Number of individuals: 135
| Significantly mutated genes (q≤0.1) | 278 |
| Mutations seen in COSMIC | 0 |
| Significantly mutated genes on COSMIC territory | 0 |
| Genes with clustered mutations (≤3 aa apart) | 1 |
| Significantly mutated genesets | 89 |
| . . . . . . . . . (excluding sig.mutated genes) | 1 |
Read 150 MAFs of type "Broad"
Total number of mutations in input MAFs: 49189
After removing 5553 noncoding mutations: 43636
After collapsing adjacent/redundant mutations: 43634
Please see MutSigPreprocess "stdout.txt" for full details.
Number of mutations before filtering: 43634
After removing 46 mutations outside patient set: 43588
After removing 87 mutations outside gene set: 43501
After removing 12445 mutations outside category set: 31056
After removing 21260 "impossible" mutations in
gene-patient-category bins of zero coverage: 9796
Please see MutSigRun "stdout.txt" for full details.
Final set of mutations used in analysis
| type | count |
|---|---|
| De_novo_Start_InFrame | 2 |
| De_novo_Start_OutOfFrame | 21 |
| Missense_Mutation | 6814 |
| Nonsense_Mutation | 482 |
| Read-through | 8 |
| Splice_Site_DNP | 17 |
| Splice_Site_SNP | 184 |
| Synonymous | 2268 |
| Total | 9796 |
| category | n | N | rate | rate_per_mb | relative_rate |
|---|---|---|---|---|---|
| A->T | 1783 | 49,414,217 | 0.000036 | 36.1 | 0.51 |
| *Np(A/C/T)->nonflip | 3900 | 83,231,156 | 0.000047 | 46.9 | 0.67 |
| *NpG->nonflip | 548 | 23,965,121 | 0.000023 | 22.9 | 0.33 |
| C->G | 583 | 57,782,060 | 0.000010 | 10.1 | 0.14 |
| indel+null | 714 | 107,196,277 | 6.66e-06 | 6.66 | 0.095 |
| Total | 7528 | 107,196,277 | 0.000070 | 70.2 | 1.00 |

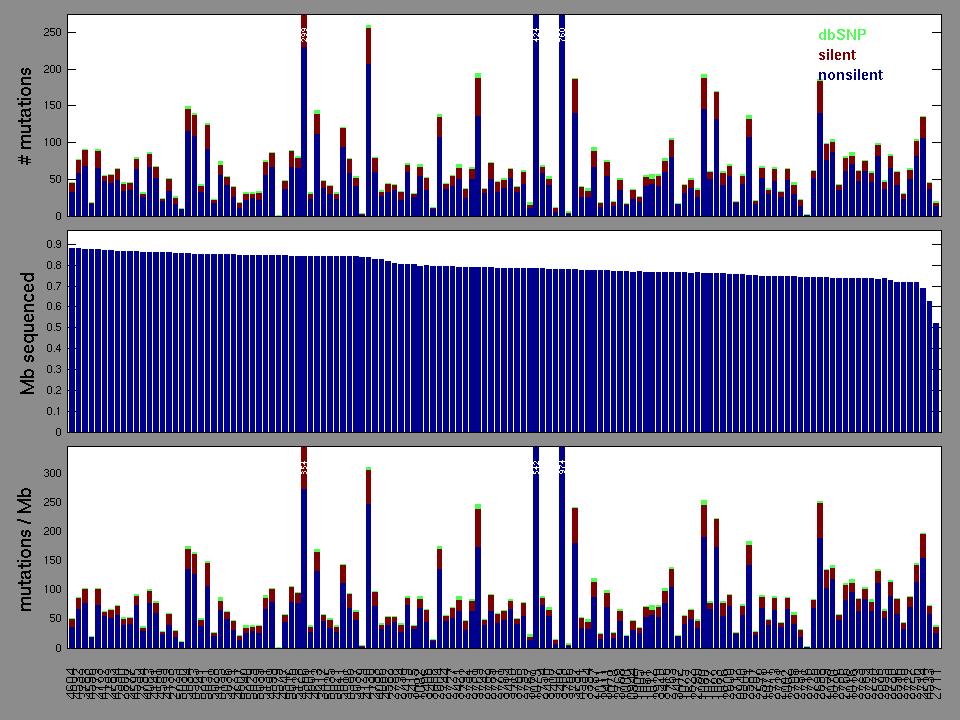
| rank | gene | description | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | USH2A | Usher syndrome 2A (autosomal recessive, mild) | 133 | 13 | 13 | 13 | 3 | 9 | 0 | 0 | 0 | 4 | <1.00e-11 | <7.86e-09 |
| 2 | ADAMTS12 | ADAM metallopeptidase with thrombospondin type 1 motif, 12 | 1350 | 20 | 20 | 19 | 5 | 7 | 9 | 3 | 1 | 0 | <1.00e-11 | <7.86e-09 |
| 3 | SLIT3 | slit homolog 3 (Drosophila) | 540 | 8 | 6 | 8 | 3 | 1 | 4 | 1 | 1 | 1 | <1.00e-11 | <7.86e-09 |
| 4 | ADAM22 | ADAM metallopeptidase domain 22 | 670 | 8 | 8 | 8 | 1 | 3 | 3 | 1 | 1 | 0 | <1.00e-11 | <7.86e-09 |
| 5 | CNBD1 | cyclic nucleotide binding domain containing 1 | 659 | 6 | 6 | 6 | 0 | 2 | 2 | 0 | 0 | 2 | <1.00e-11 | <7.86e-09 |
