This pipeline computes the correlation between APOBRC groups and selected clinical features.
Testing the association between APOBEC groups identified by 2 different apobec score and 17 clinical features across 108 patients, no significant finding detected with P value < 0.05 and Q value < 0.25.
-
3 subtypes identified in current cancer cohort by 'APOBEC MUTLOAD MINESTIMATE'. These subtypes do not correlate to any clinical features.
-
3 subtypes identified in current cancer cohort by 'APOBEC ENRICH'. These subtypes do not correlate to any clinical features.
Table 1. Get Full Table Overview of the association between APOBEC groups by 2 different APOBEC scores and 17 clinical features. Shown in the table are P values (Q values). Thresholded by P value < 0.05 and Q value < 0.25, no significant finding detected.
|
Clinical Features |
Statistical Tests |
APOBEC MUTLOAD MINESTIMATE |
APOBEC ENRICH |
| DAYS TO DEATH OR LAST FUP | logrank test |
0.926 (0.984) |
0.232 (0.838) |
| HISTOLOGICAL TYPE | Fisher's exact test |
0.399 (0.848) |
0.396 (0.848) |
| HISTOLOGIC GRADE | Fisher's exact test |
0.779 (0.966) |
0.805 (0.966) |
| KARNOFSKY PERFORMANCE SCORE | Kruskal-Wallis (anova) |
0.24 (0.838) |
0.131 (0.838) |
| PATHOLOGY T STAGE | Fisher's exact test |
0.553 (0.907) |
0.755 (0.966) |
| PATHOLOGY N STAGE | Fisher's exact test |
0.733 (0.966) |
0.321 (0.838) |
| PATHOLOGIC STAGE | Fisher's exact test |
0.51 (0.907) |
0.801 (0.966) |
| YEARS TO BIRTH | Kruskal-Wallis (anova) |
0.286 (0.838) |
0.012 (0.409) |
| ETHNICITY | Fisher's exact test |
1 (1.00) |
1 (1.00) |
| GENDER | Fisher's exact test |
0.182 (0.838) |
0.283 (0.838) |
| RACE | Fisher's exact test |
0.691 (0.966) |
0.853 (0.966) |
| RADIATION THERAPY | Fisher's exact test |
0.56 (0.907) |
0.474 (0.907) |
| BMI | Fisher's exact test |
0.38 (0.848) |
0.55 (0.907) |
| NUMBER PACK YEARS SMOKED | Kruskal-Wallis (anova) |
0.312 (0.838) |
0.224 (0.838) |
| SMOKER | Fisher's exact test |
0.877 (0.966) |
0.648 (0.966) |
| COUNTRY OF ORIGIN | Fisher's exact test |
0.142 (0.838) |
0.104 (0.838) |
| ORIGIN ASIA | Fisher's exact test |
0.881 (0.966) |
0.0583 (0.838) |
Table S1. Description of APOBEC group #1: 'APOBEC MUTLOAD MINESTIMATE'
| Cluster Labels | 0 | HIGH | LOW |
|---|---|---|---|
| Number of samples | 71 | 19 | 18 |
P value = 0.926 (logrank test), Q value = 0.98
Table S2. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #1: 'DAYS_TO_DEATH_OR_LAST_FUP'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 108 | 11 | 0.2 - 35.0 (13.0) |
| 0 | 69 | 8 | 0.2 - 35.0 (13.1) |
