), Q value = 0.14
Table S9. Gene #13: '7p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 7P GAIN MUTATED | 11 | 17 | 30 |
| 7P GAIN WILD-TYPE | 22 | 10 | 20 |
Figure S9. Get High-res Image Gene #13: '7p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
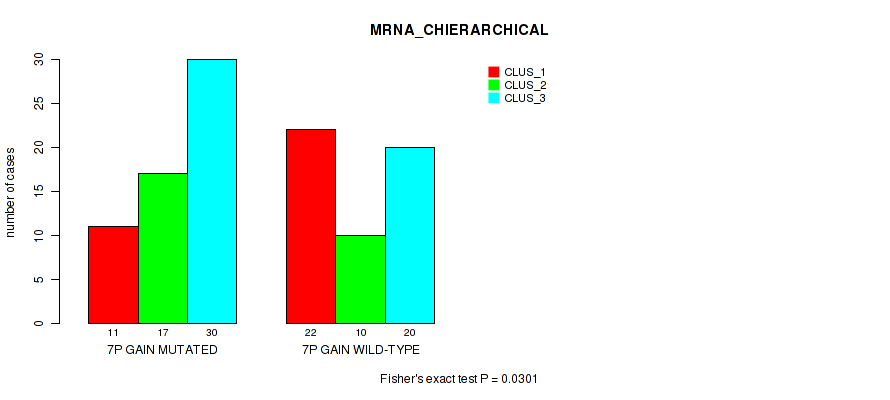
P value = 0.00974 (Fisher's exact test), Q value = 0.084
Table S10. Gene #13: '7p gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 7P GAIN MUTATED | 10 | 19 | 29 |
| 7P GAIN WILD-TYPE | 23 | 11 | 18 |
Figure S10. Get High-res Image Gene #13: '7p gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
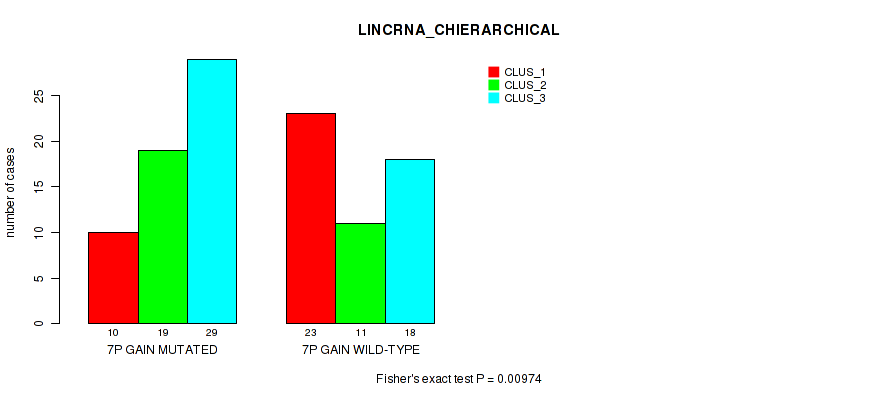
P value = 0.0205 (Fisher's exact test), Q value = 0.12
Table S11. Gene #14: '7q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 7Q GAIN MUTATED | 9 | 17 | 22 |
| 7Q GAIN WILD-TYPE | 24 | 10 | 28 |
Figure S11. Get High-res Image Gene #14: '7q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
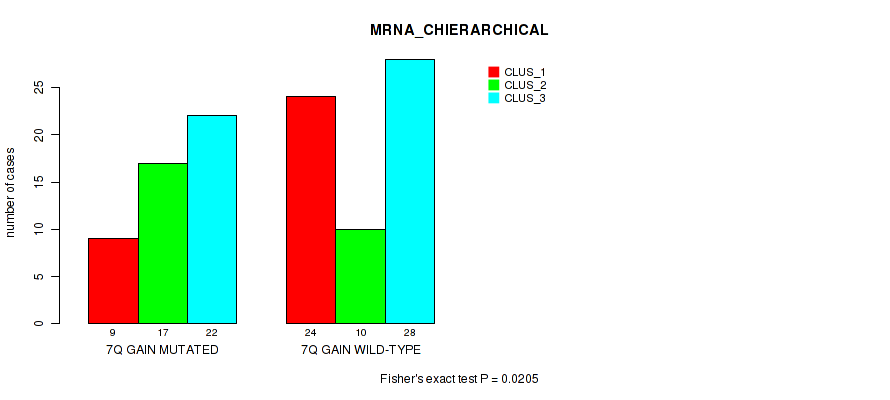
P value = 0.0323 (Fisher's exact test), Q value = 0.15
Table S12. Gene #14: '7q gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 7Q GAIN MUTATED | 9 | 18 | 21 |
| 7Q GAIN WILD-TYPE | 24 | 12 | 26 |
Figure S12. Get High-res Image Gene #14: '7q gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
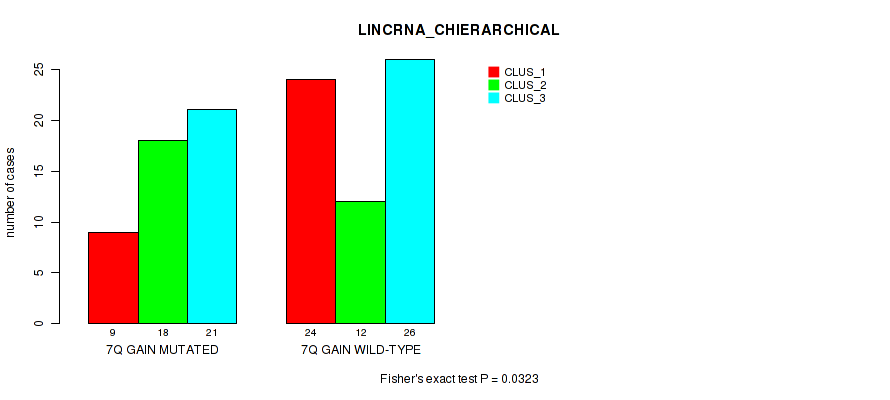
P value = 0.0408 (Fisher's exact test), Q value = 0.16
Table S13. Gene #20: '10q gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 10Q GAIN MUTATED | 5 | 0 | 2 |
| 10Q GAIN WILD-TYPE | 28 | 30 | 45 |
Figure S13. Get High-res Image Gene #20: '10q gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
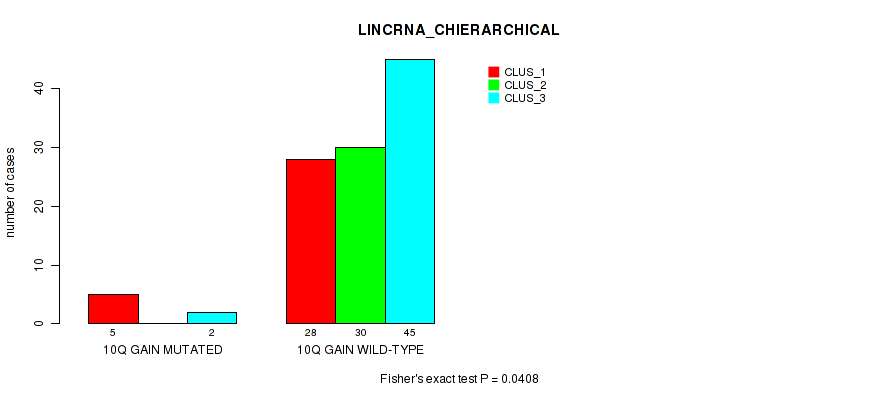
P value = 0.0398 (Fisher's exact test), Q value = 0.16
Table S14. Gene #23: '12p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 12P GAIN MUTATED | 12 | 3 | 8 |
| 12P GAIN WILD-TYPE | 21 | 24 | 42 |
Figure S14. Get High-res Image Gene #23: '12p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
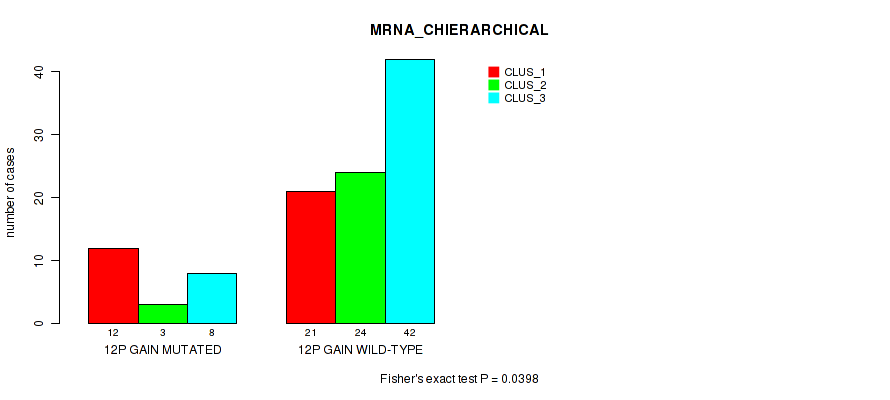
P value = 0.00023 (Fisher's exact test), Q value = 0.036