| 6 | VWF | von Willebrand factor | 1517 | 12 | 12 | 12 | 1 | 4 | 5 | 0 | 1 | 2 | <1.00e-11 | <7.86e-09 |
| 7 | LRP1B | low density lipoprotein-related protein 1B (deleted in tumors) | 16534 | 52 | 39 | 52 | 11 | 16 | 26 | 1 | 3 | 6 | <1.00e-11 | <7.86e-09 |
| 8 | COL22A1 | collagen, type XXII, alpha 1 | 5772 | 16 | 15 | 16 | 4 | 3 | 7 | 0 | 0 | 6 | <1.00e-11 | <7.86e-09 |
| 9 | SVEP1 | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | 4185 | 10 | 8 | 10 | 3 | 4 | 4 | 1 | 0 | 1 | <1.00e-11 | <7.86e-09 |
| 10 | SPAG17 | sperm associated antigen 17 | 8370 | 17 | 13 | 17 | 1 | 3 | 9 | 2 | 0 | 3 | <1.00e-11 | <7.86e-09 |
| 11 | CSMD1 | CUB and Sushi multiple domains 1 | 13866 | 28 | 25 | 28 | 7 | 8 | 16 | 0 | 2 | 2 | <1.00e-11 | <7.86e-09 |
| 12 | PAPPA2 | pappalysin 2 | 11978 | 24 | 20 | 23 | 10 | 8 | 14 | 1 | 0 | 1 | <1.00e-11 | <7.86e-09 |
| 13 | CSMD3 | CUB and Sushi multiple domains 3 | 28648 | 52 | 43 | 52 | 10 | 15 | 23 | 3 | 4 | 7 | <1.00e-11 | <7.86e-09 |
| 14 | MYH13 | myosin, heavy chain 13, skeletal muscle | 6075 | 11 | 9 | 11 | 2 | 2 | 5 | 3 | 0 | 1 | <1.00e-11 | <7.86e-09 |
| 15 | C6 | complement component 6 | 6615 | 11 | 11 | 11 | 1 | 2 | 3 | 2 | 0 | 4 | <1.00e-11 | <7.86e-09 |
| 16 | RYR3 | ryanodine receptor 3 | 16065 | 24 | 23 | 24 | 8 | 11 | 10 | 1 | 0 | 2 | <1.00e-11 | <7.86e-09 |
| 17 | LRP2 | low density lipoprotein-related protein 2 | 17623 | 22 | 19 | 22 | 8 | 7 | 11 | 1 | 2 | 1 | <1.00e-11 | <7.86e-09 |
| 18 | SCN1A | sodium channel, voltage-gated, type I, alpha subunit | 17275 | 18 | 13 | 18 | 3 | 8 | 8 | 0 | 0 | 2 | <1.00e-11 | <7.86e-09 |
| 19 | PCDH15 | protocadherin 15 | 26729 | 22 | 19 | 22 | 6 | 4 | 14 | 0 | 1 | 3 | <1.00e-11 | <7.86e-09 |
| 20 | RYR2 | ryanodine receptor 2 (cardiac) | 75777 | 60 | 38 | 59 | 14 | 11 | 33 | 7 | 2 | 7 | <1.00e-11 | <7.86e-09 |
| 21 | SPHKAP | SPHK1 interactor, AKAP domain containing | 36302 | 21 | 17 | 21 | 3 | 6 | 12 | 1 | 0 | 2 | <1.00e-11 | <7.86e-09 |
| 22 | HEATR7B2 | 35164 | 20 | 16 | 20 | 2 | 6 | 8 | 1 | 0 | 5 | <1.00e-11 | <7.86e-09 | |
| 23 | TTN | titin | 336559 | 173 | 79 | 172 | 46 | 37 | 93 | 16 | 15 | 12 | <1.00e-11 | <7.86e-09 |
| 24 | MUC16 | mucin 16, cell surface associated | 174250 | 68 | 41 | 68 | 30 | 17 | 37 | 5 | 7 | 2 | <1.00e-11 | <7.86e-09 |
| 25 | AMPH | amphiphysin | 5805 | 10 | 10 | 10 | 3 | 4 | 4 | 0 | 0 | 2 | 2.84e-11 | 2.15e-08 |