| HIGH | 18 | 2 | 0.5 - 27.4 (13.3) |
| LOW | 13 | 1 | 0.2 - 26.0 (12.1) |
Figure S1. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #1: 'DAYS_TO_DEATH_OR_LAST_FUP'
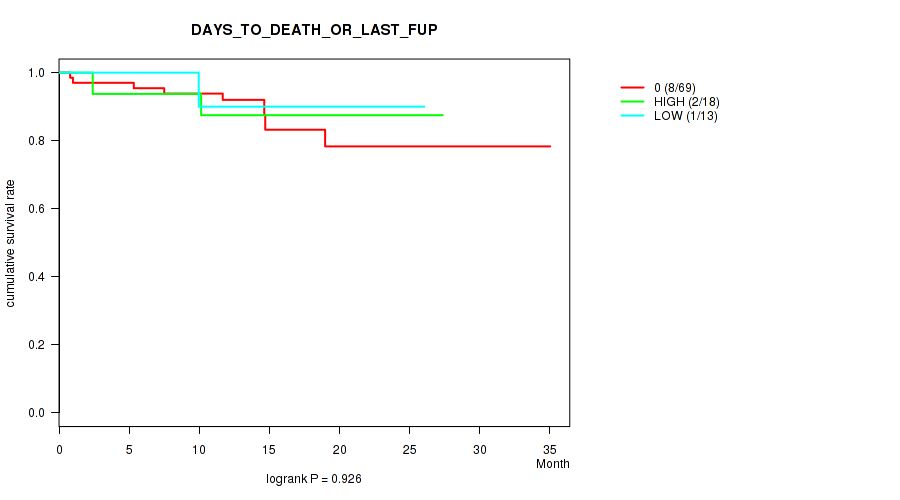
P value = 0.399 (Fisher's exact test), Q value = 0.85
Table S3. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #2: 'HISTOLOGICAL_TYPE'
| nPatients | ACINAR ADENOCARCINOMA | ADENOCARCINOMA | BASALOID SQUAMOUS CELL CARCINOMA | INVASIVE MUCINOUS ADENOCARCINOMA | LEPIDIC ADENOCARCINOMA | MICROPAPILLARY ADENOCARCINOMA | OTHER | PAPILLARY ADENOCARCINOMA | SOLID ADENOCARCINOMA | SQUAMOUS CELL CARCINOMA |
|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 13 | 67 | 1 | 3 | 1 | 2 | 7 | 4 | 8 | 2 |
| 0 | 11 | 40 | 0 | 3 | 1 | 1 | 5 | 4 | 6 | 0 |
| HIGH | 0 | 14 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 |
| LOW | 2 | 13 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
Figure S2. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #2: 'HISTOLOGICAL_TYPE'
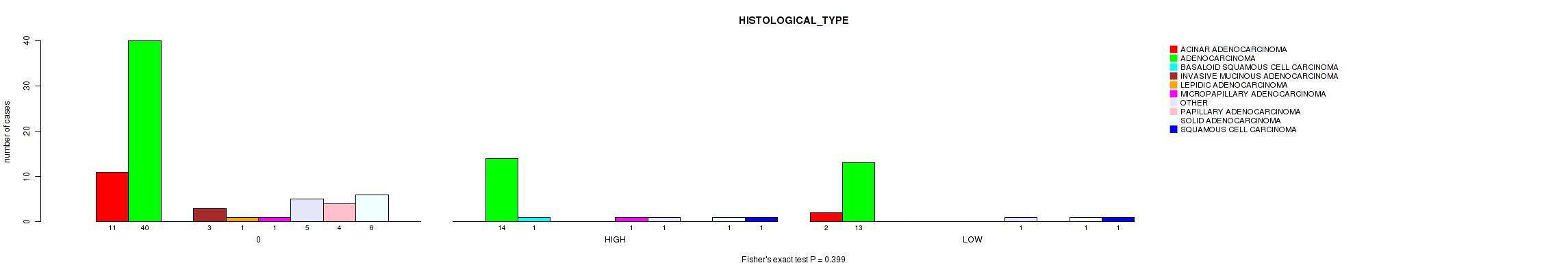
P value = 0.779 (Fisher's exact test), Q value = 0.97
Table S4. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #3: 'HISTOLOGIC_GRADE'
| nPatients | G1 | G2 | G3 | G4 | GX |
|---|---|---|---|---|---|
| ALL | 7 | 63 | 31 | 1 | 6 |
| 0 | 5 | 41 | 21 | 1 | 3 |
| HIGH | 1 | 9 | 7 | 0 | 2 |
| LOW | 1 | 13 | 3 | 0 | 1 |
Figure S3. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #3: 'HISTOLOGIC_GRADE'
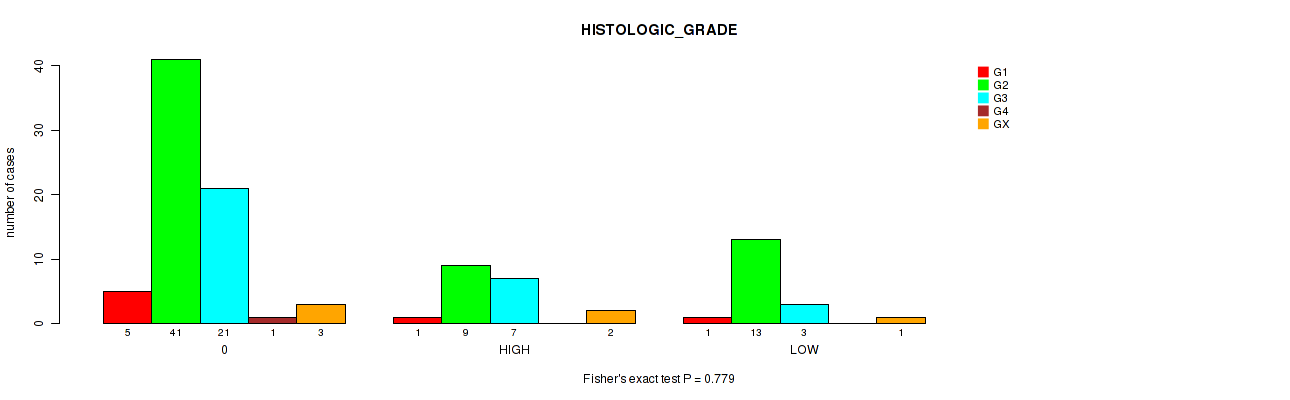
P value = 0.24 (Kruskal-Wallis (anova)), Q value = 0.84
Table S5. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #4: 'KARNOFSKY_PERFORMANCE_SCORE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 69 | 81.7 (9.1) |
| 0 | 44 | 80.2 (8.5) |
| HIGH | 14 | 85.0 (10.2) |
| LOW | 11 | 83.6 (9.2) |
Figure S4. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #4: 'KARNOFSKY_PERFORMANCE_SCORE'
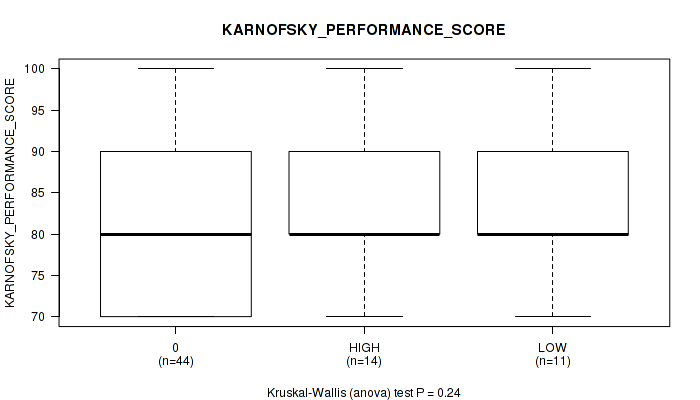
P value = 0.553 (Kruskal-Wallis (anova)), Q value = 0.91
Table S6. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #5: 'PATHOLOGY_T_STAGE'
| T1 | T2 | T3 | |
|---|---|---|---|
| ALL | 27 | 69 | 12 |
| 0 | 20 | 44 | 7 |
| HIGH | 3 | 12 | 4 |
| LOW | 4 | 13 | 1 |
Figure S5. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #5: 'PATHOLOGY_T_STAGE'
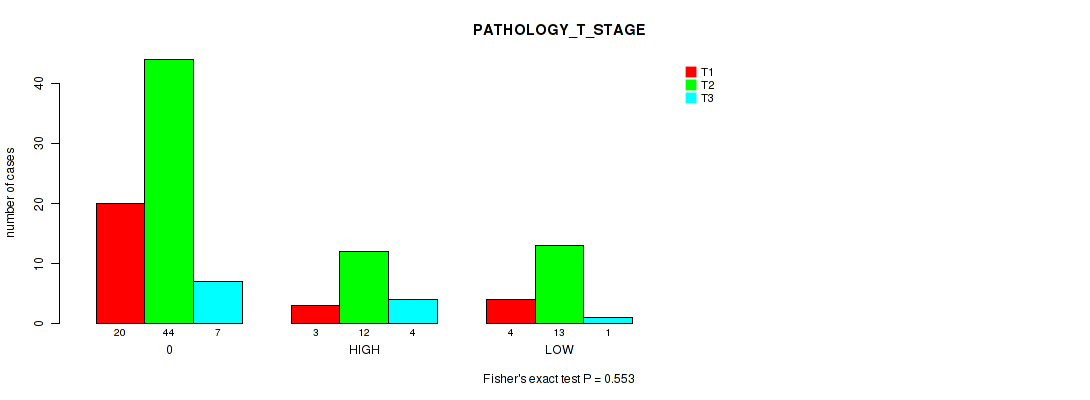
P value = 0.733 (Kruskal-Wallis (anova)), Q value = 0.97
Table S7. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #6: 'PATHOLOGY_N_STAGE'
| N0 | N1 | N2 | |
|---|---|---|---|
| ALL | 71 | 17 | 19 |
| 0 | 45 | 10 | 15 |
| HIGH | 14 | 3 | 2 |