Table S15. Gene #30: '16p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 16P GAIN MUTATED | 5 | 5 | 27 |
| 16P GAIN WILD-TYPE | 28 | 22 | 23 |
Figure S15. Get High-res Image Gene #30: '16p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
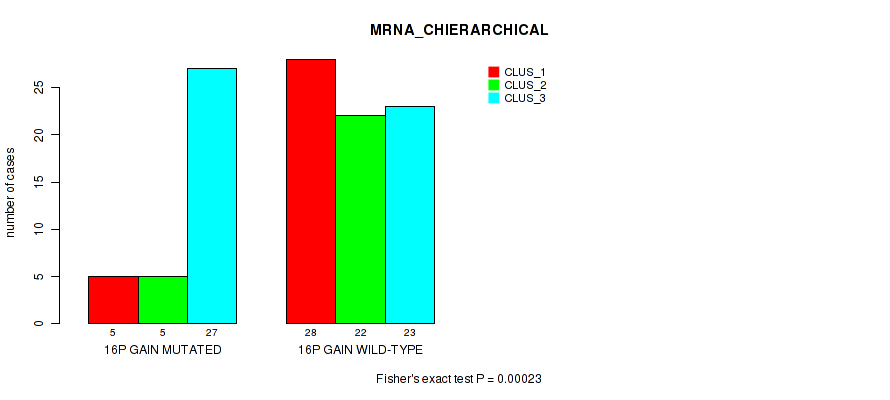
P value = 0.00248 (Fisher's exact test), Q value = 0.058
Table S16. Gene #30: '16p gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 16P GAIN MUTATED | 5 | 8 | 24 |
| 16P GAIN WILD-TYPE | 28 | 22 | 23 |
Figure S16. Get High-res Image Gene #30: '16p gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
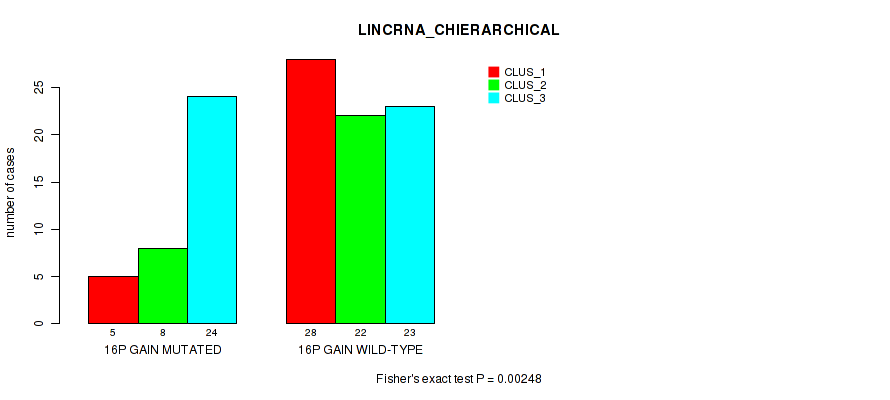
P value = 0.026 (Fisher's exact test), Q value = 0.14
Table S17. Gene #31: '16q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 16Q GAIN MUTATED | 5 | 4 | 19 |
| 16Q GAIN WILD-TYPE | 28 | 23 | 31 |
Figure S17. Get High-res Image Gene #31: '16q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
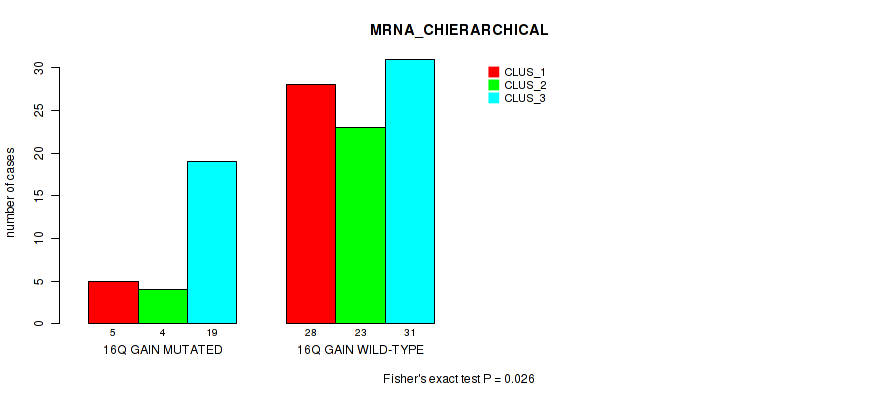
P value = 0.0169 (Fisher's exact test), Q value = 0.11
Table S18. Gene #34: '18p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 18P GAIN MUTATED | 6 | 1 | 1 |
| 18P GAIN WILD-TYPE | 27 | 26 | 49 |
Figure S18. Get High-res Image Gene #34: '18p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
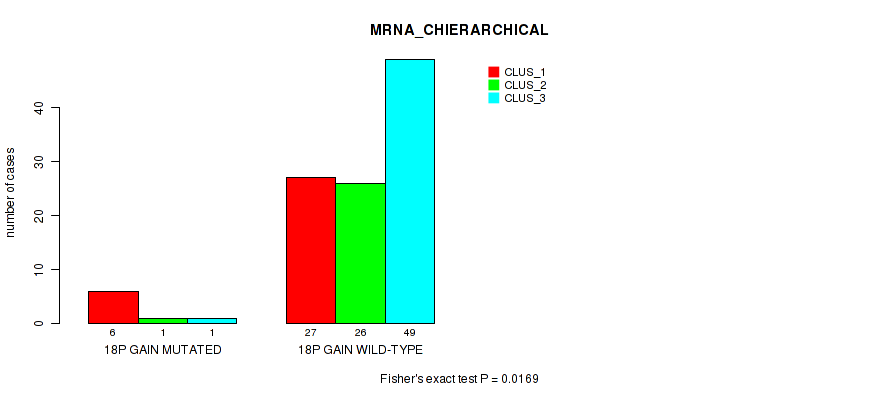
P value = 0.0433 (Fisher's exact test), Q value = 0.16
Table S19. Gene #35: '18q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 18Q GAIN MUTATED | 5 | 1 | 1 |
| 18Q GAIN WILD-TYPE | 28 | 26 | 49 |
Figure S19. Get High-res Image Gene #35: '18q gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
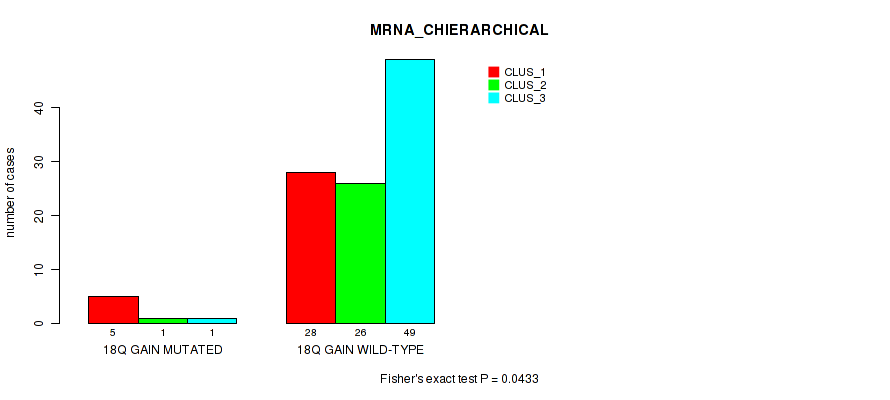
P value = 0.0472 (Fisher's exact test), Q value = 0.17
Table S20. Gene #38: '20p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 20P GAIN MUTATED | 11 | 7 | 6 |
| 20P GAIN WILD-TYPE | 22 | 20 | 44 |
Figure S20. Get High-res Image Gene #38: '20p gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
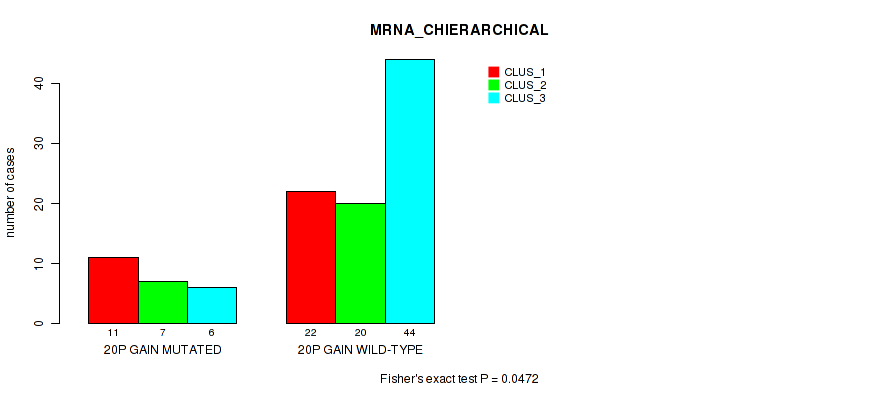
P value = 0.04 (Fisher's exact test), Q value = 0.16