| 26 | GPR112 | G protein-coupled receptor 112 | 9578 | 11 | 11 | 11 | 0 | 5 | 2 | 0 | 1 | 3 | 3.74e-11 | 2.71e-08 |
| 27 | SMG1 | 1350 | 6 | 6 | 6 | 2 | 0 | 4 | 2 | 0 | 0 | 7.58e-11 | 5.30e-08 | |
| 28 | SELP | selectin P (granule membrane protein 140kDa, antigen CD62) | 945 | 6 | 6 | 6 | 1 | 1 | 3 | 0 | 0 | 2 | 1.14e-10 | 7.70e-08 |
| 29 | PPFIA2 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | 132 | 4 | 4 | 4 | 0 | 2 | 0 | 1 | 0 | 1 | 1.28e-10 | 8.31e-08 |
| 30 | DNAH11 | dynein, axonemal, heavy chain 11 | 23377 | 16 | 16 | 16 | 4 | 3 | 9 | 0 | 1 | 3 | 1.55e-10 | 9.72e-08 |
Notes
N = number of sequenced bases in this gene across the individual set
n = number of (nonsilent) mutations in this gene across the individual set
npat = number of patients (individuals) with at least one nonsilent mutation
nsite = number of unique sites having a nonsilent mutation
nsil = number of silent mutations in this gene across the individual set
n1 = number of nonsilent mutations of type "A->T"
n2 = number of nonsilent mutations of type "*Np(A/C/T)->nonflip"
n3 = number of nonsilent mutations of type "*NpG->nonflip"
n4 = number of nonsilent mutations of type "C->G"
n5 = number of nonsilent mutations of type "indel+null"
null = mutation category that includes nonsense, frameshift, splice-site mutations
p_classic = p-value for the observed amount of nonsilent mutations being elevated in this gene
p_ns_s = p-value for the observed nonsilent/silent ratio being elevated in this gene
p = p-value (overall)
q = q-value, False Discovery Rate (Benjamini-Hochberg procedure)
Two distinct analyses are reported using the COSMIC database:
(COS1) COSMIC used as a filter to increase power by restricting the territory of each gene
(COS2) COSMIC used as a prior for the importance of a gene
| rank | gene | description | n | cos | n_cos | N_cos | cos_ev | p | q |
|---|
Notes
n = number of mutations in this gene in the individual set
cos = number of unique mutated sites in this gene in COSMIC
n_cos = overlap between n and cos
N_cos = number of individuals × cos
cos_ev = total evidence: number of reports in COSMIC for mutations seen in this gene
p = p-value for seeing the observed amount of overlap in this gene
q = q-value, False Discovery Rate (Benjamini-Hochberg procedure)
| rank | gene | description | n | cos | n_cos | cos_ev |
|---|
Notes
n = number of mutations in this gene in the individual set
cos = number of unique mutated sites in this gene in COSMIC