| LOW | 12 | 4 | 2 |
Figure S6. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #6: 'PATHOLOGY_N_STAGE'
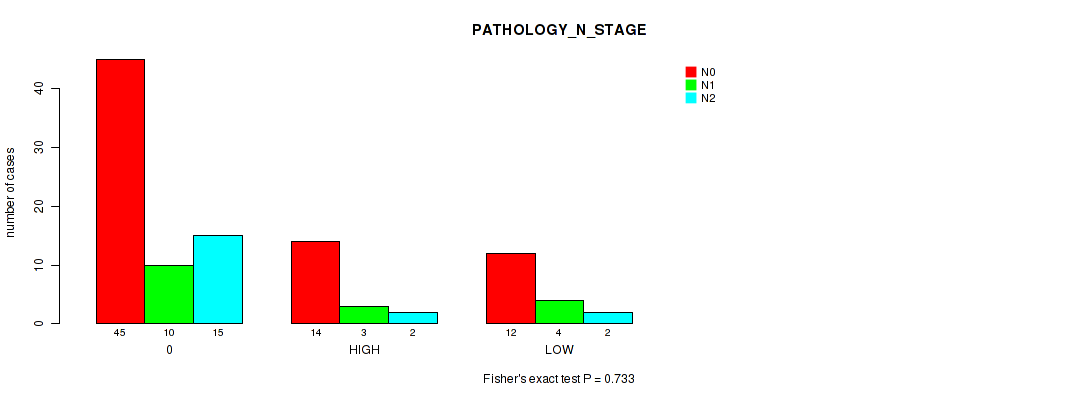
P value = 0.51 (Fisher's exact test), Q value = 0.91
Table S8. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #7: 'PATHOLOGIC_STAGE'
| nPatients | STAGE I | STAGE IA | STAGE IB | STAGE IIA | STAGE IIB | STAGE III | STAGE IIIA | STAGE IV |
|---|---|---|---|---|---|---|---|---|
| ALL | 2 | 22 | 33 | 17 | 12 | 1 | 20 | 1 |
| 0 | 2 | 15 | 20 | 13 | 5 | 1 | 14 | 1 |
| HIGH | 0 | 3 | 8 | 0 | 5 | 0 | 3 | 0 |
| LOW | 0 | 4 | 5 | 4 | 2 | 0 | 3 | 0 |
Figure S7. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #7: 'PATHOLOGIC_STAGE'
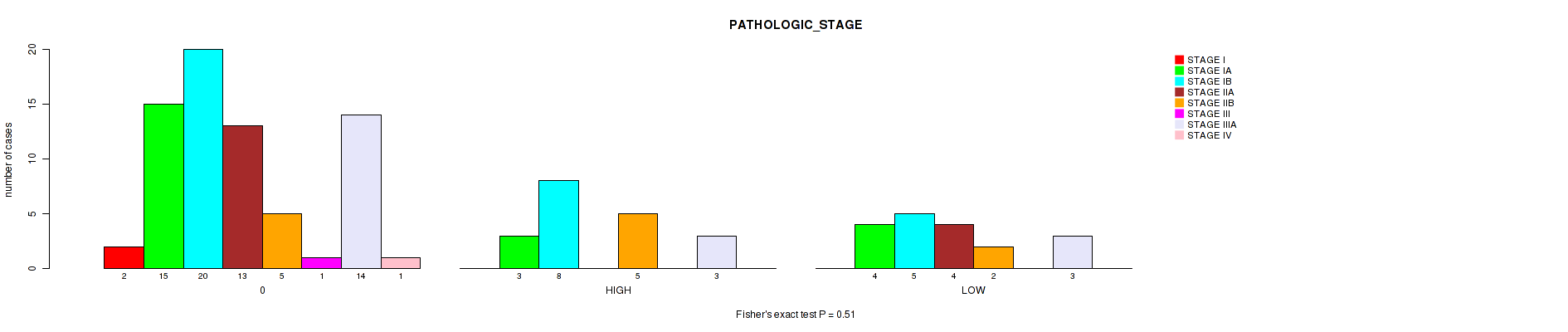
P value = 0.286 (Kruskal-Wallis (anova)), Q value = 0.84
Table S9. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #8: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 108 | 62.5 (9.7) |
| 0 | 71 | 61.5 (10.4) |
| HIGH | 19 | 63.4 (8.1) |
| LOW | 18 | 65.3 (7.9) |
Figure S8. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #8: 'YEARS_TO_BIRTH'
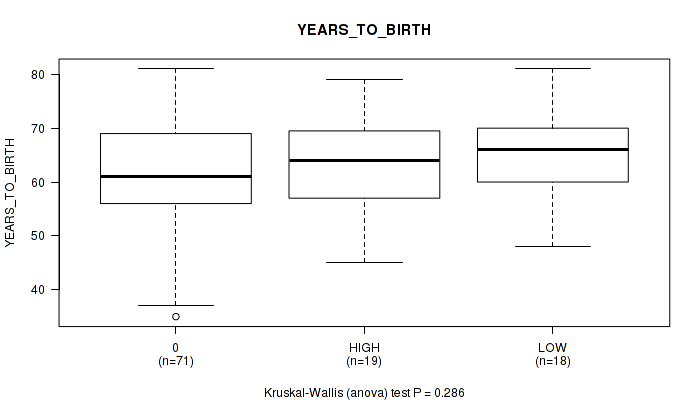
P value = 1 (Fisher's exact test), Q value = 1
Table S10. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #9: 'ETHNICITY'
| nPatients | HISPANIC OR LATINO | NOT HISPANIC OR LATINO |
|---|---|---|
| ALL | 3 | 33 |
| 0 | 3 | 22 |
| HIGH | 0 | 5 |
| LOW | 0 | 6 |
Figure S9. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #9: 'ETHNICITY'
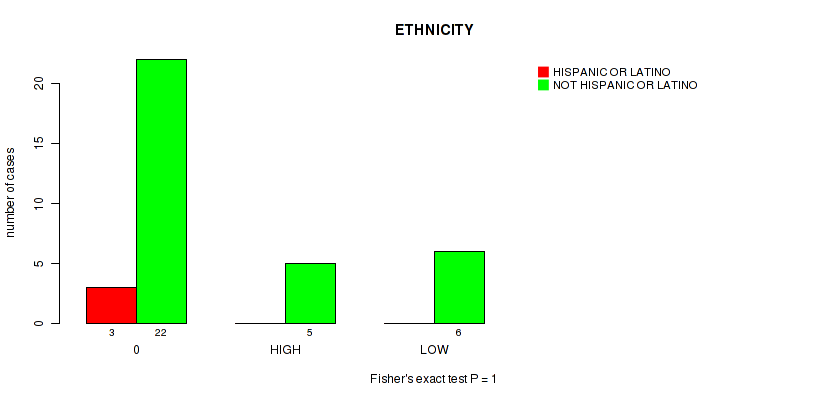
P value = 0.182 (Fisher's exact test), Q value = 0.84
Table S11. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #10: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 36 | 72 |
| 0 | 27 | 44 |
| HIGH | 3 | 16 |
| LOW | 6 | 12 |
Figure S10. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #10: 'GENDER'
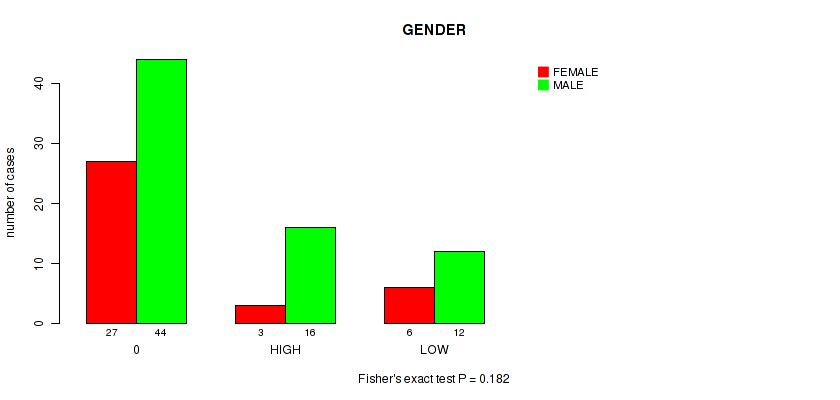
P value = 0.691 (Fisher's exact test), Q value = 0.97
Table S12. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #11: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | WHITE |
|---|---|---|---|---|
| ALL | 1 | 1 | 1 | 32 |
| 0 | 1 | 1 | 0 | 22 |
| HIGH | 0 | 0 | 0 | 5 |
| LOW | 0 | 0 | 1 | 5 |
Figure S11. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #11: 'RACE'
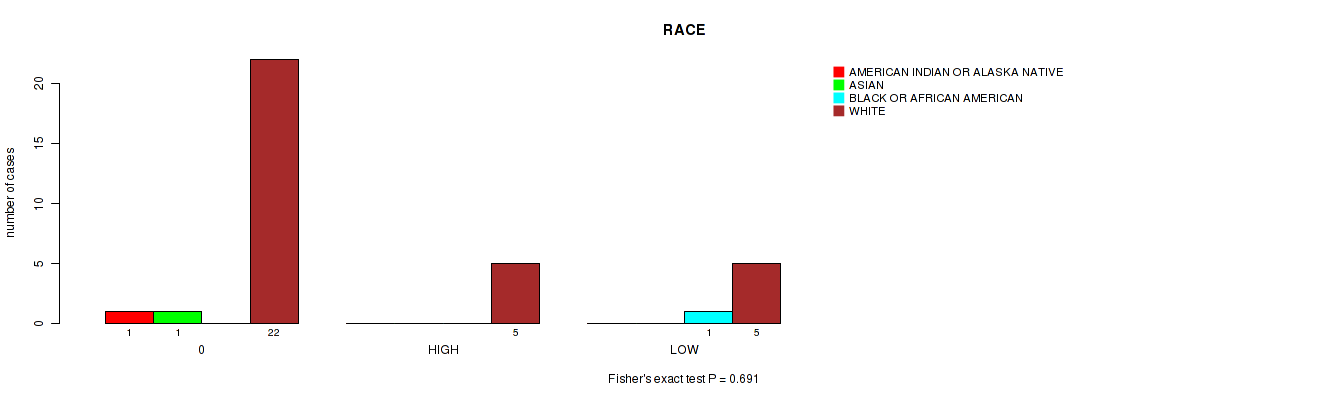
P value = 0.56 (Fisher's exact test), Q value = 0.91
Table S13. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #12: 'RADIATION_THERAPY'
| nPatients | NO | YES |
|---|---|---|
| ALL | 69 | 22 |
| 0 | 49 | 13 |
| HIGH | 12 | 5 |
| LOW | 8 | 4 |