Table S21. Gene #38: '20p gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 20P GAIN MUTATED | 10 | 9 | 5 |
| 20P GAIN WILD-TYPE | 23 | 21 | 42 |
Figure S21. Get High-res Image Gene #38: '20p gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
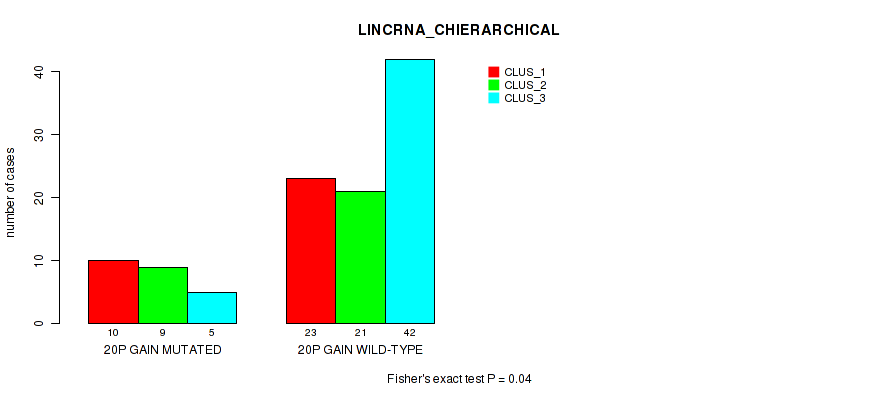
P value = 0.0495 (Fisher's exact test), Q value = 0.17
Table S22. Gene #41: '21q gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 21Q GAIN MUTATED | 1 | 5 | 10 |
| 21Q GAIN WILD-TYPE | 32 | 25 | 37 |
Figure S22. Get High-res Image Gene #41: '21q gain' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
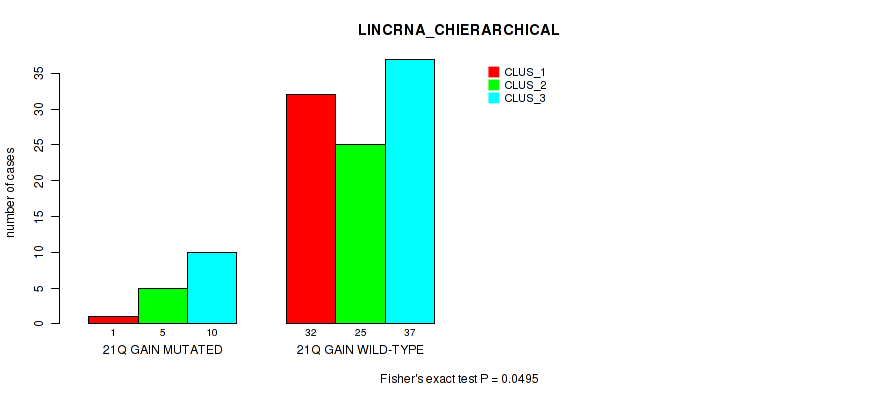
P value = 0.0365 (Fisher's exact test), Q value = 0.16
Table S23. Gene #44: 'xp gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| XP GAIN MUTATED | 18 | 6 | 22 |
| XP GAIN WILD-TYPE | 15 | 21 | 28 |
Figure S23. Get High-res Image Gene #44: 'xp gain' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
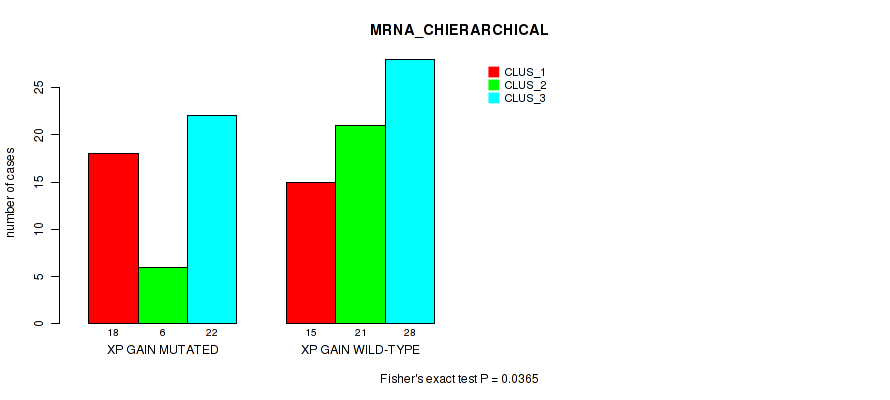
P value = 0.0266 (Fisher's exact test), Q value = 0.14
Table S24. Gene #48: '1p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 1P LOSS MUTATED | 9 | 4 | 3 |
| 1P LOSS WILD-TYPE | 24 | 23 | 47 |
Figure S24. Get High-res Image Gene #48: '1p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
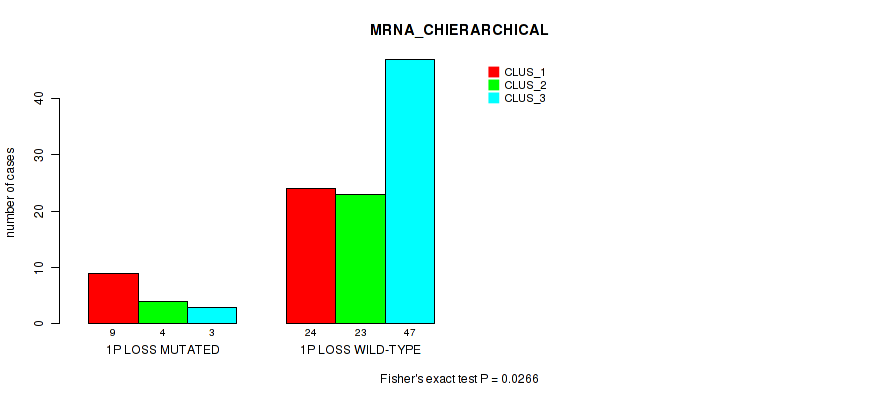
P value = 0.0123 (Fisher's exact test), Q value = 0.09
Table S25. Gene #48: '1p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 1P LOSS MUTATED | 9 | 5 | 2 |
| 1P LOSS WILD-TYPE | 24 | 25 | 45 |
Figure S25. Get High-res Image Gene #48: '1p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
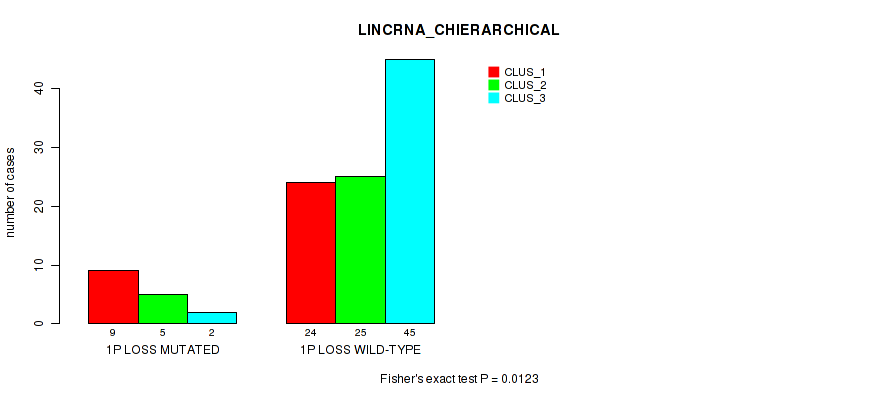
P value = 0.00372 (Fisher's exact test), Q value = 0.062
Table S26. Gene #52: '3p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 3P LOSS MUTATED | 17 | 3 | 16 |
| 3P LOSS WILD-TYPE | 16 | 24 | 34 |
Figure S26. Get High-res Image Gene #52: '3p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
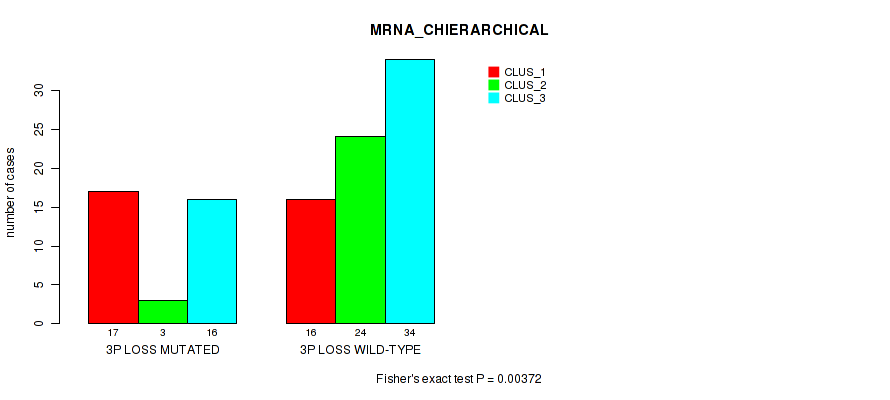
P value = 0.0299 (Fisher's exact test), Q value = 0.14
Table S27. Gene #52: '3p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 3P LOSS MUTATED | 16 | 5 | 15 |