n_cos = overlap between n and cos
cos_ev = total evidence: number of reports in COSMIC for mutations seen in this gene
| gene | patient | chr | start | end | type | n_cos |
|---|
| rank | gene | desc | n | mindist | npairs3 | npairs12 |
|---|---|---|---|---|---|---|
| 1 | C20orf26 | chromosome 20 open reading frame 26 | 10 | 0 | 1 | 1 |
| 2 | DNAH8 | dynein, axonemal, heavy chain 8 | 26 | 3 | 0 | 1 |
| 3 | ABCA13 | ATP-binding cassette, sub-family A (ABC1), member 13 | 16 | 8 | 0 | 1 |
| 4 | SCN5A | sodium channel, voltage-gated, type V, alpha subunit | 6 | 19 | 0 | 0 |
| 5 | GBF1 | golgi-specific brefeldin A resistance factor 1 | 4 | 21 | 0 | 0 |
| 6 | CUBN | cubilin (intrinsic factor-cobalamin receptor) | 18 | 30 | 0 | 0 |
| 7 | PKHD1L1 | polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1 | 29 | 84 | 0 | 0 |
| 8 | CELSR3 | cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) | 6 | 161 | 0 | 0 |
| 9 | MXRA5 | matrix-remodelling associated 5 | 10 | 686 | 0 | 0 |
| 10 | GPR98 | G protein-coupled receptor 98 | 23 | 850 | 0 | 0 |
| 11 | SYNE1 | spectrin repeat containing, nuclear envelope 1 | 27 | 851 | 0 | 0 |
| 12 | A2ML1 | alpha-2-macroglobulin-like 1 | 4 | Inf | 0 | 0 |
| 13 | AACS | acetoacetyl-CoA synthetase | 2 | Inf | 0 | 0 |
| 14 | AAK1 | AP2 associated kinase 1 | 4 | Inf | 0 | 0 |
| 15 | AARS2 | alanyl-tRNA synthetase 2, mitochondrial (putative) | 4 | Inf | 0 | 0 |
| 16 | ABCA10 | ATP-binding cassette, sub-family A (ABC1), member 10 | 4 | Inf | 0 | 0 |
| 17 | ABCA4 | ATP-binding cassette, sub-family A (ABC1), member 4 | 8 | Inf | 0 | 0 |
| 18 | ABCA6 | ATP-binding cassette, sub-family A (ABC1), member 6 | 5 | Inf | 0 | 0 |
| 19 | ABCB1 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 | 9 | Inf | 0 | 0 |
| 20 | ABCB5 | ATP-binding cassette, sub-family B (MDR/TAP), member 5 | 4 | Inf | 0 | 0 |
| 21 | ABCC1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 | 4 | Inf | 0 | 0 |
| 22 | ABCC12 | ATP-binding cassette, sub-family C (CFTR/MRP), member 12 | 9 | Inf | 0 | 0 |
| 23 | ABCC2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | 2 | Inf | 0 | 0 |
| 24 | ABCC8 | ATP-binding cassette, sub-family C (CFTR/MRP), member 8 | 6 | Inf | 0 | 0 |
| 25 | ABCC9 | ATP-binding cassette, sub-family C (CFTR/MRP), member 9 | 5 | Inf | 0 | 0 |
| 26 | ABCF2 | ATP-binding cassette, sub-family F (GCN20), member 2 | 2 | Inf | 0 | 0 |