Figure S12. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #12: 'RADIATION_THERAPY'
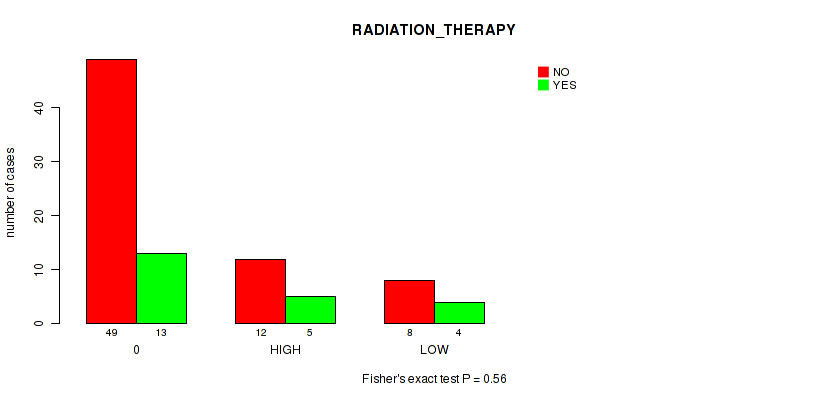
P value = 0.38 (Fisher's exact test), Q value = 0.85
Table S14. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #13: 'BMI'
| nPatients | NORMAL | OBESE | OVERWEIGHT | SEVERELY OBESE | UNDERWEIGHT |
|---|---|---|---|---|---|
| ALL | 53 | 10 | 27 | 3 | 15 |
| 0 | 39 | 6 | 16 | 2 | 8 |
| HIGH | 5 | 2 | 7 | 0 | 5 |
| LOW | 9 | 2 | 4 | 1 | 2 |
Figure S13. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #13: 'BMI'
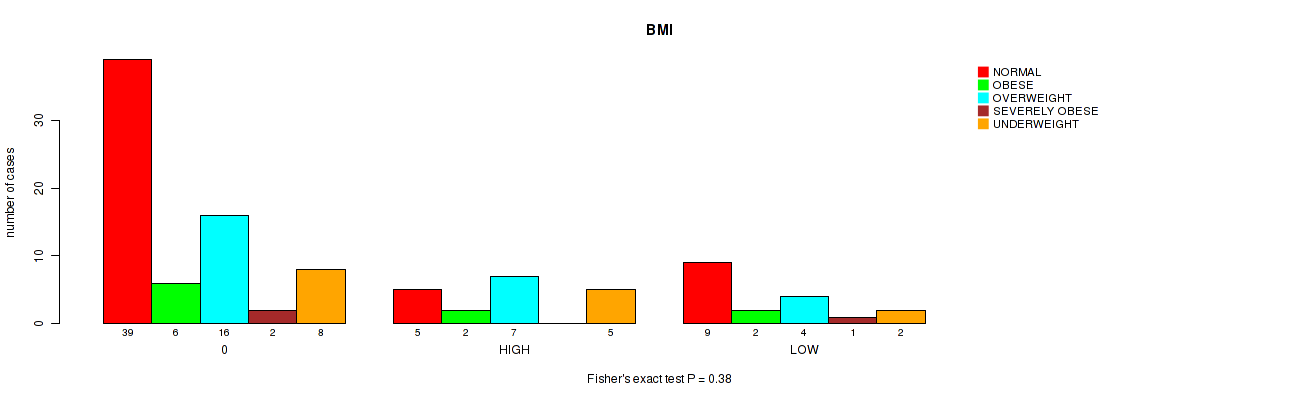
P value = 0.312 (Kruskal-Wallis (anova)), Q value = 0.84
Table S15. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #14: 'NUMBER_PACK_YEARS_SMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 54 | 27.3 (23.6) |
| 0 | 33 | 23.0 (19.3) |
| HIGH | 11 | 39.1 (35.1) |
| LOW | 10 | 28.5 (18.7) |
Figure S14. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #14: 'NUMBER_PACK_YEARS_SMOKED'
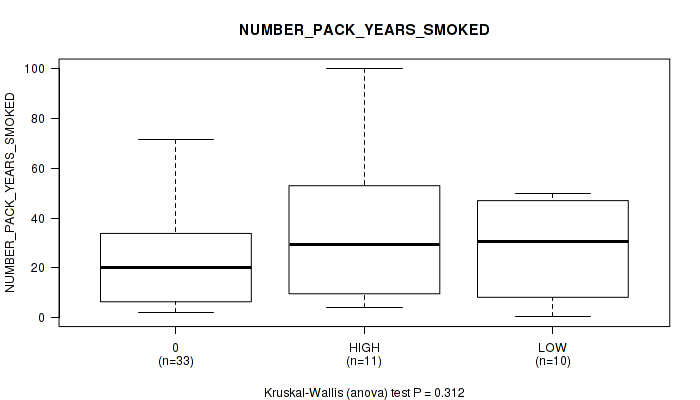
P value = 0.877 (Fisher's exact test), Q value = 0.97
Table S16. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #15: 'SMOKER'
| nPatients | NON-SMOKER | SMOKER |
|---|---|---|
| ALL | 45 | 60 |
| 0 | 31 | 38 |
| HIGH | 7 | 11 |
| LOW | 7 | 11 |
Figure S15. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #15: 'SMOKER'
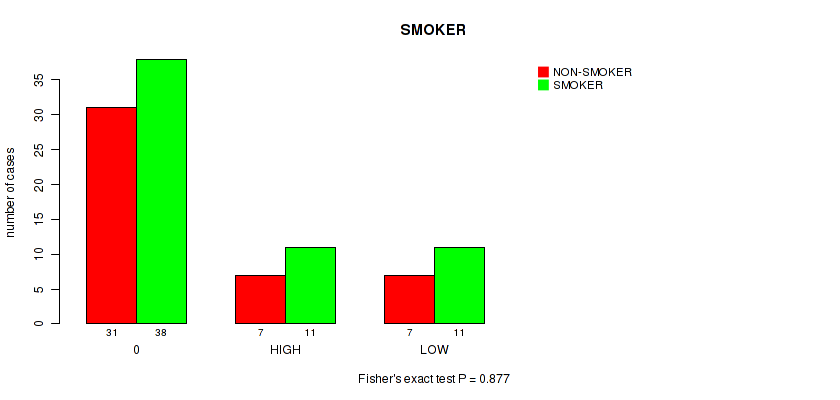
P value = 0.142 (Fisher's exact test), Q value = 0.84
Table S17. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #16: 'COUNTRY_OF_ORIGIN'
| nPatients | BULGARIA | CANADA | CHINA | JAMAICA | MEXICO | POLAND | RUSSIA | UKRAINE | UNITED STATES | VIETNAM |
|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 2 | 1 | 18 | 1 | 2 | 2 | 1 | 3 | 32 | 44 |
| 0 | 1 | 1 | 12 | 0 | 2 | 0 | 1 | 0 | 23 | 30 |
| HIGH | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 1 | 5 | 8 |
| LOW | 1 | 0 | 3 | 1 | 0 | 0 | 0 | 2 | 4 | 6 |
Figure S16. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #16: 'COUNTRY_OF_ORIGIN'
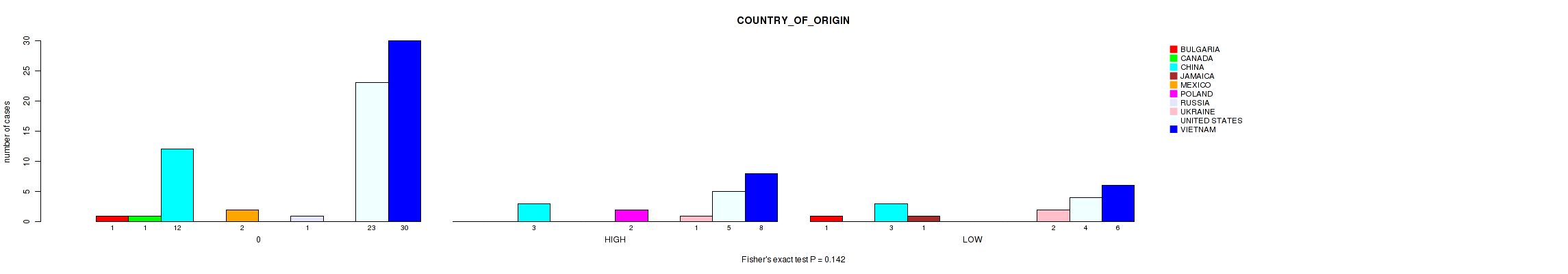
P value = 0.881 (Fisher's exact test), Q value = 0.97
Table S18. Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #17: 'ORIGIN_ASIA'
| nPatients | ASIAN | WESTERN |
|---|---|---|
| ALL | 62 | 44 |
| 0 | 42 | 28 |
| HIGH | 11 | 8 |
| LOW | 9 | 8 |
Figure S17. Get High-res Image Clustering Approach #1: 'APOBEC MUTLOAD MINESTIMATE' versus Clinical Feature #17: 'ORIGIN_ASIA'
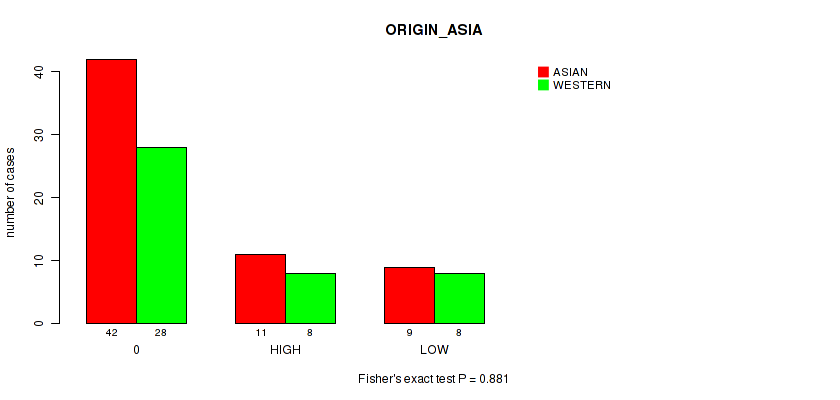
Table S19. Description of APOBEC group #2: 'APOBEC ENRICH'
| Cluster Labels | FC.HIGH.SIG | FC.LOW.NONSIG | FC.NEUTRAL |