| 3P LOSS WILD-TYPE | 17 | 25 | 32 |
Figure S27. Get High-res Image Gene #52: '3p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
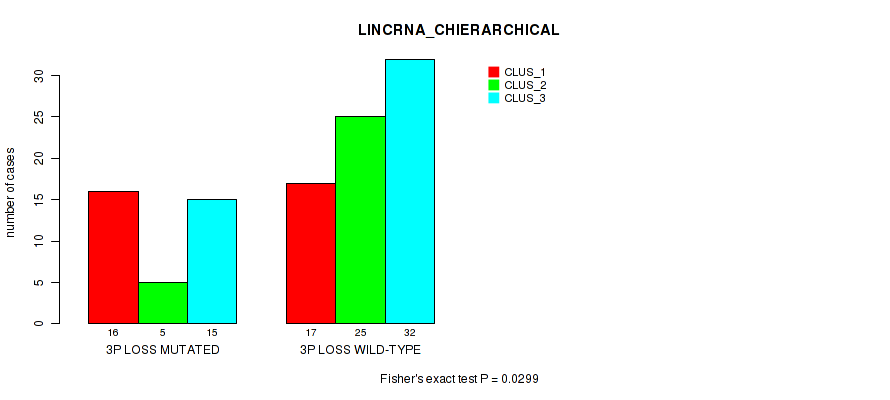
P value = 0.00628 (Fisher's exact test), Q value = 0.066
Table S28. Gene #53: '3q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 3Q LOSS MUTATED | 14 | 2 | 11 |
| 3Q LOSS WILD-TYPE | 19 | 25 | 39 |
Figure S28. Get High-res Image Gene #53: '3q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
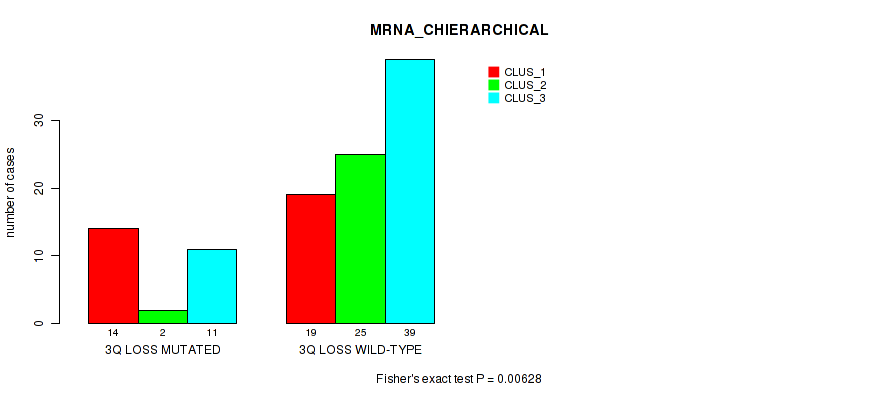
P value = 0.00301 (Fisher's exact test), Q value = 0.058
Table S29. Gene #53: '3q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 3Q LOSS MUTATED | 15 | 3 | 9 |
| 3Q LOSS WILD-TYPE | 18 | 27 | 38 |
Figure S29. Get High-res Image Gene #53: '3q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
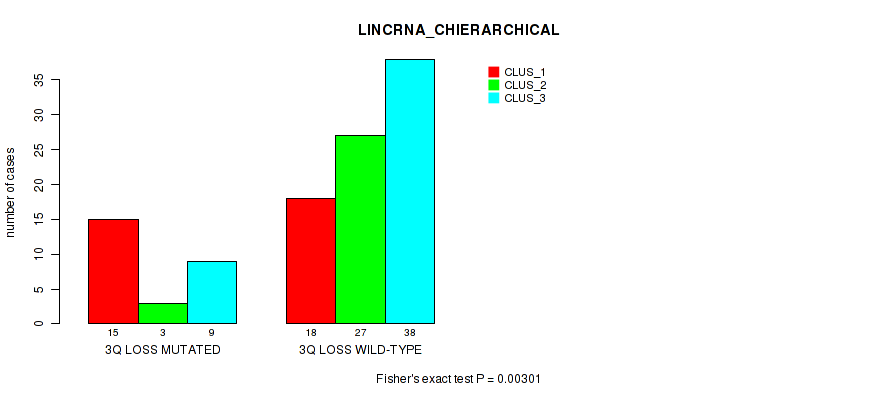
P value = 0.00163 (Fisher's exact test), Q value = 0.058
Table S30. Gene #57: '5q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 5Q LOSS MUTATED | 9 | 8 | 2 |
| 5Q LOSS WILD-TYPE | 24 | 19 | 48 |
Figure S30. Get High-res Image Gene #57: '5q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
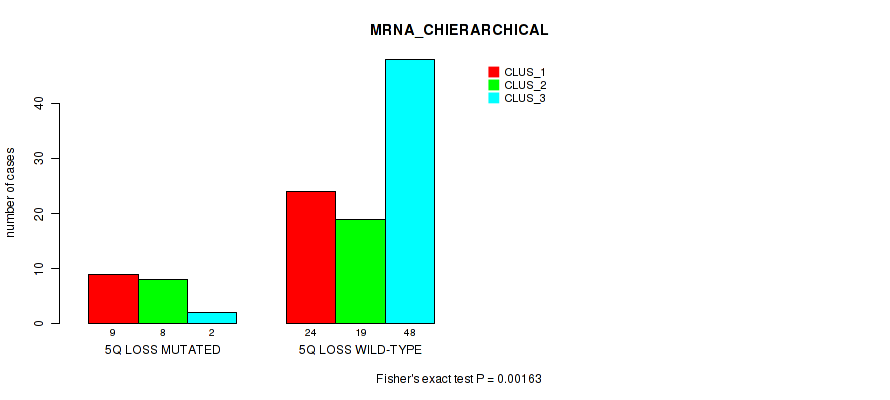
P value = 0.00243 (Fisher's exact test), Q value = 0.058
Table S31. Gene #57: '5q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 5Q LOSS MUTATED | 7 | 10 | 2 |
| 5Q LOSS WILD-TYPE | 26 | 20 | 45 |
Figure S31. Get High-res Image Gene #57: '5q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
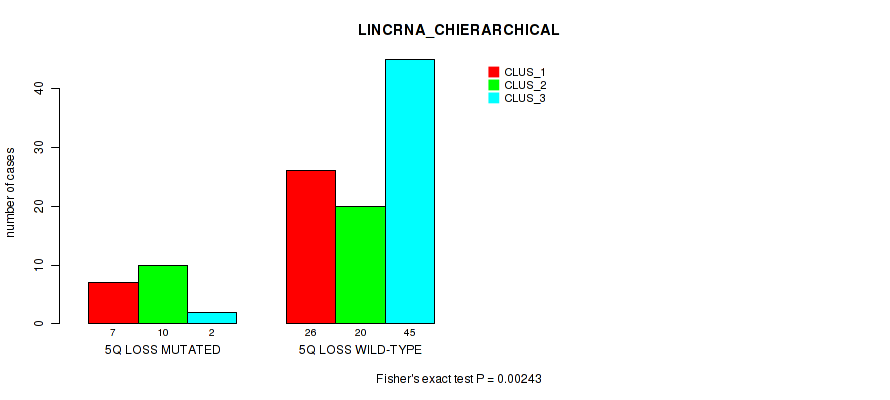
P value = 0.0496 (Fisher's exact test), Q value = 0.17
Table S32. Gene #67: '10q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 10Q LOSS MUTATED | 4 | 6 | 18 |
| 10Q LOSS WILD-TYPE | 29 | 21 | 32 |
Figure S32. Get High-res Image Gene #67: '10q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
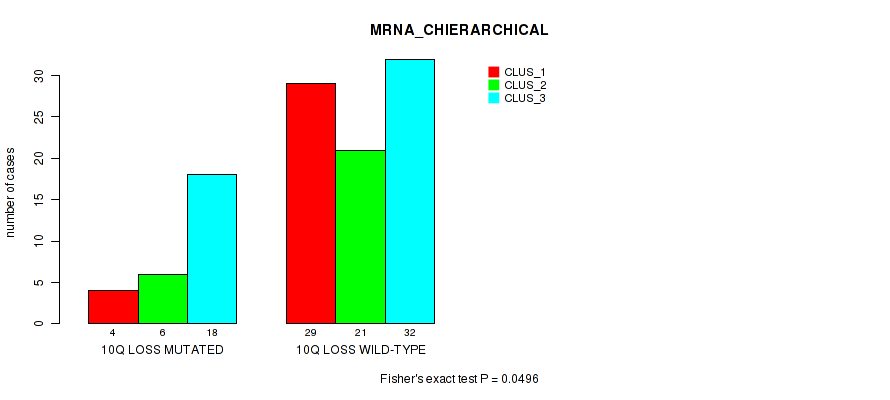
P value = 0.0271 (Fisher's exact test), Q value = 0.14
Table S33. Gene #67: '10q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 10Q LOSS MUTATED | 3 | 9 | 16 |
| 10Q LOSS WILD-TYPE | 30 | 21 | 31 |