| 27 | ABCF3 | ATP-binding cassette, sub-family F (GCN20), member 3 | 2 | Inf | 0 | 0 |
| 28 | ABCG8 | ATP-binding cassette, sub-family G (WHITE), member 8 (sterolin 2) | 5 | Inf | 0 | 0 |
| 29 | ABLIM1 | actin binding LIM protein 1 | 2 | Inf | 0 | 0 |
| 30 | ABLIM3 | actin binding LIM protein family, member 3 | 2 | Inf | 0 | 0 |
Notes
n = number of mutations in this gene in the individual set
mindist = distance (in aa) between closest pair of mutations in this gene
npairs3 = how many pairs of mutations are within 3 aa of each other
npairs12 = how many pairs of mutations are within 12 aa of each other
| rank | geneset | description | N_genes | mut_tally | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GPCRDB_CLASS_B_SECRETIN_ LIKE | 20 | CALCR(2), CRHR1(3), CRHR2(1), ELTD1(9), EMR1(5), EMR2(3), GLP2R(2), GPR64(3) | 70035 | 28 | 22 | 28 | 7 | 6 | 13 | 1 | 4 | 4 | <1.00e-11 | <7.70e-10 | |
| 2 | STRIATED_MUSCLE_ CONTRACTION | 37 | ACTN3(2), DMD(10), MYBPC3(1), MYH6(8), MYH7(6), MYH8(14), MYOM1(2), NEB(9), TNNT3(1), TTN(173) | 823331 | 226 | 91 | 225 | 69 | 52 | 114 | 20 | 19 | 21 | <1.00e-11 | <7.70e-10 | |
| 3 | NO1PATHWAY | Shear stress in endothelial cells increases cytoplasmic calcium, which activates nitric oxide synthase III to release NO, which in turn regulates cardiac contractions. | 28 | CHRM1(1), CHRNA1(2), FLT4(1), NOS3(1), RYR2(60), SLC7A1(2) | 268724 | 67 | 41 | 66 | 20 | 12 | 38 | 7 | 3 | 7 | <1.00e-11 | <7.70e-10 |
| 4 | HSA04720_LONG_TERM_ POTENTIATION | Genes involved in long-term potentiation | 67 | ADCY8(8), ARAF(1), BRAF(4), CACNA1C(14), CAMK2A(2), CAMK2D(1), CAMK4(1), CREBBP(10), EP300(4), GRIA1(5), GRIN2B(11), GRM1(8), ITPR1(1), ITPR2(5), ITPR3(3), KRAS(2), MAPK1(1), MAPK3(1), PLCB1(6), PLCB4(3), PPP3CC(1), PRKCA(3), RAF1(1), RPS6KA2(2) | 461187 | 98 | 57 | 98 | 27 | 29 | 39 | 4 | 10 | 16 | <1.00e-11 | <7.70e-10 |
| 5 | HSA04730_LONG_TERM_ DEPRESSION | Genes involved in long-term depression | 74 | ARAF(1), BRAF(4), CACNA1A(6), CRHR1(3), GNAS(2), GRIA1(5), GRM1(8), GUCY2C(3), GUCY2D(1), ITPR1(1), ITPR2(5), ITPR3(3), KRAS(2), LYN(1), MAPK1(1), MAPK3(1), NOS3(1), PLA2G4A(4), PLCB1(6), PLCB4(3), PPP2R2A(1), PRKCA(3), RAF1(1), RYR1(23) | 551618 | 89 | 53 | 89 | 25 | 15 | 50 | 3 | 10 | 11 | <1.00e-11 | <7.70e-10 |