|---|---|---|---|
| Number of samples | 25 | 60 | 23 |
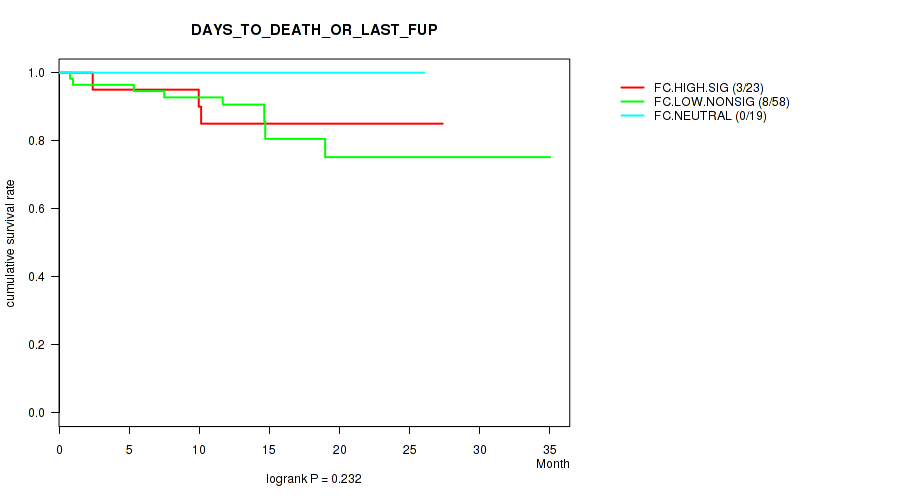
P value = 0.232 (logrank test), Q value = 0.84
Table S20. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #1: 'DAYS_TO_DEATH_OR_LAST_FUP'
| nPatients | nDeath | Duration Range (Median), Month | |
|---|---|---|---|
| ALL | 108 | 11 | 0.2 - 35.0 (13.0) |
| FC.HIGH.SIG | 23 | 3 | 0.2 - 27.4 (12.4) |
| FC.LOW.NONSIG | 58 | 8 | 0.2 - 35.0 (13.1) |
| FC.NEUTRAL | 19 | 0 | 1.2 - 26.0 (13.3) |
Figure S18. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #1: 'DAYS_TO_DEATH_OR_LAST_FUP'
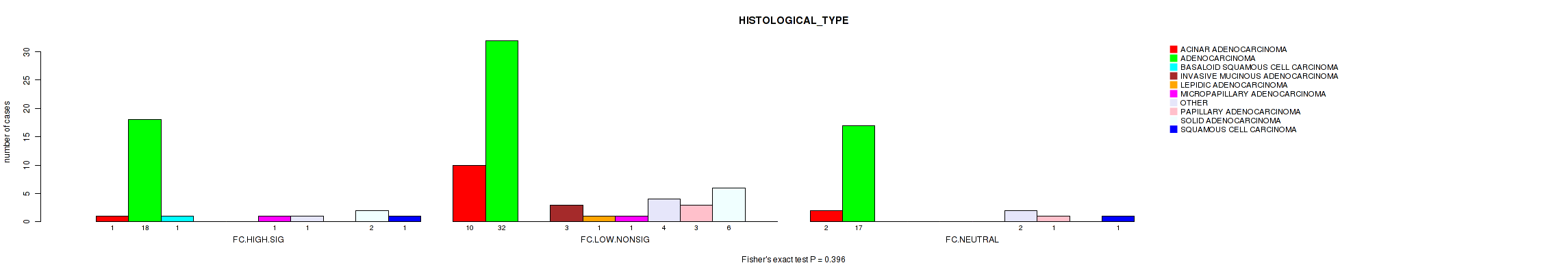
P value = 0.396 (Fisher's exact test), Q value = 0.85
Table S21. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #2: 'HISTOLOGICAL_TYPE'
| nPatients | ACINAR ADENOCARCINOMA | ADENOCARCINOMA | BASALOID SQUAMOUS CELL CARCINOMA | INVASIVE MUCINOUS ADENOCARCINOMA | LEPIDIC ADENOCARCINOMA | MICROPAPILLARY ADENOCARCINOMA | OTHER | PAPILLARY ADENOCARCINOMA | SOLID ADENOCARCINOMA | SQUAMOUS CELL CARCINOMA |
|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 13 | 67 | 1 | 3 | 1 | 2 | 7 | 4 | 8 | 2 |
| FC.HIGH.SIG | 1 | 18 | 1 | 0 | 0 | 1 | 1 | 0 | 2 | 1 |
| FC.LOW.NONSIG | 10 | 32 | 0 | 3 | 1 | 1 | 4 | 3 | 6 | 0 |
| FC.NEUTRAL | 2 | 17 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 1 |
Figure S19. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #2: 'HISTOLOGICAL_TYPE'
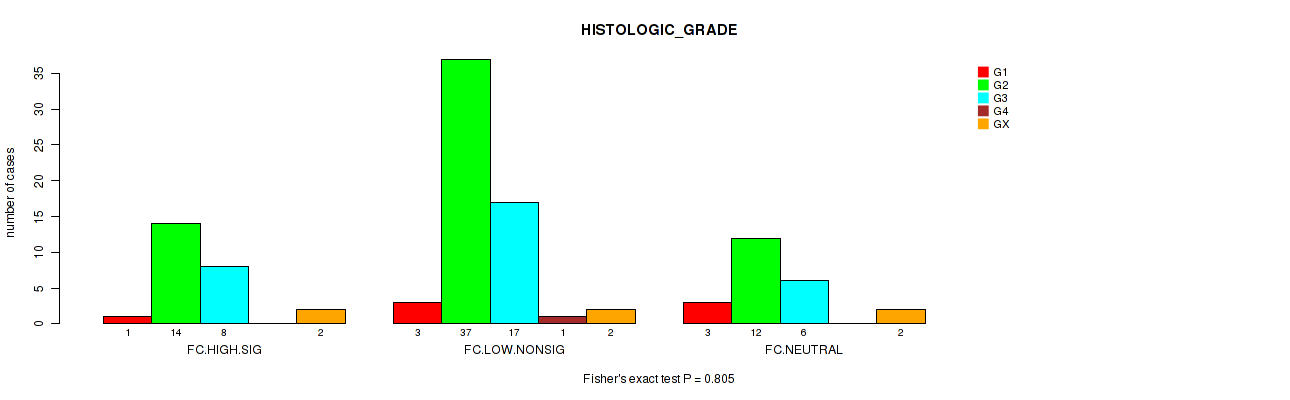
P value = 0.805 (Fisher's exact test), Q value = 0.97
Table S22. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #3: 'HISTOLOGIC_GRADE'
| nPatients | G1 | G2 | G3 | G4 | GX |
|---|---|---|---|---|---|
| ALL | 7 | 63 | 31 | 1 | 6 |
| FC.HIGH.SIG | 1 | 14 | 8 | 0 | 2 |
| FC.LOW.NONSIG | 3 | 37 | 17 | 1 | 2 |
| FC.NEUTRAL | 3 | 12 | 6 | 0 | 2 |
Figure S20. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #3: 'HISTOLOGIC_GRADE'
P value = 0.131 (Kruskal-Wallis (anova)), Q value = 0.84
Table S23. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #4: 'KARNOFSKY_PERFORMANCE_SCORE'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 69 | 81.7 (9.1) |
| FC.HIGH.SIG | 20 | 85.0 (10.0) |
| FC.LOW.NONSIG | 39 | 79.7 (8.7) |
| FC.NEUTRAL | 10 | 83.0 (6.7) |
Figure S21. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #4: 'KARNOFSKY_PERFORMANCE_SCORE'
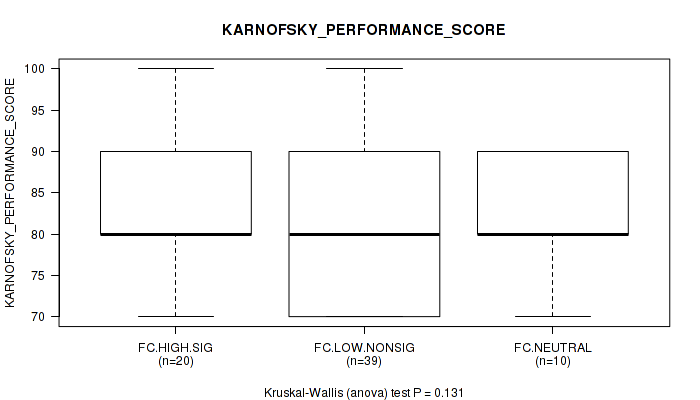
P value = 0.755 (Kruskal-Wallis (anova)), Q value = 0.97
Table S24. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #5: 'PATHOLOGY_T_STAGE'
| T1 | T2 | T3 | |
|---|---|---|---|
| ALL | 27 | 69 | 12 |