Figure S33. Get High-res Image Gene #67: '10q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
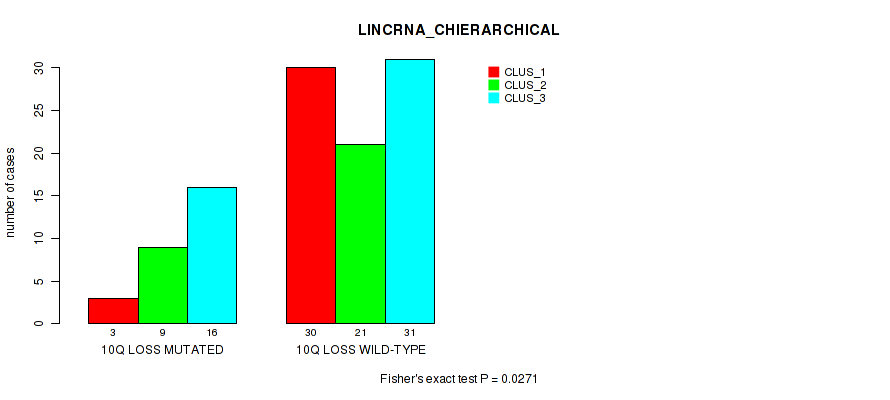
P value = 0.0102 (Fisher's exact test), Q value = 0.084
Table S34. Gene #68: '11p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 11P LOSS MUTATED | 10 | 11 | 6 |
| 11P LOSS WILD-TYPE | 23 | 16 | 44 |
Figure S34. Get High-res Image Gene #68: '11p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
P value = 0.011 (Fisher's exact test), Q value = 0.087
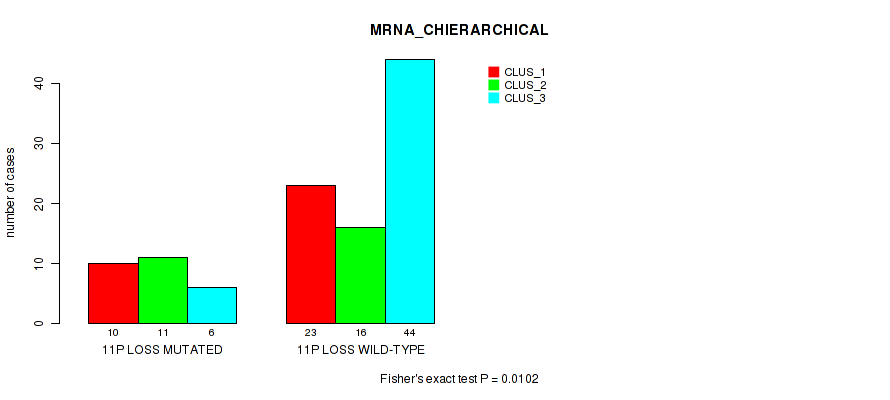
Table S35. Gene #68: '11p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 11P LOSS MUTATED | 8 | 13 | 6 |
| 11P LOSS WILD-TYPE | 25 | 17 | 41 |
Figure S35. Get High-res Image Gene #68: '11p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
P value = 0.00201 (Fisher's exact test), Q value = 0.058
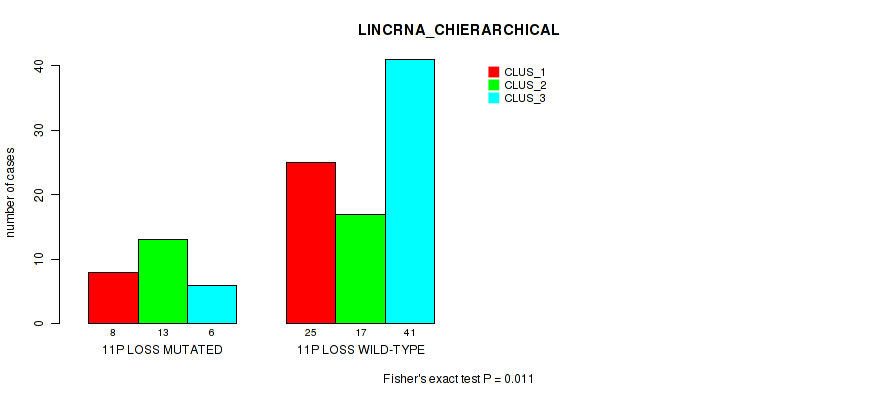
Table S36. Gene #70: '12p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 12P LOSS MUTATED | 1 | 7 | 16 |
| 12P LOSS WILD-TYPE | 32 | 23 | 31 |
Figure S36. Get High-res Image Gene #70: '12p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
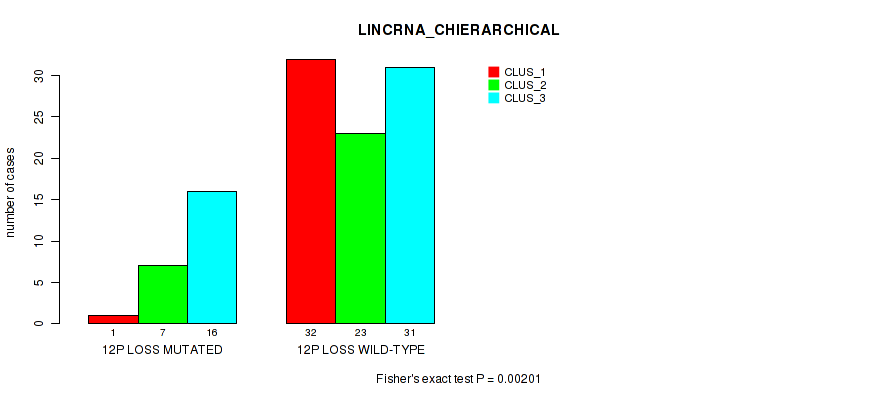
P value = 0.00172 (Fisher's exact test), Q value = 0.058
Table S37. Gene #71: '12q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 12Q LOSS MUTATED | 1 | 6 | 16 |
| 12Q LOSS WILD-TYPE | 32 | 24 | 31 |
Figure S37. Get High-res Image Gene #71: '12q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
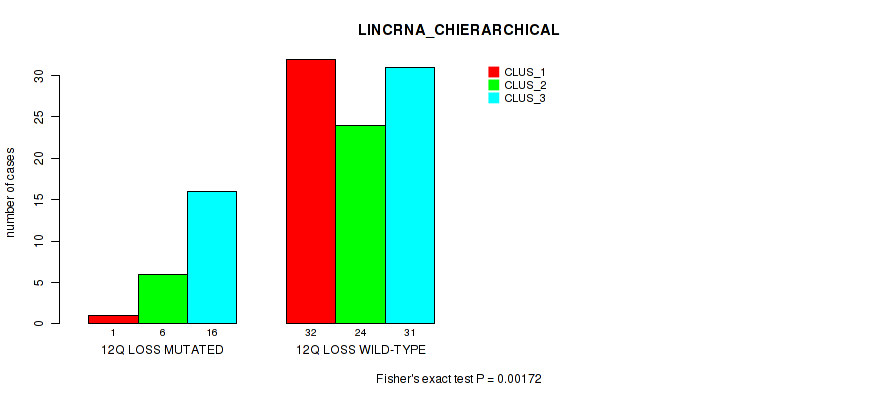
P value = 0.00975 (Fisher's exact test), Q value = 0.084
Table S38. Gene #77: '15q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 15Q LOSS MUTATED | 9 | 14 | 29 |
| 15Q LOSS WILD-TYPE | 24 | 16 | 18 |
Figure S38. Get High-res Image Gene #77: '15q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
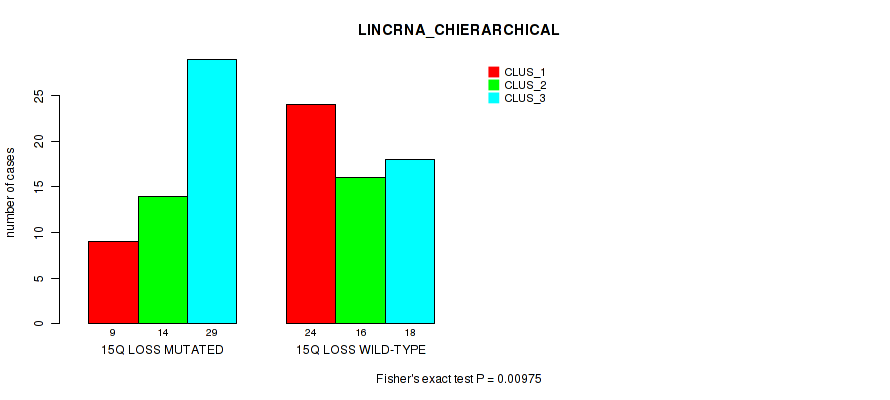
P value = 0.00668 (Fisher's exact test), Q value = 0.066
Table S39. Gene #78: '16p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 16P LOSS MUTATED | 5 | 8 | 2 |
| 16P LOSS WILD-TYPE | 28 | 19 | 48 |