| 6 | HSA04020_CALCIUM_ SIGNALING_PATHWAY | Genes involved in calcium signaling pathway | 169 | ADCY2(11), ADCY3(3), ADCY4(3), ADCY8(8), ATP2A3(2), ATP2B1(4), ATP2B2(4), ATP2B4(1), CACNA1A(6), CACNA1C(14), CACNA1D(1), CACNA1E(14), CACNA1F(3), CACNA1G(4), CACNA1H(1), CACNA1I(7), CAMK2A(2), CAMK2D(1), CAMK4(1), CHRM1(1), EGFR(2), ERBB3(2), GNAL(1), GNAS(2), GRM1(8), ITPR1(1), ITPR2(5), ITPR3(3), MYLK(1), NOS3(1), P2RX4(1), PDE1A(2), PDGFRB(5), PHKG1(1), PHKG2(1), PLCB1(6), PLCB4(3), PLCE1(6), PPP3CC(1), PRKCA(3), RYR1(23), RYR2(60), RYR3(24), SLC8A3(5), TBXA2R(1) | 1662500 | 259 | 92 | 258 | 73 | 60 | 141 | 15 | 18 | 25 | <1.00e-11 | <7.70e-10 |
| 7 | SMOOTH_MUSCLE_CONTRACTION | 138 | ADCY2(11), ADCY3(3), ADCY4(3), ADCY5(2), ADCY8(8), ATP2A3(2), CAMK2A(2), CAMK2D(1), CREB3(1), CRHR1(3), GRK4(1), IL1B(1), ITPR1(1), ITPR2(5), ITPR3(3), NOS3(1), PRKCA(3), PRKCE(2), PRKCZ(1), RGS6(3), RYR1(23), RYR2(60), RYR3(24), TNXB(12), USP5(1) | 1159895 | 177 | 77 | 176 | 52 | 51 | 89 | 11 | 11 | 15 | <1.00e-11 | <7.70e-10 | |
| 8 | CALCIUM_REGULATION_ IN_CARDIAC_CELLS | 139 | ADCY2(11), ADCY3(3), ADCY4(3), ADCY5(2), ADCY8(8), ATP1B2(1), ATP2A3(2), ATP2B1(4), ATP2B2(4), CACNA1A(6), CACNA1C(14), CACNA1D(1), CACNA1E(14), CAMK2A(2), CAMK2D(1), CAMK4(1), CHRM1(1), GJB1(1), GRK4(1), ITPR1(1), ITPR2(5), ITPR3(3), PEA15(1), PRKCA(3), PRKCE(2), PRKCZ(1), RGS6(3), RYR1(23), RYR2(60), RYR3(24), SLC8A3(5), USP5(1) | 1466343 | 212 | 88 | 211 | 58 | 59 | 109 | 11 | 13 | 20 | <1.00e-11 | <7.70e-10 | |
| 9 | HSA04512_ECM_RECEPTOR_ INTERACTION | Genes involved in ECM-receptor interaction | 87 | AGRN(1), CD36(1), CHAD(1), COL11A2(6), COL1A1(4), COL4A1(6), COL4A4(6), COL4A6(2), COL5A3(7), FNDC3A(1), GP1BA(3), GP5(3), HSPG2(4), ITGA1(1), ITGA2(5), ITGA5(1), ITGA7(2), ITGA8(8), ITGB4(2), ITGB6(1), LAMA1(7), LAMA2(10), LAMA3(6), LAMA5(5), LAMB1(5), LAMB2(3), LAMB4(3), RELN(27), TNC(5), TNXB(12), VWF(12) | 1365132 | 160 | 78 | 160 | 51 | 48 | 77 | 5 | 12 | 18 | 1.60e-10 | 1.10e-08 |
| 10 | LAIRPATHWAY | The local acute inflammatory response is mediated by activated macrophages and mast cells or by complement activation. | 16 | C3(1), C6(11), C7(4), ITGAL(2), SELP(6) | 74494 | 24 | 23 | 24 | 8 | 5 | 10 | 3 | 0 | 6 | 6.75e-10 | 4.16e-08 |
Notes: (Please see notes under significantly mutated gene table)
| rank | geneset | description | N_genes | mut_tally | N | n | npat | nsite | nsil | n1 | n2 | n3 | n4 | n5 | p | q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GPCRDB_CLASS_B_SECRETIN_ LIKE | 19 | CALCR(2), CRHR1(3), CRHR2(1), EMR1(5), EMR2(3), GLP2R(2), GPR64(3) | 63050 | 19 | 17 | 19 | 7 | 5 | 10 | 1 | 2 | 1 | 2.17e-06 | 0.0013 | |