| FC.HIGH.SIG | 5 | 16 | 4 |
| FC.LOW.NONSIG | 15 | 38 | 7 |
| FC.NEUTRAL | 7 | 15 | 1 |
Figure S22. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #5: 'PATHOLOGY_T_STAGE'
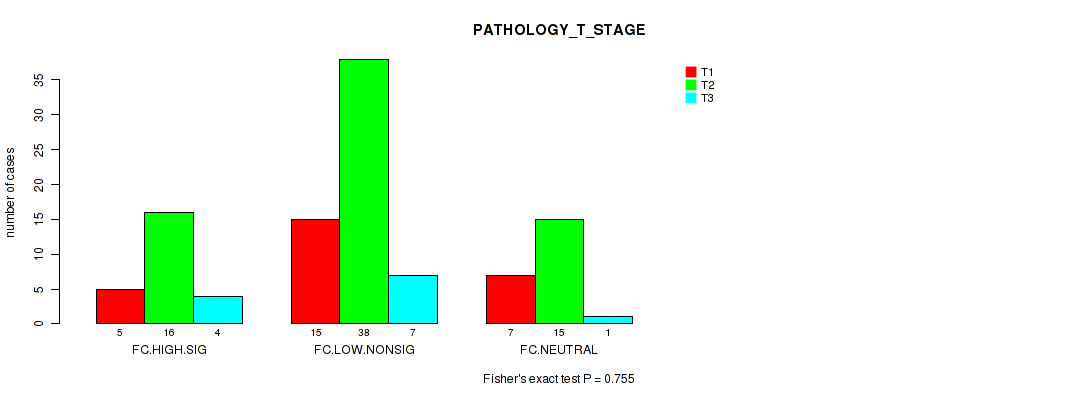
P value = 0.321 (Kruskal-Wallis (anova)), Q value = 0.84
Table S25. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #6: 'PATHOLOGY_N_STAGE'
| N0 | N1 | N2 | |
|---|---|---|---|
| ALL | 71 | 17 | 19 |
| FC.HIGH.SIG | 17 | 5 | 3 |
| FC.LOW.NONSIG | 35 | 10 | 14 |
| FC.NEUTRAL | 19 | 2 | 2 |
Figure S23. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #6: 'PATHOLOGY_N_STAGE'
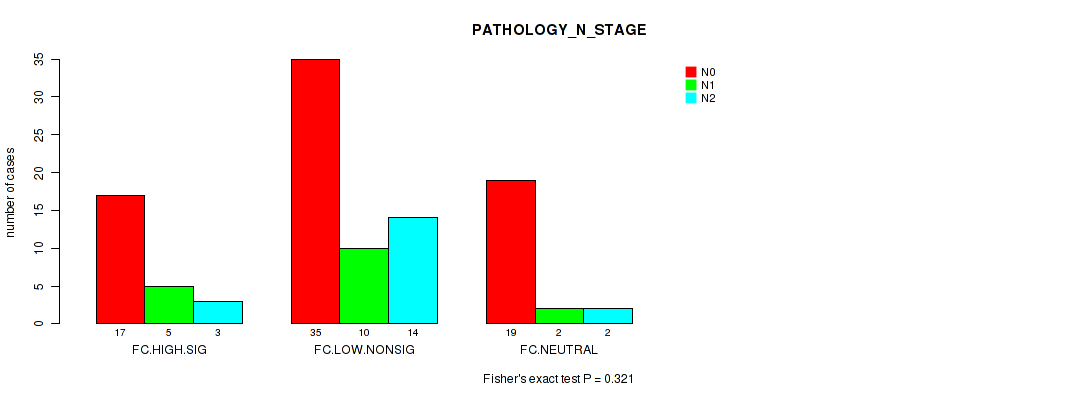
P value = 0.801 (Fisher's exact test), Q value = 0.97
Table S26. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #7: 'PATHOLOGIC_STAGE'
| nPatients | STAGE I | STAGE IA | STAGE IB | STAGE IIA | STAGE IIB | STAGE III | STAGE IIIA | STAGE IV |
|---|---|---|---|---|---|---|---|---|
| ALL | 2 | 22 | 33 | 17 | 12 | 1 | 20 | 1 |
| FC.HIGH.SIG | 0 | 5 | 8 | 3 | 5 | 0 | 4 | 0 |
| FC.LOW.NONSIG | 2 | 10 | 17 | 12 | 4 | 1 | 13 | 1 |
| FC.NEUTRAL | 0 | 7 | 8 | 2 | 3 | 0 | 3 | 0 |
Figure S24. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #7: 'PATHOLOGIC_STAGE'
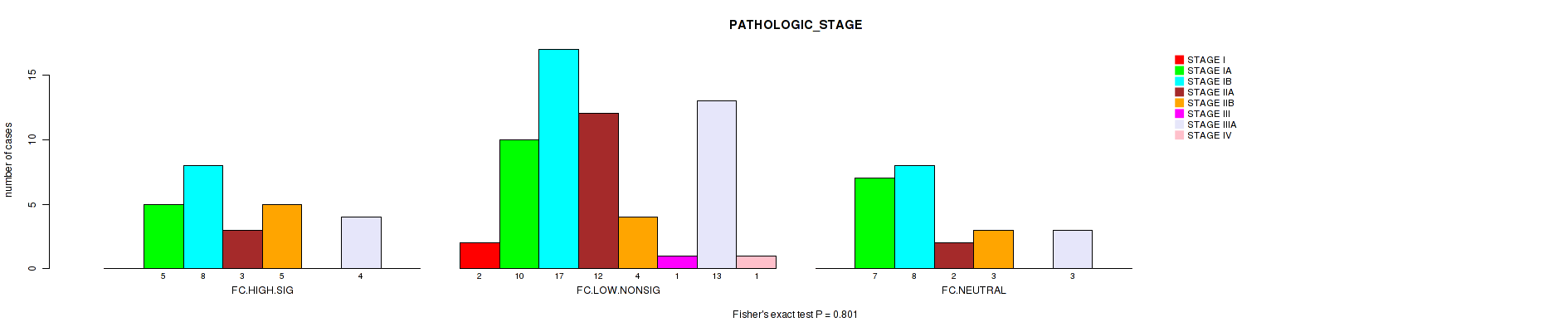
P value = 0.012 (Kruskal-Wallis (anova)), Q value = 0.41
Table S27. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #8: 'YEARS_TO_BIRTH'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 108 | 62.5 (9.7) |
| FC.HIGH.SIG | 25 | 64.6 (8.5) |
| FC.LOW.NONSIG | 60 | 60.0 (10.0) |
| FC.NEUTRAL | 23 | 66.7 (8.4) |
Figure S25. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #8: 'YEARS_TO_BIRTH'
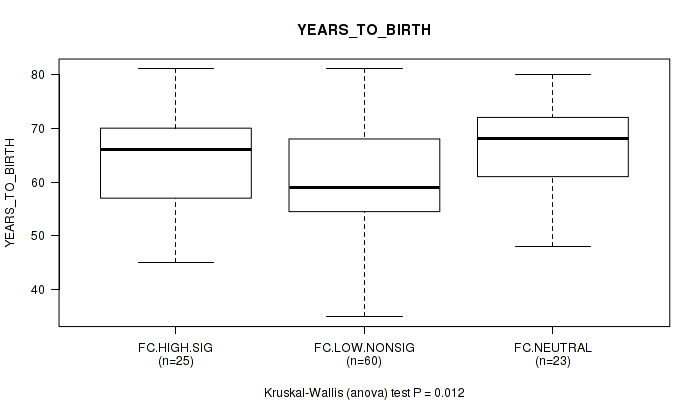
P value = 1 (Fisher's exact test), Q value = 1
Table S28. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #9: 'ETHNICITY'
| nPatients | HISPANIC OR LATINO | NOT HISPANIC OR LATINO |
|---|---|---|
| ALL | 3 | 33 |
| FC.HIGH.SIG | 0 | 5 |
| FC.LOW.NONSIG | 2 | 18 |
| FC.NEUTRAL | 1 | 10 |
Figure S26. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #9: 'ETHNICITY'
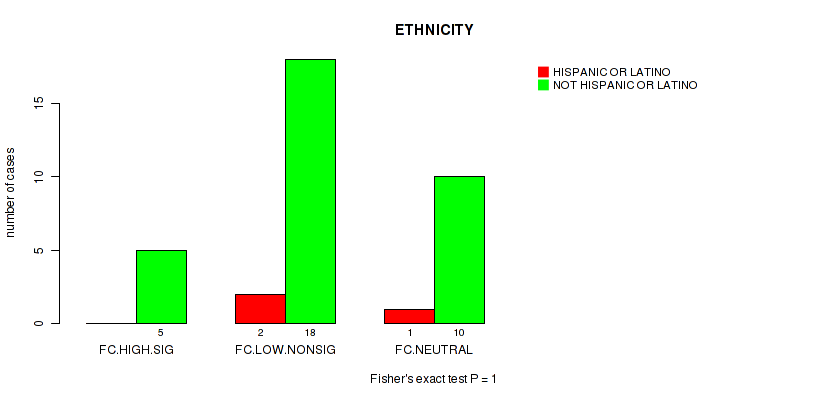
P value = 0.283 (Fisher's exact test), Q value = 0.84
Table S29. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #10: 'GENDER'
| nPatients | FEMALE | MALE |
|---|---|---|
| ALL | 36 | 72 |
| FC.HIGH.SIG | 5 | 20 |
| FC.LOW.NONSIG | 23 | 37 |
| FC.NEUTRAL | 8 | 15 |
Figure S27. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #10: 'GENDER'
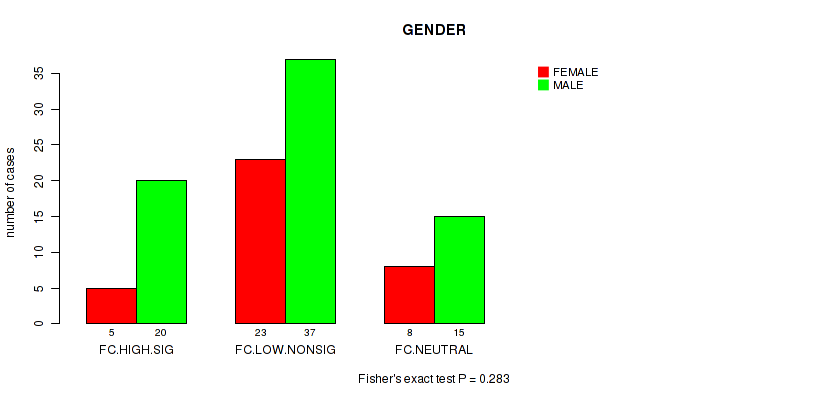
P value = 0.853 (Fisher's exact test), Q value = 0.97