Figure S39. Get High-res Image Gene #78: '16p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
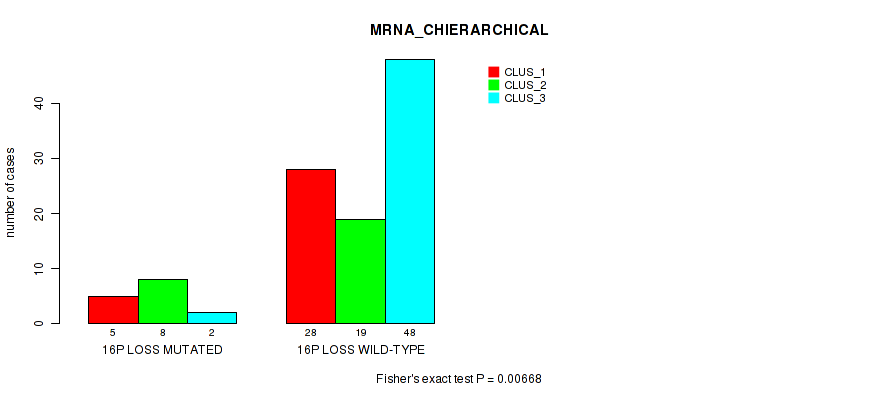
P value = 0.0414 (Fisher's exact test), Q value = 0.16
Table S40. Gene #78: '16p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 16P LOSS MUTATED | 4 | 8 | 3 |
| 16P LOSS WILD-TYPE | 29 | 22 | 44 |
Figure S40. Get High-res Image Gene #78: '16p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
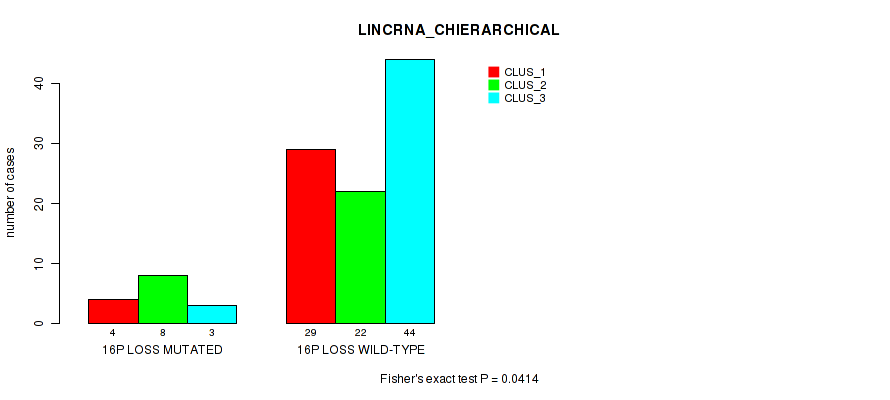
P value = 0.0209 (Fisher's exact test), Q value = 0.12
Table S41. Gene #82: '18p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 18P LOSS MUTATED | 6 | 10 | 24 |
| 18P LOSS WILD-TYPE | 27 | 17 | 26 |
Figure S41. Get High-res Image Gene #82: '18p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
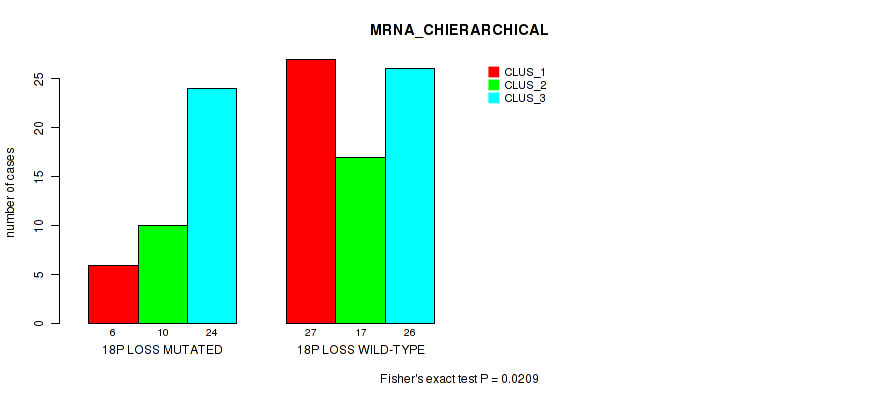
P value = 0.00536 (Fisher's exact test), Q value = 0.066
Table S42. Gene #82: '18p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 18P LOSS MUTATED | 5 | 12 | 23 |
| 18P LOSS WILD-TYPE | 28 | 18 | 24 |
Figure S42. Get High-res Image Gene #82: '18p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
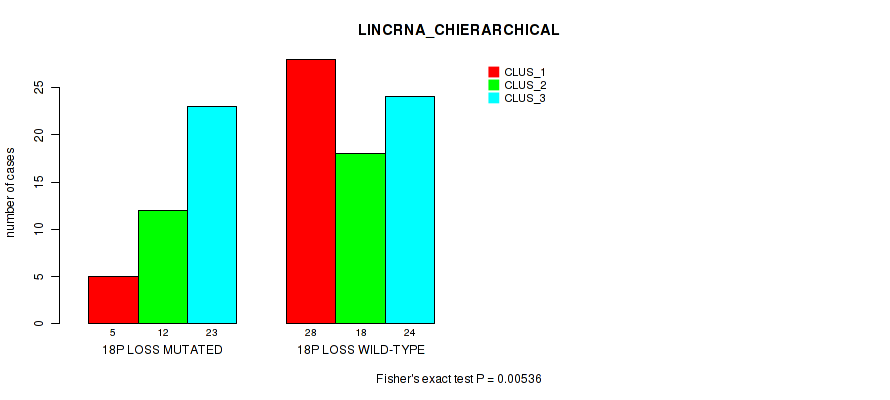
P value = 0.0369 (Fisher's exact test), Q value = 0.16
Table S43. Gene #83: '18q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 18Q LOSS MUTATED | 8 | 12 | 26 |
| 18Q LOSS WILD-TYPE | 25 | 15 | 24 |
Figure S43. Get High-res Image Gene #83: '18q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
P value = 0.012 (Fisher's exact test), Q value = 0.09
Table S44. Gene #83: '18q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 18Q LOSS MUTATED | 7 | 14 | 25 |
| 18Q LOSS WILD-TYPE | 26 | 16 | 22 |
Figure S44. Get High-res Image Gene #83: '18q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
P value = 0.0174 (Fisher's exact test), Q value = 0.11
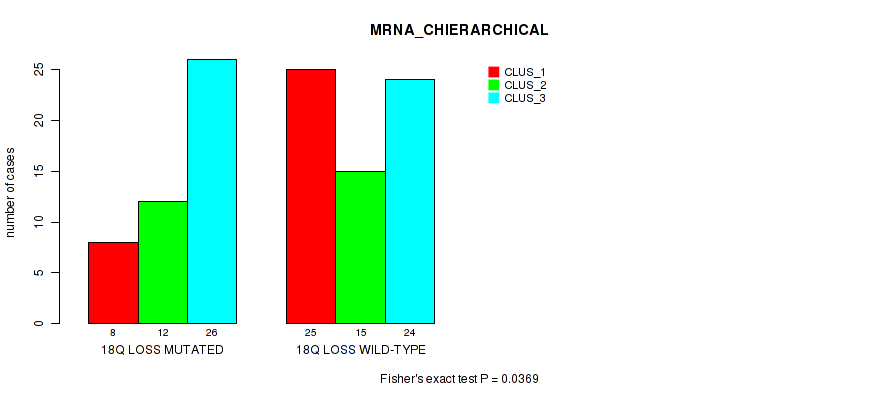
Table S45. Gene #88: '21p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 21P LOSS MUTATED | 19 | 10 | 13 |
| 21P LOSS WILD-TYPE | 14 | 17 | 37 |
Figure S45. Get High-res Image Gene #88: '21p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