| 2 | GPCRDB_CLASS_C_METABOTROPIC_ GLUTAMATE_PHEROMONE | 12 | CASR(5), GRM2(3) | 19889 | 8 | 8 | 8 | 3 | 2 | 5 | 0 | 1 | 0 | 0.00037 | 0.11 | |
| 3 | ST_MYOCYTE_AD_PATHWAY | Cardiac myocytes have a variety of adrenergic receptors that induce subtype-specific signaling effects. | 22 | APC(7), EPHB2(3), ITPR1(1), ITPR2(5), ITPR3(3), KCNJ9(3), MAPK1(1), RHO(1) | 145209 | 24 | 22 | 24 | 3 | 5 | 11 | 2 | 2 | 4 | 0.00069 | 0.12 |
| 4 | HSA04664_FC_EPSILON_ RI_SIGNALING_PATHWAY | Genes involved in Fc epsilon RI signaling pathway | 72 | BTK(2), IL4(1), INPP5D(4), LYN(1), MAP2K6(2), MAPK1(1), MAPK3(1), MS4A2(1), PIK3R5(4), PLA2G4A(4), PRKCA(3), PRKCE(2), RAF1(1), SOS1(3) | 207363 | 30 | 25 | 30 | 7 | 6 | 17 | 0 | 3 | 4 | 0.00079 | 0.12 |
| 5 | HSA00950_ALKALOID_ BIOSYNTHESIS_I | Genes involved in alkaloid biosynthesis I | 5 | DDC(3), TAT(3) | 13232 | 6 | 6 | 6 | 1 | 2 | 4 | 0 | 0 | 0 | 0.0011 | 0.14 |
| 6 | IL12PATHWAY | IL12 and Stat4 Dependent Signaling Pathway in Th1 Development | 20 | CD3E(1), CXCR3(1), IL12RB2(2), JAK2(3), MAP2K6(2) | 40623 | 9 | 8 | 9 | 2 | 1 | 4 | 0 | 1 | 3 | 0.0015 | 0.15 |
| 7 | TRKAPATHWAY | Nerve growth factor (NGF) promotes neuronal survival and proliferation by binding its receptor TrkA, which activates PI3K/AKT, Ras, and the MAP kinase pathway. | 12 | NTRK1(5), PRKCA(3), SOS1(3) | 61862 | 11 | 11 | 11 | 1 | 0 | 7 | 0 | 2 | 2 | 0.0020 | 0.18 |
| 8 | GLYCOLYSISPATHWAY | Glycolysis is an evolutionarily conserved pathway by which one glucose molecule is converted to two pyruvate molecules for a gain of 2 ATP. | 9 | GPI(3), HK1(3), PGK1(2) | 37507 | 8 | 7 | 8 | 0 | 5 | 2 | 0 | 0 | 1 | 0.0023 | 0.18 |
| 9 | TPOPATHWAY | Thrombopoietin binds to its receptor and activates cell growth through the Erk and JNK MAP kinase pathways, protein kinase C, and JAK/STAT activation. | 22 | JAK2(3), MAPK3(1), PRKCA(3), RAF1(1), RASA1(2), SOS1(3), STAT1(1), STAT5B(1) | 108948 | 15 | 15 | 15 | 1 | 1 | 8 | 0 | 2 | 4 | 0.0030 | 0.21 |
| 10 | ST_INTERLEUKIN_4_ PATHWAY | Like IL-13, IL-4 is produced by Th2 cells on activation of the T cell antigen receptor, and by mast and basophil cells on activation of the IgE receptor. | 26 | IL4(1), INPP5D(4), JAK2(3), SOS1(3) | 56800 | 11 | 11 | 11 | 0 | 1 | 7 | 0 | 2 | 1 | 0.0042 | 0.25 |
Notes: (Please see notes under significantly mutated gene table)