Table S30. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #11: 'RACE'
| nPatients | AMERICAN INDIAN OR ALASKA NATIVE | ASIAN | BLACK OR AFRICAN AMERICAN | WHITE |
|---|---|---|---|---|
| ALL | 1 | 1 | 1 | 32 |
| FC.HIGH.SIG | 0 | 0 | 0 | 5 |
| FC.LOW.NONSIG | 1 | 1 | 0 | 17 |
| FC.NEUTRAL | 0 | 0 | 1 | 10 |
Figure S28. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #11: 'RACE'
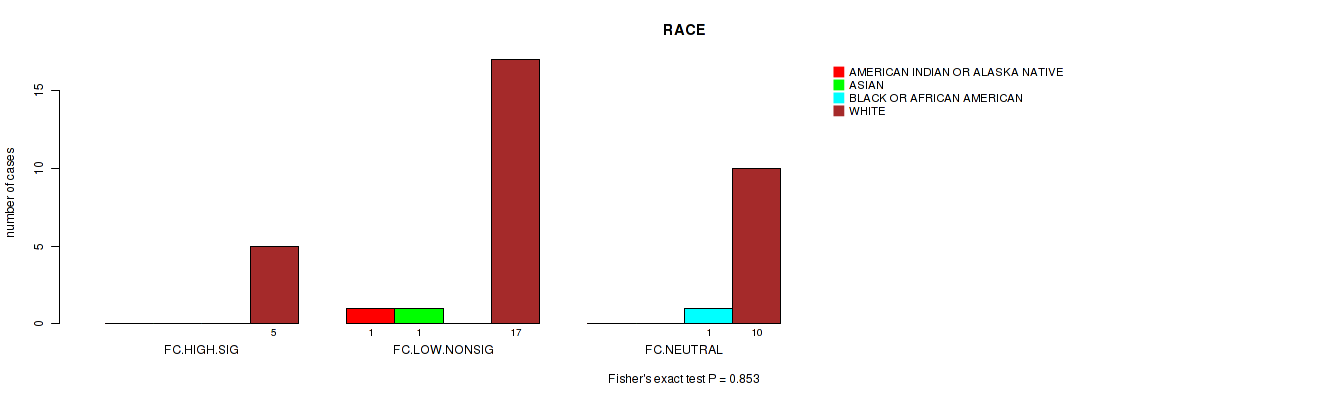
P value = 0.474 (Fisher's exact test), Q value = 0.91
Table S31. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #12: 'RADIATION_THERAPY'
| nPatients | NO | YES |
|---|---|---|
| ALL | 69 | 22 |
| FC.HIGH.SIG | 14 | 7 |
| FC.LOW.NONSIG | 40 | 12 |
| FC.NEUTRAL | 15 | 3 |
Figure S29. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #12: 'RADIATION_THERAPY'
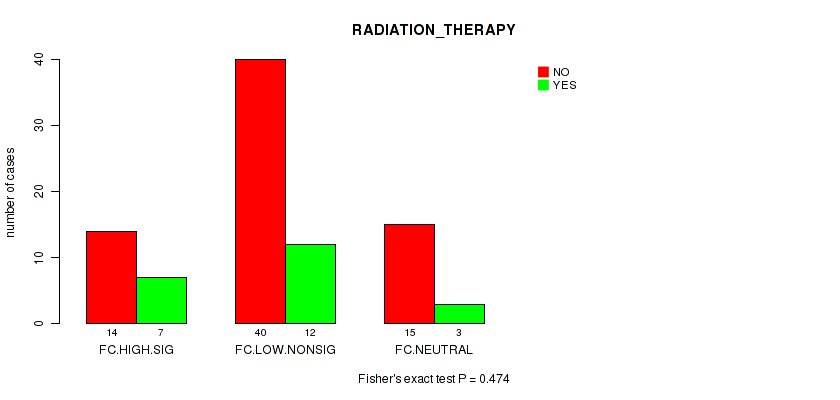
P value = 0.55 (Fisher's exact test), Q value = 0.91
Table S32. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #13: 'BMI'
| nPatients | NORMAL | OBESE | OVERWEIGHT | SEVERELY OBESE | UNDERWEIGHT |
|---|---|---|---|---|---|
| ALL | 53 | 10 | 27 | 3 | 15 |
| FC.HIGH.SIG | 9 | 2 | 9 | 0 | 5 |
| FC.LOW.NONSIG | 30 | 5 | 15 | 2 | 8 |
| FC.NEUTRAL | 14 | 3 | 3 | 1 | 2 |
Figure S30. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #13: 'BMI'
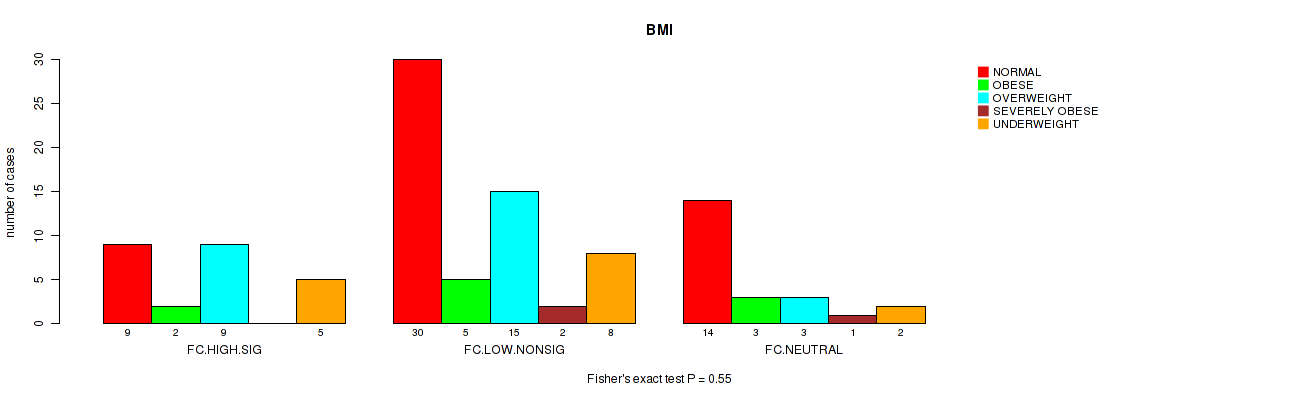
P value = 0.224 (Kruskal-Wallis (anova)), Q value = 0.84
Table S33. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #14: 'NUMBER_PACK_YEARS_SMOKED'
| nPatients | Mean (Std.Dev) | |
|---|---|---|
| ALL | 54 | 27.3 (23.6) |
| FC.HIGH.SIG | 14 | 30.9 (34.0) |
| FC.LOW.NONSIG | 28 | 23.1 (20.1) |
| FC.NEUTRAL | 12 | 32.9 (15.2) |
Figure S31. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #14: 'NUMBER_PACK_YEARS_SMOKED'
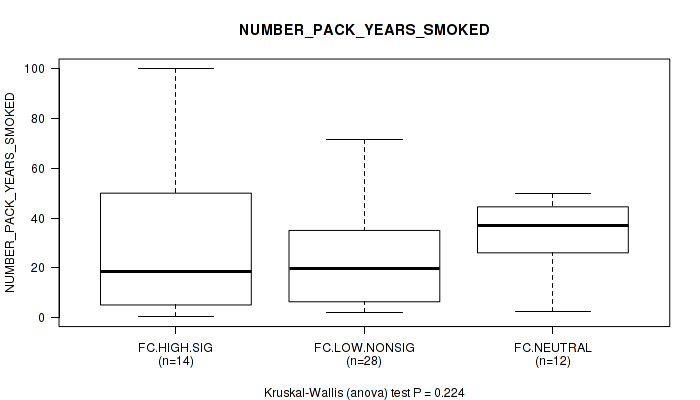
P value = 0.648 (Fisher's exact test), Q value = 0.97
Table S34. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #15: 'SMOKER'
| nPatients | NON-SMOKER | SMOKER |
|---|---|---|
| ALL | 45 | 60 |
| FC.HIGH.SIG | 10 | 14 |
| FC.LOW.NONSIG | 27 | 31 |
| FC.NEUTRAL | 8 | 15 |
Figure S32. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #15: 'SMOKER'
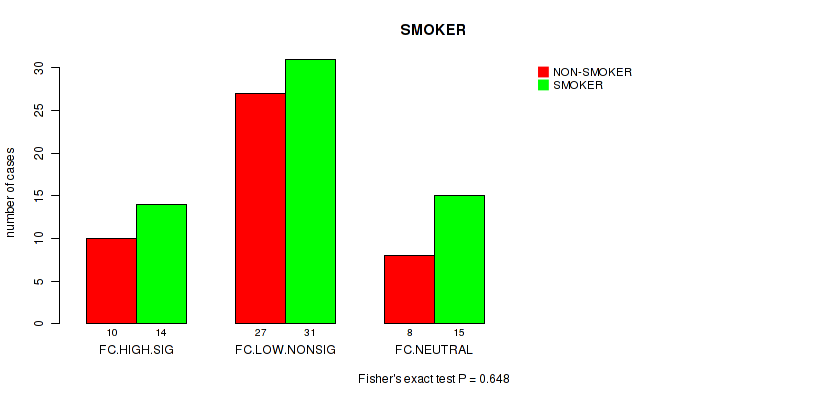
P value = 0.104 (Fisher's exact test), Q value = 0.84
Table S35. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #16: 'COUNTRY_OF_ORIGIN'
| nPatients | BULGARIA | CANADA | CHINA | JAMAICA | MEXICO | POLAND | RUSSIA | UKRAINE | UNITED STATES | VIETNAM |
|---|---|---|---|---|---|---|---|---|---|---|
| ALL | 2 | 1 | 18 | 1 | 2 | 2 | 1 | 3 | 32 | 44 |