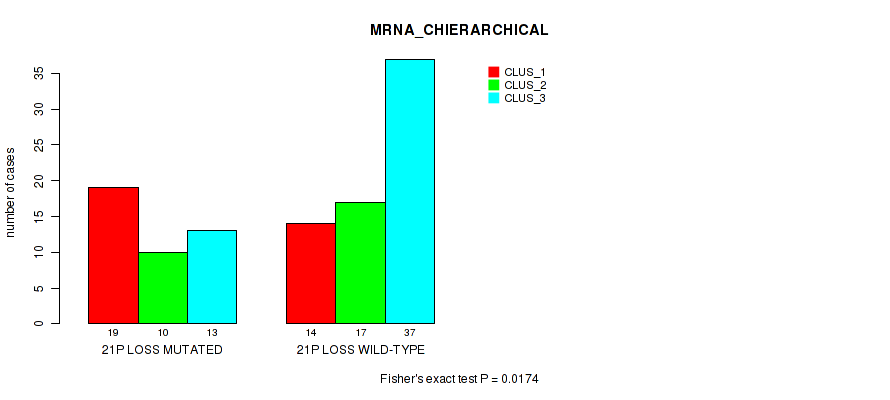
P value = 0.0065 (Fisher's exact test), Q value = 0.066
Table S46. Gene #88: '21p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 21P LOSS MUTATED | 17 | 15 | 10 |
| 21P LOSS WILD-TYPE | 16 | 15 | 37 |
Figure S46. Get High-res Image Gene #88: '21p loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
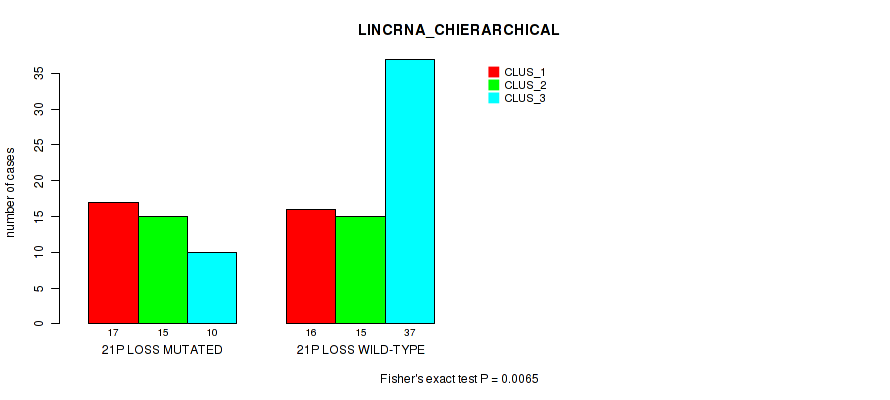
P value = 0.0201 (Fisher's exact test), Q value = 0.12
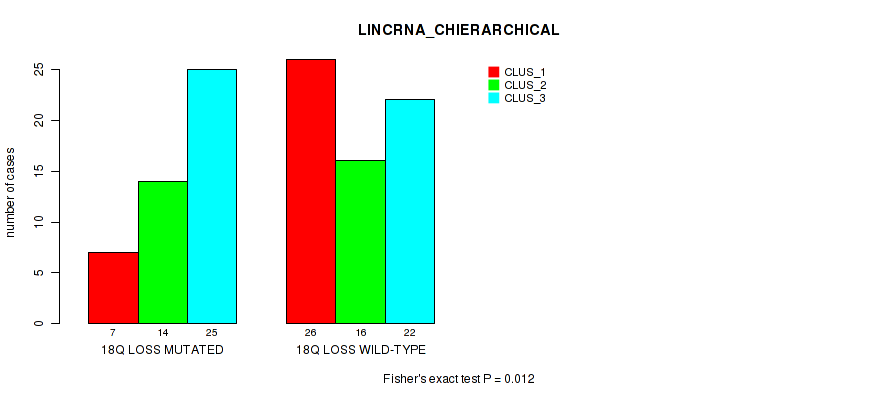
Table S47. Gene #89: '21q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 21Q LOSS MUTATED | 17 | 10 | 11 |
| 21Q LOSS WILD-TYPE | 16 | 17 | 39 |
Figure S47. Get High-res Image Gene #89: '21q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
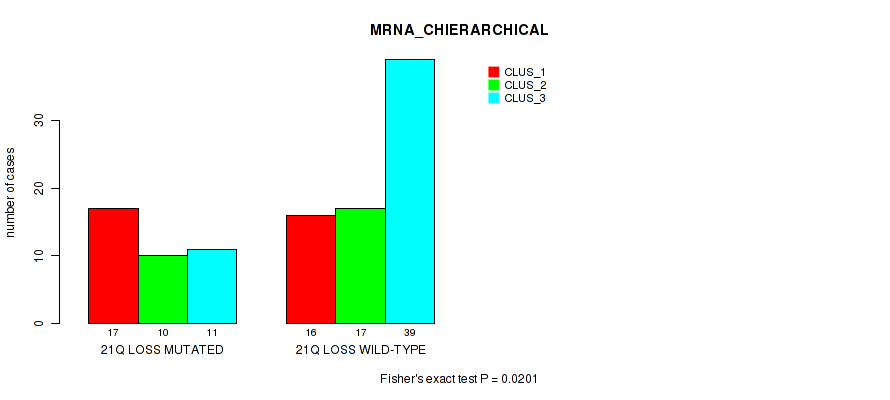
P value = 0.00306 (Fisher's exact test), Q value = 0.058
Table S48. Gene #89: '21q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| 21Q LOSS MUTATED | 16 | 14 | 8 |
| 21Q LOSS WILD-TYPE | 17 | 16 | 39 |
Figure S48. Get High-res Image Gene #89: '21q loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
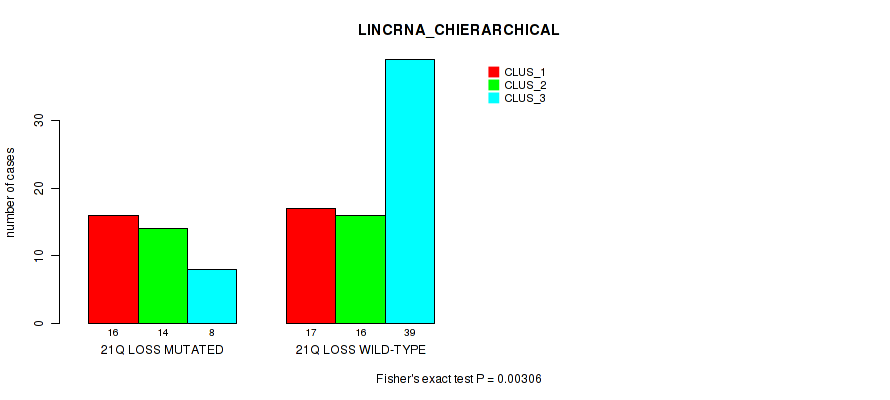
P value = 0.00553 (Fisher's exact test), Q value = 0.066
Table S49. Gene #90: '22p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 22P LOSS MUTATED | 21 | 12 | 14 |
| 22P LOSS WILD-TYPE | 12 | 15 | 36 |
Figure S49. Get High-res Image Gene #90: '22p loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
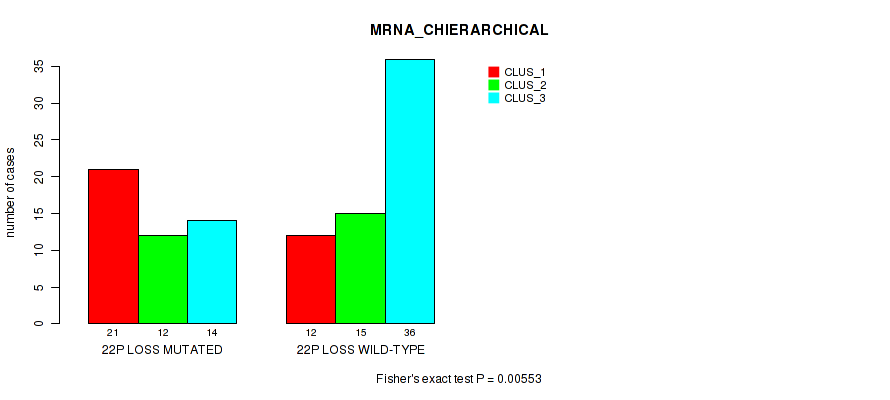
P value = 0.00697 (Fisher's exact test), Q value = 0.066
Table S50. Gene #91: '22q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| 22Q LOSS MUTATED | 22 | 11 | 16 |
| 22Q LOSS WILD-TYPE | 11 | 16 | 34 |
Figure S50. Get High-res Image Gene #91: '22q loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
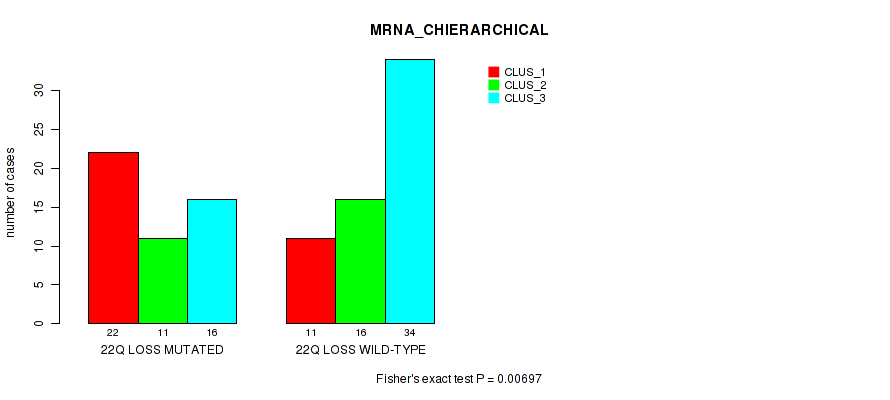
P value = 0.00668 (Fisher's exact test), Q value = 0.066
Table S51. Gene #94: 'yp loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| YP LOSS MUTATED | 25 | 17 | 21 |
| YP LOSS WILD-TYPE | 8 | 10 | 29 |