| FC.HIGH.SIG | 0 | 0 | 6 | 0 | 0 | 2 | 0 | 1 | 5 | 11 |
| FC.LOW.NONSIG | 1 | 1 | 10 | 0 | 1 | 0 | 1 | 0 | 18 | 27 |
| FC.NEUTRAL | 1 | 0 | 2 | 1 | 1 | 0 | 0 | 2 | 9 | 6 |
Figure S33. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #16: 'COUNTRY_OF_ORIGIN'
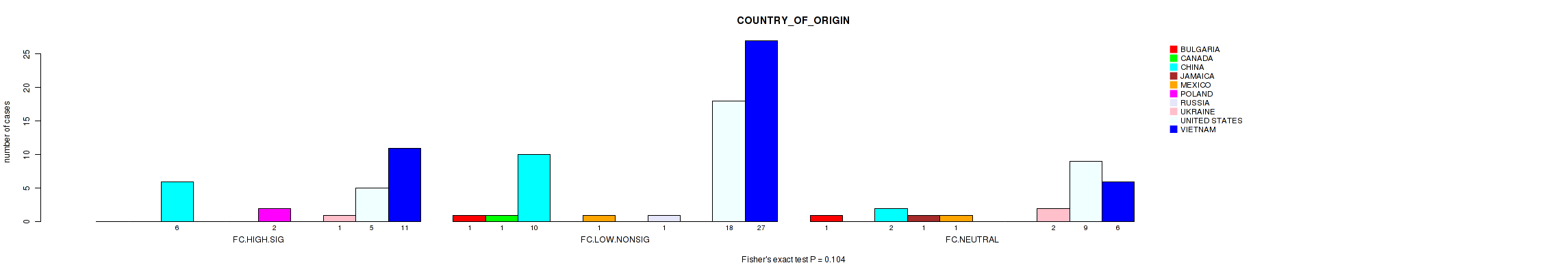
P value = 0.0583 (Fisher's exact test), Q value = 0.84
Table S36. Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #17: 'ORIGIN_ASIA'
| nPatients | ASIAN | WESTERN |
|---|---|---|
| ALL | 62 | 44 |
| FC.HIGH.SIG | 17 | 8 |
| FC.LOW.NONSIG | 37 | 22 |
| FC.NEUTRAL | 8 | 14 |
Figure S34. Get High-res Image Clustering Approach #2: 'APOBEC ENRICH' versus Clinical Feature #17: 'ORIGIN_ASIA'
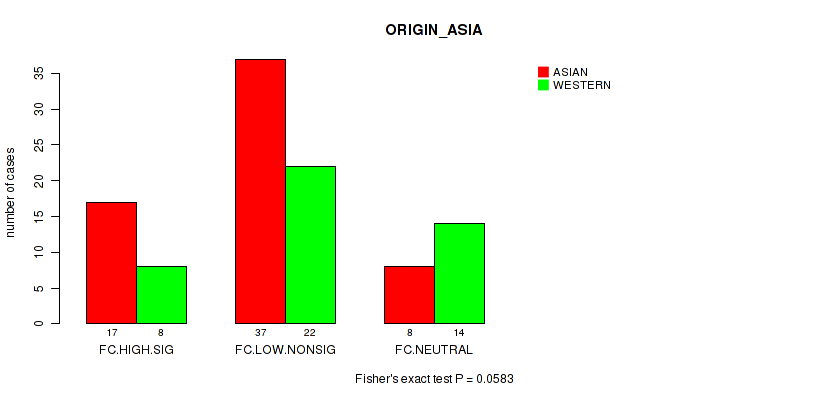
-
APOBEC groups file = /cromwell_root/fc-e7058367-eaa6-44b5-aab5-1ec08acf146a/53b59b4a-b38d-4dfd-9ded-fd5171ac2ee1/mutation_apobec/ca6ef43d-ce49-4015-b418-a9b28606bbcb/call-preprocess_clinical_apobec/APOBEC_for_clinical.correlaion.input.categorical.txt
-
Clinical data file = /cromwell_root/fc-e7058367-eaa6-44b5-aab5-1ec08acf146a/39eab10a-1791-41cf-866c-13e6472ce02e/normalize_clinical_cptac/df4eac3a-77b6-4ce7-961f-e1027c1ad609/call-normalize_clinical_cptac_task_1/CPTAC3-LUAD-TP.clin.merged.picked.txt
-
Number of patients = 108
-
Number of selected clinical features = 17
APOBEC classification based on APOBEC_MutLoad_MinEstimate : a. APOBEC non group -- samples with zero value, b. APOBEC high group -- samples above median value in non zero samples, c. APOBEC low group -- samples below median value in non zero samples.
APOBEC classification based on APOBEC_enrich : a. No Enrichmment group -- all samples with BH_Fisher_p-value_tCw > 0.05, b. Low enrichment group -- samples with BH_Fisher_p-value_tCw = < 0.05 and APOBEC_enrich=<2, c. High enrichment group -- samples with BH_Fisher_p-value_tCw =< 0.05 and APOBEC_enrich>2.
For survival clinical features, the Kaplan-Meier survival curves of tumors with and without gene mutations were plotted and the statistical significance P values were estimated by logrank test (Bland and Altman 2004) using the 'survdiff' function in R
For binary clinical features, two-tailed Fisher's exact tests (Fisher 1922) were used to estimate the P values using the 'fisher.test' function in R
For multiple hypothesis correction, Q value is the False Discovery Rate (FDR) analogue of the P value (Benjamini and Hochberg 1995), defined as the minimum FDR at which the test may be called significant. We used the 'Benjamini and Hochberg' method of 'p.adjust' function in R to convert P values into Q values.