Figure S51. Get High-res Image Gene #94: 'yp loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
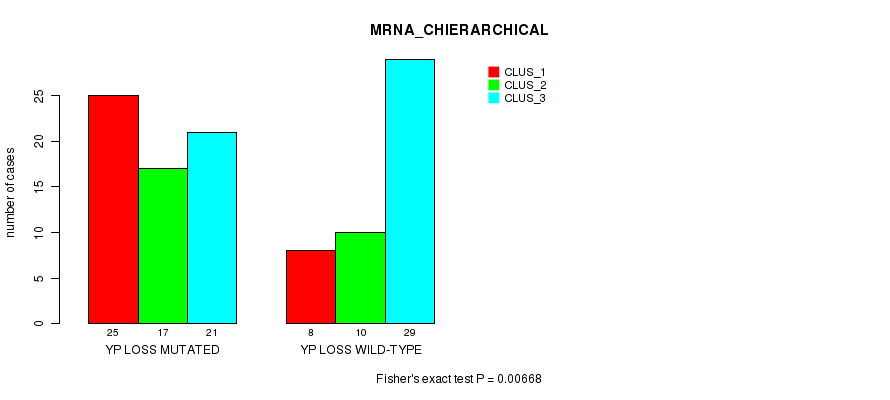
P value = 0.00219 (Fisher's exact test), Q value = 0.058
Table S52. Gene #94: 'yp loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| YP LOSS MUTATED | 25 | 20 | 18 |
| YP LOSS WILD-TYPE | 8 | 10 | 29 |
Figure S52. Get High-res Image Gene #94: 'yp loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
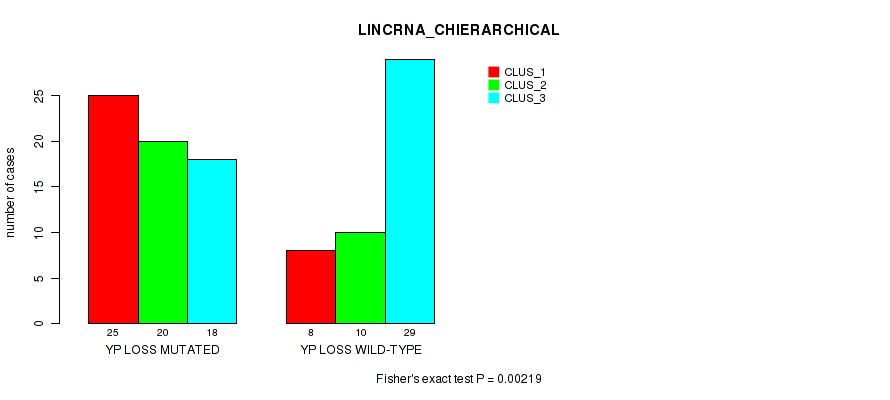
P value = 0.0497 (Fisher's exact test), Q value = 0.17
Table S53. Gene #95: 'yq loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 27 | 50 |
| YQ LOSS MUTATED | 24 | 17 | 23 |
| YQ LOSS WILD-TYPE | 9 | 10 | 27 |
Figure S53. Get High-res Image Gene #95: 'yq loss' versus Molecular Subtype #1: 'MRNA_CHIERARCHICAL'
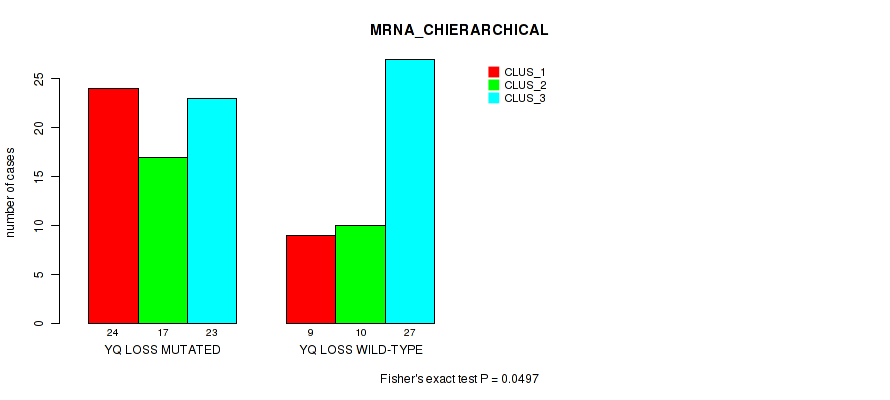
P value = 0.00393 (Fisher's exact test), Q value = 0.062
Table S54. Gene #95: 'yq loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
| nPatients | CLUS_1 | CLUS_2 | CLUS_3 |
|---|---|---|---|
| ALL | 33 | 30 | 47 |
| YQ LOSS MUTATED | 25 | 20 | 19 |
| YQ LOSS WILD-TYPE | 8 | 10 | 28 |
Figure S54. Get High-res Image Gene #95: 'yq loss' versus Molecular Subtype #2: 'LINCRNA_CHIERARCHICAL'
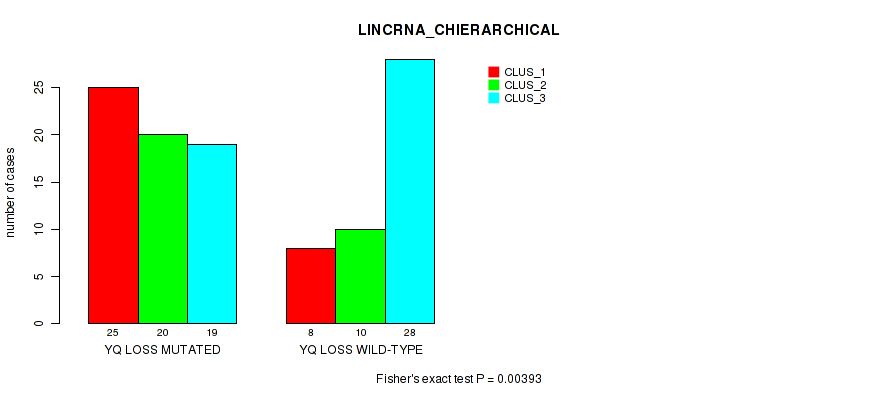
-
Copy number data file = broad_values_by_arm.txt from GISTIC pipeline
-
Processed Copy number data file = /cromwell_root/fc-e7058367-eaa6-44b5-aab5-1ec08acf146a/038fcf2f-32c7-437e-beb7-ded16a017a25/correlate_genomic_events_all/d2cdaeae-8d2e-46f5-b7bd-6cd908ffe652/call-preprocess_genomic_event/transformed.cor.cli.txt
-
Molecular subtypes file = /cromwell_root/fc-e7058367-eaa6-44b5-aab5-1ec08acf146a/69dd18b3-97fe-4143-b0f4-32e5d55e929d/aggregate_clusters_workflow/d44c315a-761b-4b72-be1f-cb969be36102/call-aggregate_clusters/CPTAC3-LUAD-TP.transposedmergedcluster.txt
-
Number of patients = 110
-
Number of significantly arm-level cnvs = 95
-
Number of molecular subtypes = 2
-
Exclude genes that fewer than K tumors have mutations, K = 3
For binary or multi-class clinical features (nominal or ordinal), two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.